Abstract
Exonic splicing enhancers (ESEs) activate pre-mRNA splicing by promoting the use of the flanking splice sites. They are recognized by members of the serine/arginine-rich (SR) family of proteins, such as splicing factor 2/alternative splicing factor (SF2/ASF), which recruit basal splicing factors to form the initial complexes during spliceosome assembly. The in vitro splicing kinetics of an ESE-dependent IgM pre-mRNA suggested that an SF2/ASF-specific ESE has additional functions later in the splicing reaction, after the completion of the first catalytic step. A bimolecular exon ligation assay, which physically uncouples the first and second catalytic steps of splicing in a trans-splicing reaction, was adapted to test the function of the ESE after the first step. A 3′ exon containing the SF2/ASF-specific ESE underwent bimolecular exon ligation, whereas 3′ exons without the ESE or with control sequences did not. The ESE-dependent trans-splicing reaction occurred after inactivation of U1 or U2 small nuclear ribonucleoprotein particles, compatible with a functional assay for events after the first step of splicing. The ESE-dependent step appears to take place before the ATP-independent part of the second catalytic step. Bimolecular exon ligation also occurred in an S100 cytosolic extract, requiring both the SF2/ASF-dependent ESE and complementation with SF2/ASF. These data suggest that some ESEs can act late in the splicing reaction, together with appropriate SR proteins, to enhance the second catalytic step of splicing.
The removal of introns from pre-mRNA involves two transesterification steps. Many intronic and exonic sequence elements function as splicing signals in constitutive and regulated splicing. One class of element, exonic splicing enhancers (ESEs), stimulates processing of upstream introns. Members of the serine/arginine-rich (SR) protein family have been directly implicated in the mechanisms of ESE function (1). SR proteins are also essential splicing factors required for spliceosome assembly and progression through the first transesterification step. ESEs appear to function as specific binding sites for SR proteins, which then recruit other factors to the splice sites, promoting assembly of the earliest prespliceosomal complexes (2). However, some SR proteins remain associated with the mRNA throughout the splicing reaction and probably during mRNA transport through the nuclear pores (3–7). Thus, ESEs and SR proteins may have additional functions beyond the early stages of splicing.
One of the best characterized ESE-containing pre-mRNAs consists of the last intron and last two membrane-isoform exons (M1 and M2) of the Ig μ heavy chain (IgM) gene (8). An ESE located in the M2 exon promotes early spliceosome assembly, leading up to the first catalytic step (8). This ESE can be replaced by other enhancer sequences to activate splicing (9, 10). However, an effect of this or other ESEs on the second step of splicing had not been reported. While studying splicing of this substrate, we observed an unexpected splicing pattern with kinetics suggestive of ESE function in the second step of splicing. We exploited a bimolecular exon ligation reaction that uncouples the two transesterification steps of splicing (11) to further explore a requirement for ESE action late in the splicing reaction, after the completion of the first catalytic step.
MATERIALS AND METHODS
RNA Substrates.
IgM transcription templates were made by PCR using primers P and A (10), with pμMΔ (8) or splicing factor 2/alternative splicing factor (SF2/ASF)-winner clone B1 (10) as DNA templates. The adenovirus major late (AdML) transcription template for cis-splicing was the BamHI-linearized plasmid pAdML PAR (17). For the 5′ fragment AdML ΔAG transcription template, PCR was performed with a T7 RNA polymerase promoter primer and primer ΔAG-A (AGAGAGAGAGGAAAAAAAAGGGA), using pAdML ΔAG as a template. 3′ M2 exons were made by PCR with primer SP6-PPT-IgM-S (ATTTAGGTGACACTATAGCTGTCTCTGTCACCTGCAGGTGAAATGACTCTCAGCAT) or SP6-TGCAG-IgM-S (ATTTAGGTGACACTATAGTGCAGGTGAAATGACTCTCAGCAT) and primer A. The template for 3′ exon PCR was SF2/ASF winner clone B1 (10) or an enhancer-deleted construct initially made with primer RΔ (GTGAAATGACTCTCAGCATTCTAGTAAACTTATTCTTACGT) and primer A. The 3′ fragment M2-Δ differs from the downstream exon of μMΔ by including 10 nucleotides (neutral in cis-splicing assays) immediately upstream of the wild-type enhancer (8). In vitro-transcribed RNA was labeled with [32P]GTP and then gel-purified and quantitated, or labeled with [3H]GTP and then DNase-treated and quantitated. The 3′ M2 exons with deletions of the exonic splicing silencer element (12) (M2-SF2W-δsil and Δ-δsil) were made by running off transcripts at a BfaI site.
Splicing Reactions.
