Abstract
The highly conserved SMC (Structural Maintenance of Chromosomes) proteins function in chromosome condensation, segregation, and other aspects of chromosome dynamics in both eukaryotes and prokaryotes. A null mutation in the Caulobacter crescentus smc gene is conditionally lethal and causes a cell cycle arrest at the predivisional cell stage. Chromosome segregation in wild-type and smc null mutant cells was examined by monitoring the intracellular localization of the replication origin and terminus by using fluorescence in situ hybridization. In wild-type cells, the origin is located at the flagellated pole of swarmer cells and, immediately after the initiation of DNA replication in stalked cells, one of the origins moves to the opposite pole, giving a bipolar localization of the origins. The terminus moves from the end of the swarmer cell opposite the origin to midcell. A subpopulation of the smc null mutant cells had mislocalized origins or termini, showing that the smc null mutation gives DNA segregation defects. Nucleoid morphology was also abnormal. Thus, we propose that the Caulobacter chromosomal origins have specific cellular addresses and that the SMC protein plays important roles in maintaining chromosome structure and in partitioning. The specific cell cycle arrest in the smc null mutant indicates the presence of a cell cycle checkpoint that senses perturbations in chromosome organization or segregation.
Most bacterial cells contain a single circular chromosome that is folded and compacted into a structure called the nucleoid. The newly replicated chromosomes are segregated to daughter cells with very high fidelity (1–3). Despite its essential function in the bacterial cell cycle, little is known about the mechanism ensuring accurate chromosome segregation. However, recent work has shown that the bacterial nucleoid is highly organized and that the organization may be important for efficient chromosome segregation (4, 5). In Bacillus subtilis and Escherichia coli, the origin proximal regions of the chromosomes tend to be oriented toward opposite ends of the cells (6–10). Newly replicated origins are separated rapidly (7, 9), suggesting that a mitotic-like apparatus might function in bacteria. The terminus proximal region of the chromosome generally locates toward midcell, and regions in between the origin and the terminus locate at intermediate positions (6–10).
Proteins that are involved in efficient chromosome partitioning include Spo0J of B. subtilis and ParA and ParB of Caulobacter crescentus, which are homologs of plasmid partitioning proteins. Deletion of spo0J results in an increase in the number of anucleate cells (2), indicating that it has a role in chromosome segregation. Spo0J binds to at least eight sites in the origin-proximal part of the chromosome (11) and is found in discrete foci at the same intracellular positions as the origin (10, 12–15). The Caulobacter ParA and ParB proteins are essential, and overexpression causes chromosome segregation defects. The ParB protein binds to a region close to the origin, and the ParA and ParB proteins localize in foci at the poles (3).
Another protein that may have a role in chromosome segregation is SMC (Structural Maintenance of Chromosomes). SMC proteins are present in bacteria, archaea, and eukaryotes, and are involved in many different aspects of chromosome maintenance and structure, including chromosome condensation, sister chromatid cohesion, dosage compensation, and DNA repair (16–18). The SMC homolog of B. subtilis has a role in maintaining chromosome structure and in partitioning, because a smc null mutant exhibits decondensed nucleoids and produces anucleate cells (19, 20). The B. subtilis SMC protein is associated with the nucleoid and localizes in discrete foci near the cell poles (19, 21). A SMC homolog is not present in E. coli, but the MukB protein may be a functional analog of SMC; the properties of MukB are remarkably like those of SMC, and the phenotypes of smc and mukB mutations are similar (22–24).
Caulobacter is a valuable bacterial model system for studying chromosome segregation because chromosome replication is initiated only once per cell cycle at a well defined time, making it easy to separate and analyze cell cycle events in synchronized populations (25). Differentiation is an integral part of the Caulobacter cell cycle. A motile swarmer cell differentiates into a stalked cell by shedding the flagellum, synthesizing a stalk at the same pole, and initiating chromosome replication (see Fig. 5). The stalked cell then differentiates into an asymmetric predivisional cell by synthesizing a flagellum at the pole opposite the stalk. Cell division yields a new swarmer cell and a stalked cell.