Totals of 20 fmol of 5′ RNA and 100 fmol of 3′ RNA were used in 25-μl reactions containing 32% nuclear extract or S100 cytosolic extract and 3.2 mM MgCl2 (13). Totals of 5, 10, or 20 pmol of purified Escherichia coli-expressed SF2/ASF (14), or baculovirus-expressed SC35 or SRp55, were used in S100 complementation reactions. Anti-U1 (nucleotides 2–11) and anti-U2 (nucleotides 1–15) oligonucleotides were used at 16 μM for RNase H experiments (15). ATP was depleted by addition of glucose to 5 mM and hexokinase to 50 units/ml for 15 min at 30°C (16). Splicing reaction products were separated on denaturing 5.5%, 10%, or 12% polyacrylamide gels. The trans-spliced mRNA product was excised from the gel, eluted, reverse transcribed with primer A by using Superscript II (Life Technologies), and PCR-amplified with primer Ad5p (CGTTCGTCCTCACTCTCTT) and primer A by using Pfu DNA polymerase (Stratagene). The product was cloned into pCR-blunt (Invitrogen) and sequenced.
RESULTS
Splicing Kinetics of an ESE-Dependent IgM pre-mRNA Suggest an Additional Enhancement of the Second Catalytic Step.
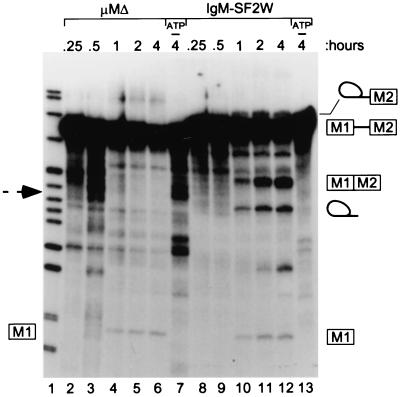
Splicing was compared between an IgM substrate with an in vitro-selected SF2/ASF-specific ESE (10) (IgM-SF2W) in place of the wild-type M2 exon enhancer, and a substrate in which the wild-type ESE had been removed (μMΔ) (8). Both IgM-SF2W and μMΔ gave first-step intermediates in nuclear extract (between 0.5 and 1 hr; lanes 4 and 10), but only IgM-SF2W RNA gave second-step products (Fig. 1). The IgM pre-mRNA is strongly (but not completely) dependent on enhancer sequences for the first catalytic step of splicing, and this is reflected in Fig. 1, as there was no increased accumulation of splicing intermediates with the μMΔ substrate compared with IgM-SF2W. Nevertheless, the kinetics of appearance of the first-step intermediates and second-step products suggested an additional effect of the ESE.
Figure 1.
In vitro splicing kinetics of IgM pre-mRNA. A time course of cis-splicing of IgM pre-mRNAs with (IgM-SF2W) or without (μMΔ) an SF2/ASF-specific ESE is shown. The positions of the pre-mRNA, 5′ exon M1 and lariat-exon M2 intermediates, and splicing products are shown by diagrams on the right. A dashed arrow shows the expected position of the μMΔ-spliced product. Lane 1, pBR322/MspI size markers; lanes 7 and 13, control reactions lacking ATP.
Bimolecular Exon Ligation of the M2 Exon Requires an Enhancer.
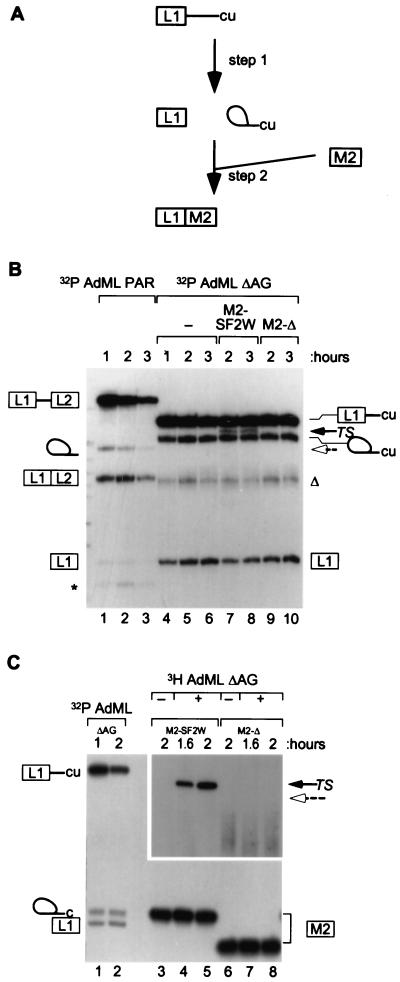
To directly test the mechanisms of ESE action on the second step of splicing, we used a trans-splicing assay developed by Anderson and Moore (ref. 11, and Fig. 2A). A 5′ RNA substrate containing an upstream exon and an intron with a strong branch site and polypyrimidine tract can efficiently undergo the first step of splicing. Then, subsequent excess addition of a 3′ RNA containing a 3′ splice site and a downstream exon allows the second step to occur, generating a trans-spliced mRNA product. This physical separation of the first and second catalytic steps makes it possible to test whether an ESE can be recognized and participate in splicing after the completion of the first step. As previously reported, a radiolabeled 5′ RNA substrate [AdML ΔAG, containing the AdML first leader exon and first intron, including the branch site and an improved 3′ polypyrimidine tract (17)] could undergo the first step of splicing in HeLa nuclear extract (Fig. 2B, lanes 4–6). As a 3′ RNA substrate, we used the M2 exon with a 20-nt in vitro-selected ESE, which was shown to be a strong SF2/ASF-specific enhancer by both splicing and binding assays (10). 3H-labeled 3′ RNA substrates with (M2-SF2W) or without (M2-Δ) the ESE, were added to the reactions in 5-fold molar excess. A trans-spliced mRNA was reproducibly seen when M2-SF2W was added (Fig. 2B, lanes 7 and 8), but not with M2-Δ (lanes 9 and 10). In a reciprocal labeling experiment (11), again, a trans-spliced mRNA product was reproducibly seen with M2-SF2W (Fig. 2C, lanes 4 and 5) and not with M2-Δ (lanes 7 and 8). The lack of trans-splicing with M2-Δ was not an artifact of deletion and exon shortening, as four additional variants of the M2 exon, containing control sequences (C3, C4, C6, and C7 in ref. 10) instead of an SF2/ASF-ESE, did not show trans-splicing in nuclear extract (data not shown). The M2-SF2W and M2-Δ exons were equally stable during incubation for up to 2 hr in the splicing reaction (Fig. 2C, lanes 4 and 5 compared with lanes 7 and 8). The trans-spliced band was verified as a correctly spliced mRNA by sequencing.
Figure 2.
ESE dependence of bimolecular exon ligation. (A) A scheme of the trans-splicing assay is shown, with the 5′ leader exon denoted by the boxed L1 and the intron by a line ending with “cu” (17). After incubation, to allow the first step of splicing to occur, IgM M2 exon (with a 3′ splice site) was added as a 3′ substrate. (B) AdML ΔAG RNA was 32P-labeled and incubated in nuclear extract for the times shown (lanes 4–6). 3H-labeled 3′ exons were added for the second and third hours of the reactions (lanes 7–10). The trans-spliced mRNA (TS) is shown by a black arrow. The expected position of trans-splicing with M2-Δ is marked by a dashed open arrow. Control splicing in cis between AdML PAR exons L1 and L2 is shown in lanes 1–3. (C) In a reciprocal labeling experiment, the 3′ exons were 32P-labeled, and the 5′ fragment was made with [3H]GTP. Controls with 32P-labeled AdML ΔAG pre-mRNA are shown in lanes 1 and 2. Other controls omitting the 3H-labeled 5′ RNA are shown in lanes 3 and 6. A longer exposure of the trans-spliced products is shown. In B (and in subsequent figures), the asterisk indicates an artifactual band characteristic of the AdML PAR substrate (11, 17); the triangle indicates an artifactual product that appears to derive from the AdML ΔAG lariat intermediate or from use of an alternative branch site.
SF2/ASF Is Required for Enhancer-Dependent Bimolecular Exon Ligation.
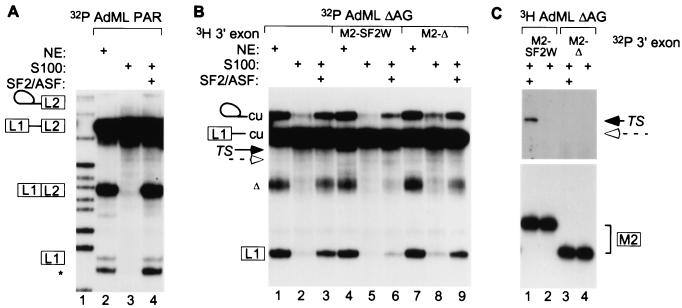
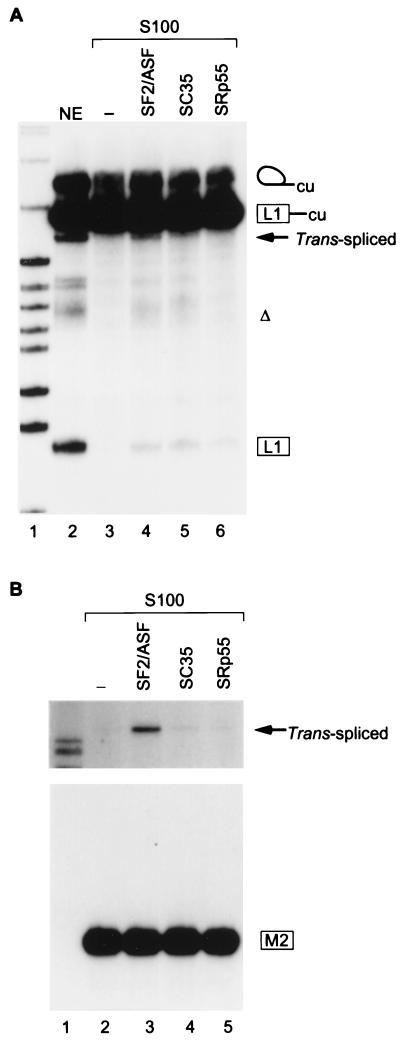
To test the mechanism of action of the SF2W enhancer, we performed trans-splicing in an S100 extract complementation assay (18). As a cis-splicing control, the full-length AdML PAR RNA (17) spliced efficiently in S100 extract complemented with recombinant SF2/ASF, but not in S100 alone (Fig. 3A, lanes 3 and 4). This result suggests that either AdML pre-mRNA splicing is activated by a general action of SF2/ASF (19), or there are one or more SF2/ASF-dependent enhancers within the AdML substrate, as described for other constitutive exons (20, 21). To carry out bimolecular exon ligation assays, the 5′ RNA AdML ΔAG substrate was first shown to undergo the first splicing step in S100 extract complemented with SF2/ASF (Fig. 3B, lane 3). It did so only slightly less efficiently than in nuclear extract (lane 1), whereas the reaction was extremely inefficient in S100 extract alone (lane 2). A trans-spliced mRNA product was seen when the M2-SF2W exon was added to nuclear extract (lane 4) and, faintly, in S100 complemented with SF2/ASF (lane 6), but not when M2-Δ exon was added (lanes 7–9). Reciprocal labeling was used to improve the electrophoretic separation and detection of the trans-spliced product in S100 complemented with SF2/ASF (Fig. 3C, lane 1) and to confirm the absence of a product with the M2-Δ exon (lane 3). The trans-spliced product accumulated to lower levels in S100 extract complemented with SF2/ASF, compared with the nuclear extract, in contrast to the cis-splicing substrate, which was spliced with comparable efficiency in both extracts (Fig. 3A); however, the amount of intermediates in the trans-splicing reaction was also reduced. Four other in vitro-selected SF2/ASF-dependent ESEs (B2-B5 in ref. 10) were also tested, and all enhanced trans-splicing in S100 extract complemented with SF2/ASF (data not shown).
Figure 3.
Trans-splicing requirement for an ESE and SF2/ASF. (A) Control AdML PAR pre-mRNA cis-splicing was carried out in nuclear extract (NE) or in S100 extract complemented with SF2/ASF. Lane 1, size markers. (B) Trans-splicing reactions were performed in nuclear extract or in S100 extract with or without SF2/ASF. 32P-labeled AdML ΔAG was incubated for 1 hr, and then excess 3H-labeled M2-SF2W, or M2-Δ, or water was added for the second hour. Trans-spliced mRNA (TS, lanes 4 and 6) is marked by a black arrow, and the expected position of the M2-Δ mRNA is marked with a dashed open arrow. (C) A reciprocal labeling experiment is shown. The top part of the autoradiogram is a longer exposure.
The SF2/ASF-ESE Functions in the Context of a Downstream Exonic Splicing Silencer Element.
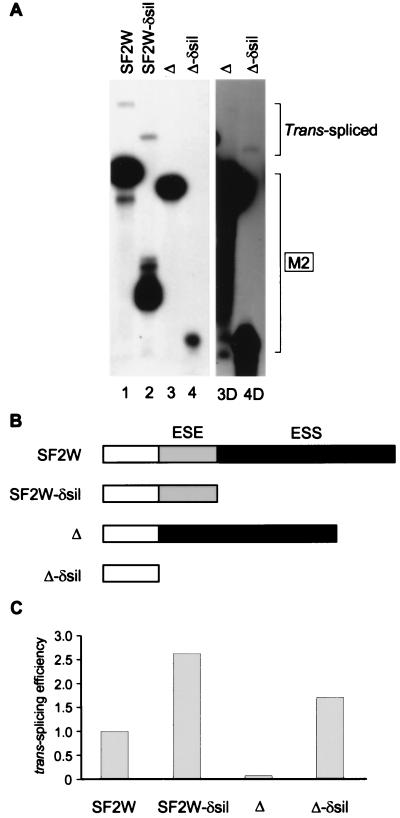
A silencer element in the IgM M2 exon was recently identified in cis-splicing experiments (12). We tested the function of this silencer, vis-à-vis an enhancer, in bimolecular exon ligation. Removal of the silencer from the M2-SF2W substrate increased the ratio of trans-spliced mRNA to unspliced 3′ exon (Fig. 4, lanes 1 and 2). With the M2-Δ substrate, which lacks the ESE, trans-splicing was nearly undetectable (lanes 3 and 3D). However, removal of the silencer in addition to the ESE resulted in a substantial increase in trans-splicing (lanes 4 and 4D). Thus, the silencer can antagonize an enhancer in both cis-splicing (12) and trans-splicing assays.
Figure 4.
Interaction between enhancer and silencer elements in trans-splicing. (A) 3H-labeled AdML ΔAG pre-mRNA was incubated for 1 hr in nuclear extract. Then, the indicated 32P-labeled 3′ M2 exons with or without an ESE or a silencer were added. The lower and upper brackets on the right show the positions of the 3′ exons and of the trans-spliced mRNAs, respectively. A longer exposure of lanes 3 and 4 is shown (lanes 3D and 4D). (B) Schematic structure of the different 3′ M2 exons. The ESE is shaded gray, and the silencer segment (ESS) is shaded black. (C) Relative trans-splicing efficiencies of the four 3′ exons. The data from A were quantitated by densitometry and normalized for loading and differential labeling of the 3′ exons. The efficiency of the SF2W 3′ exon was arbitrarily set at 1.
SF2/ASF Specificity of Bimolecular Exon Ligation Enhancement.
To test the specificity of SF2/ASF function in trans-splicing via the SF2W ESE, we performed the reactions in S100 extract complemented with one of three human recombinant SR proteins. SF2/ASF and SC35 generated first-step splicing intermediates at similar levels, whereas SRp55 did so less efficiently (Fig. 5A, lanes 4–6). However, trans-spliced mRNA was reproducibly generated more efficiently in reactions containing SF2/ASF than in those with SC35 or SRp55 (Fig. 5B, lanes 3–5). In comparisons across a range of SR protein concentrations, the ratio of trans-spliced product to unspliced 3′ exon was 11-fold higher (SE ± 3.8; n = 7) in SF2/ASF than in SC35 complementation reactions.
Figure 5.
SR protein specificity of second-step trans-splicing enhancement. (A) 32P-labeled AdML ΔAG pre-mRNA was incubated for 1 hr in S100 extract (with or without the indicated SR proteins), and 3H-labeled M2-SF2W 3′ exon was added for the second hour. Trans-spliced mRNA is shown by the black arrow. Lane 1: size markers. (B) A reciprocal labeling experiment is shown. The top part of the autoradiogram is a longer exposure. Lane 1, size markers.
The Trans-Spliced Products Are Generated from Accumulated First-Step Intermediates.
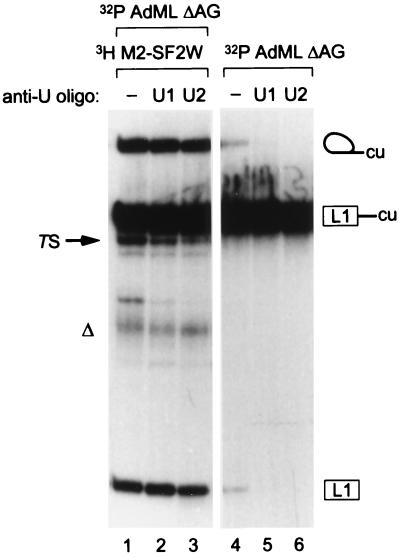
Although different 3′ exons have no effect on the efficiency with which the 5′ substrate undergoes the first step (11), it remained possible that the M2-SF2W exon underwent trans-splicing by promoting spliceosome assembly on unprocessed AdML ΔAG pre-mRNA, rather than by a direct use of the first-step upstream exon and intron lariat intermediates already assembled into a spliceosome. To rule out a first-step effect of the ESE in the free 3′ exon, processing of any pre-mRNA remaining prior to addition of the 3′ exon substrates was blocked by inactivation of U1 or U2 small nuclear RNAs via oligonucleotide-directed RNase H cleavage (15). We allowed first-step AdML ΔAG intermediates to accumulate, then inactivated U1 or U2, before finally adding the downstream M2-SF2W exon. Trans-splicing products were still detected (Fig. 6, lanes 2 and 3). Parallel control reactions were initially incubated without RNA for the first hour, then treated with or without oligonucleotides, before addition of AdML ΔAG for the final hour. Inactivation of U1 or U2 effectively inhibited the first step of splicing during the final hour of incubation (lanes 5 and 6). Note that under these conditions, the efficiency of the first step is already reduced because of the preincubation itself (lane 4). The amount of trans-spliced product was slightly reduced in the anti-U2-treated reactions (lane 3), possibly because some of the anti-U2 oligonucleotide was able to target U2 small nuclear ribonucleoprotein (snRNP) already assembled into spliceosomes. U2 small nuclear RNA is involved in the second catalytic step of splicing (22).
Figure 6.
Generation of trans-spliced products from accumulated first-step intermediates. 32P-labeled AdML ΔAG RNA was processed for 1 hr to give first-step intermediates, then incubated with buffer (lane 1), anti-U1 (lane 2), or anti-U2 (lane 3) oligonucleotides for 15 min, and finally, excess tritiated M2-SF2W 3′ exon RNA was added for another hour. Control reactions were preincubated without RNA for 1 hr, treated with buffer or oligonucleotides, and AdML ΔAG RNA was then added for a final hour of incubation.
Bimolecular Exon Ligation Is ATP-Dependent.
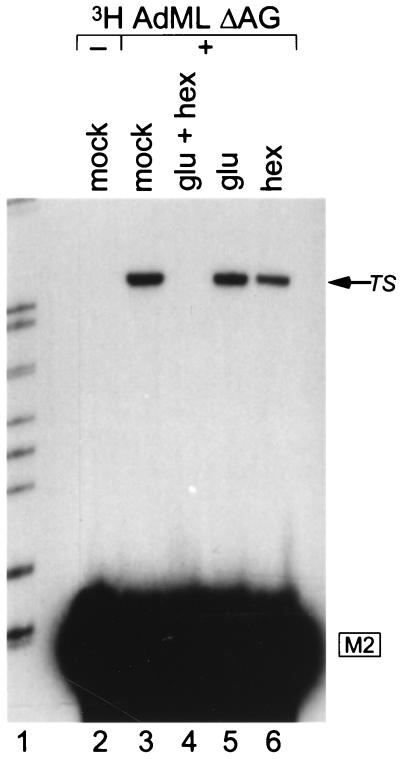
The second step of splicing includes the assembly onto the spliceosome of four protein factors dispensable for the earlier stages of the reaction (Prp16, Prp17, Slu7, and Prp18) (22). In Saccharomyces cerevisiae, the order of action of these factors has been established with respect to the ATP dependence of the second step. The identification of functional human homologues shows the mechanistic conservation of the second-step processes (23, 24). To characterize the energy requirement of ESE-dependent bimolecular exon ligation, ATP was depleted from nuclear extract. A trans-spliced mRNA product was no longer detected upon treatment with glucose and hexokinase (Fig. 7, lane 4), but was detected in control reactions (lanes 3, 5, and 6). Hexokinase alone slightly reduced the level of the product (lane 6), either nonspecifically or perhaps because of trace phosphate acceptors in the reaction.
Figure 7.
ATP dependence of ESE-stimulated bimolecular exon ligation. Tritiated AdML ΔAG RNA was incubated for 1 hr (except in lane 2), followed by glucose (glu) and hexokinase (hex; lane 3), or control treatments (lanes 3, 5, and 6), with subsequent addition of radiolabeled M2-SF2W 3′ RNA. The trans-spliced product (TS) is marked by an arrow. Lane 1, size markers.
DISCUSSION
Mammalian in vitro trans-splicing systems have provided insights into the mechanisms of cis-splicing, in particular, showing the enhancer dependence of one of the first-step trans-splicing systems (25, 26). In the second-step trans-splicing system, the first step of splicing occurs in the absence of a downstream exon (11). This reaction was adapted here to demonstrate the role of an SF2/ASF-specific ESE during, or just prior to, the second catalytic step of splicing. Also examined were the requirements of ESE-dependent bimolecular ligation for SF2/ASF, ATP (both required), and U1 or U2 snRNP (neither required).
A caveat is how closely the second step of the bimolecular exon ligation assay relates to the second step of the natural cis-splicing reaction. Although an ESE may function in the second step of cis-splicing by an indirect mechanism involving changes in a first-step component or configuration, none of the 3′ exons used affected the efficiency of the uncoupled first-step reaction (ref. 11, and this study). Moreover, the appropriate 3′ splice site was used in trans-splicing, and the spliced product was verified as having the correct exon–exon junction. There is a close correlation between the cis- and trans-splicing data in the context of the IgM M2 exon. The SR protein specificity of the SF2/ASF-ESE was recapitulated in the trans-reaction as in the cis-reaction (ref. 10; and H.-X.L., S.L.C., M. Q. Zhang, and A.R.K., unpublished work). Some cis- and trans-splicing was observed with the SF2/ASF-ESE in S100 extract complemented with SC35, but both reactions were less efficient than those with the cognate SR protein, SF2/ASF. Trans-splicing also recapitulated cis-splicing in the context of the silencer element in the M2 exon. The ESE may be required, in part, to overcome the silencer in both reactions. Thus, the data are compatible with bimolecular exon ligation representing a useful model for the second step of splicing.
The 3′ exons used in the original second-step bimolecular exon ligation study (11) were derived from AdML exon L2 and chicken cardiac troponin T (cTNT) exon 5. The latter exon contained an ESE optimized by mutations (27). A reduction in trans-splicing efficiency was observed when the AdML 3′ L2 exon was replaced by intronic sequences, compared with coding sequences from the opposite viral DNA strand, suggesting that trans-splicing efficiency may correlate with the presence of exonic sequences (11). These data may reflect the presence of as yet unmapped ESEs in the AdML L2 and antisense exons, which, like the IgM M2 ESE, might act in the second step of splicing. An implication of the present findings is that 3′ exons able to undergo efficient bimolecular exon ligation are expected to contain second-step ESEs. Conversely, 3′ exons incapable of this reaction probably lack such ESEs, or may contain second-step silencers. An uncertainty is whether the same ESE can always function to enhance both steps of splicing, as seen with the SF2/ASF-ESE. Detailed mutational analysis and systematic evolution of ligands by exponential enrichment experiments have shown that several, if not many, splicing enhancers and silencers may lie in most exons (10, 20, 21, 28). It is possible that several functional classes of ESE may be present in an exon, some perhaps being specific for a particular step of splicing.
We analyzed other ESE-containing exons to address the generality of the present findings. Cis-splicing of HIV-Tat exons 2 and 3 and β-globin exons 1 and 2 is SF2/ASF-dependent (21, 28). Tat exon 3 and β-globin exon 2 underwent trans-splicing in an SF2/ASF-dependent manner in S100 extract (data not shown), suggesting that SF2/ASF interacts with one or more second-step ESEs in these exons. In contrast, we were unable to recapitulate the well-characterized cis-splicing properties of other exons by using the second-step trans-splicing assay. IgM C3 and C4 exons are cis-spliced in an SC35-dependent manner in S100 extract (21). However, the IgM C4 3′ exon did not yield detectable trans-splicing in either nuclear extract or S100 extract complemented with SC35. The SC35-dependent ESE may be relevant only to the first step, or a second-step silencer may be present in this exon, or this 3′ exon may be intrinsically incompatible with the heterologous AdML 5′ substrate. We also analyzed mutants of cTNT exon 5 that have a clear functional hierarchy in cis-splicing enhancement (27). Unexpectedly, the different cTNT substrates were equally efficient in trans-splicing in nuclear extracts (data not shown). Again, this observation may indicate the importance of sequence context for bimolecular exon ligation (for example, the absence of a silencer sequence in cTNT exon 5), or that second-step enhancers are present in this exon, but are distinct from its well-characterized first-step enhancer.
Both the SF2/ASF-specific ESE and SF2/ASF are required for bimolecular exon ligation of the IgM M2 exon. Since SF2/ASF was added at the start of the reaction, it promotes the first splicing step. A mechanistic difference between the general and enhancer-dependent splicing functions of SF2/ASF and other SR proteins has been demonstrated (19). The formation of the first-step intermediates in the bimolecular exon ligation system may be because of a general function of SF2/ASF, perhaps mediated through protein–protein interactions, or perhaps via binding to RNA sequences other than ESEs. However, the efficient generation of first-step intermediates is not a sufficient explanation for the ESE dependence of the trans-splicing reaction. The efficiency of the first step was the same regardless of which 3′ exon was present, even exons lacking the SF2/ASF-ESE and therefore incapable of undergoing bimolecular exon ligation. In addition, the formation of first-step intermediates was also promoted by other SR proteins, but with much less trans-splicing occurring. Thus, SF2/ASF has an additional role in bimolecular exon ligation, which is enhancer-dependent.
Although SF2/ASF interacts directly with the SF2/ASF-ESE (10), it is not known whether this binding is relevant to the second-step effect. It is possible that an indirect mechanism is involved, e.g., involving protein–protein interactions. After the first transesterification step, many changes occur within the spliceosome (22). SR proteins act at later stages of spliceosome assembly, but still before the first transesterification step, by recruiting the U2 snRNP and U4/U5⋅U6 tri-snRNP (29, 30). SF2/ASF may similarly function to enhance the second step of splicing by remodeling of the spliceosome. However, purified SR proteins have also been reported to bring exons into proximity without the need for snRNPs or other factors (31). They may have a similar exon-bridging function in trans-splicing and during the second catalytic step of cis-splicing. In this scenario, SF2/ASF may bring the 3′ ESE-containing exon into the proximity of the first-step intermediates, making an engagement with the spliceosome a more likely event. Alternatively, SF2/ASF and the ESE may serve to stabilize exon–exon interactions mediated by other components. These mechanisms may or may not involve a specific interaction of SF2/ASF with the second-step catalytic machinery. Another possibility is that SF2/ASF and the SF2/ASF-ESE promote the second-step reaction by recruiting one or more second-step factors.
The ATP dependence of the bimolecular exon ligation reaction helps to order the ESE-dependent trans-splicing reaction to the initial part of the second step. It suggests that the ESE-containing exon enters the splicing reaction prior to the involvement of hPrp18 and hslu7 (whose yeast counterparts participate in the ATP-independent part of the second step). Any direct action of SF2/ASF and the SF2/ASF-ESE may involve enhancement of the recruitment of hPrp16 or hPrp17.
Here the bimolecular exon ligation reaction was used as a tool to demonstrate an effect that is presumably relevant to cis-splicing. If this assumption is correct, the late action of ESEs and SR proteins may serve a proofreading role contributing to the accuracy of cis-splicing by ensuring that the correct order of exons in constitutively or alternatively spliced multiexon pre-mRNAs is maintained throughout processing of the individual introns. On the other hand, bimolecular exon ligation may occur naturally in certain situations (32–35), perhaps as a way of joining alternative exons encoded by different genes or by different transcripts from the same gene. Such juxtaposition of exons in trans would be a diffusion-limited process, which may be facilitated by the specific interactions between the ESEs and appropriate SR proteins.
Acknowledgments
We thank R. Reed for pAdML PAR and pAdML ΔAG; Y. Shimura and A. Watakabe for pμM1–2 and pμMΔ; R.-M. Xu for SC35; X. Li and F. Rottman for SRp55; and I. Eperon for helpful discussions. This work was supported by the Wellcome Trust (045401), the National Institutes of Health (GM42699), and the U.S. Army Medical Research and Matériel Command.
ABBREVIATIONS
- ESE
exonic splicing enhancer
- SR
serine/arginine-rich
- AdML
adenovirus major late
- snRNP
small nuclear ribonucleoprotein
- SF2
splicing factor 2
- ASF
alternative splicing factor
References
- 1.Cáceres J F, Krainer A R. In: Eukaryotic mRNA Processing. Krainer A R, editor. Oxford: IRL Press; 1997. pp. 174–212. [Google Scholar]
- 2.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alzhanova-Ericsson A T, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. Genes Dev. 1996;10:2881–2893. doi: 10.1101/gad.10.22.2881. [DOI] [PubMed] [Google Scholar]
- 5.Cáceres J F, Screaton G R, Krainer A R. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanamura A, Cáceres J F, Mayeda A, Franza B R, Jr, Krainer A R. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 7.Iborra F J, Jackson D A, Cook P R. J Cell Sci. 1998;111:2269–2282. doi: 10.1242/jcs.111.15.2269. [DOI] [PubMed] [Google Scholar]
- 8.Watakabe A, Tanaka K, Shimura Y. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka K, Watakabe A, Shimura Y. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H-X, Zhang M, Krainer A R. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson K, Moore M J. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 12.Kan J L, Green M R. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayeda A, Krainer A R. Methods Mol Biol. 1999;118:315–322. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 14.Krainer A R, Mayeda A, Kozak D, Binns G. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 15.Krämer A. Methods Enzymol. 1990;181:284–292. doi: 10.1016/0076-6879(90)81129-i. [DOI] [PubMed] [Google Scholar]
- 16.Tatei K, Kimura K, Ohshima Y. J Biochem. 1989;106:372–375. doi: 10.1093/oxfordjournals.jbchem.a122860. [DOI] [PubMed] [Google Scholar]
- 17.Gozani O, Patton J G, Reed R. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krainer A, Conway G C, Kozak D. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 19.Graveley B R, Maniatis T. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 20.Schaal T D, Maniatis T. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayeda A, Screaton G R, Chandler S D, Fu X-D, Krainer A R. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umen J G, Guthrie C. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 23.Horowitz D S, Krainer A R. Genes Dev. 1997;11:139–151. doi: 10.1101/gad.11.1.139. [DOI] [PubMed] [Google Scholar]
- 24.Chua K, Reed R. Genes Dev. 1999;13:841–850. doi: 10.1101/gad.13.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiara M D, Reed R. Nature (London) 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- 26.Bruzik J P, Maniatis T. Proc Natl Acad Sci USA. 1995;92:7056–7059. doi: 10.1073/pnas.92.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaal T D, Maniatis T. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarn W Y, Steitz J A. Proc Natl Acad Sci USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roscigno R F, Garcia-Blanco M A. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 31.Stark J M, Bazett-Jones D P, Herfort M, Roth M B. Proc Natl Acad Sci USA. 1998;95:2163–2168. doi: 10.1073/pnas.95.5.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu A, Nussenzweig M C, Han H, Sanchez M, Honjo T. J Exp Med. 1991;173:1385–1393. doi: 10.1084/jem.173.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sureau A, Soret J, Vellard M, Crochet J, Perbal B. Proc Natl Acad Sci USA. 1992;89:11683–11687. doi: 10.1073/pnas.89.24.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caudavilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt F G. Proc Natl Acad Sci USA. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frantz S A, Thiara A S, Lodwick D, Ng L L, Eperon I C, Samani N J. Proc Natl Acad Sci USA. 1999;96:5400–5405. doi: 10.1073/pnas.96.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]