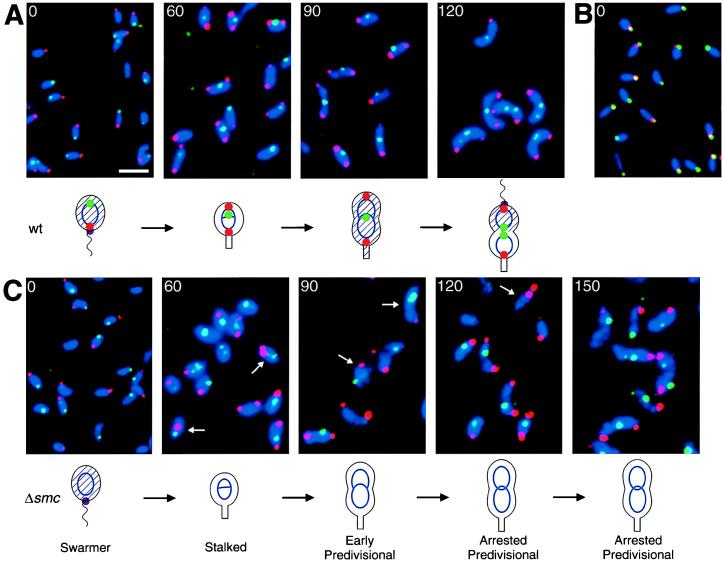
Figure 5.
Localization of the origin and terminus proximal regions of the Caulobacter chromosome by using FISH. Wild-type (A and B) and smc null mutant (C) swarmer cells were isolated and allowed to progress synchronously through the cell cycle at 32°C. At the indicated times (in minutes), the cells were fixed and hybridized simultaneously with a Cy3-labeled origin probe (red) and a FluorX-labeled terminus probe (green) (A and C). In B, the origin was localized by using a Cy3-labeled origin probe (red), and the McpA chemoreceptor was visualized by immunofluorescence staining with FITC-labeled secondary antibodies (green). The McpA protein localizes at the flagellated pole in swarmer cells (30). The nucleoids were visualized by DAPI staining (blue). Because no nucleoid-free regions are present in wild-type Caulobacter, the DAPI staining outlines the cells. Arrows indicate cells with abnormal localization of the origin or the terminus, and the white scale bar represents 2 μm. The cell cycle of each strain is diagrammed. Blue ovals and theta structures represent nonreplicating and replicating chromosomes, respectively. The intracellular localization of the origin (red dot), the terminus (green dot), and the McpA chemoreceptor (purple dot) are shown. The presence of CtrA (29) is indicated with shading of the cell.
Here, we identify the Caulobacter smc gene and show that a smc null mutation causes a conditional lethal phenotype and defects in nucleoid structure. Surprisingly, the absence of the SMC protein caused the cell cycle to arrest at the predivisional cell stage. Fluorescence in situ hybridization (FISH) analysis of the spatial disposition of the chromosomal origin and terminus showed that their normally polar and midcell locations, respectively, are disrupted in a subpopulation of smc mutant cells. Thus, SMC is important in maintaining correct nucleoid structure, and nucleoid organization defects in Caulobacter result in a cell cycle block. These results suggest that a cell cycle checkpoint in Caulobacter may monitor chromosome segregation.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Media.
The synchronizable C. crescentus strain CB15N (also named NA1000) (26) was used as the wild-type strain. C. crescentus strains were grown in peptone-yeast extract medium (PYE) or M2-glucose minimal medium (27). The chromosomal smc null allele was constructed by using a two-step knockout technique. A PCR fragment containing the ends of smc and the flanking regions was generated by double-PCR by using the primers SMC1 (5′-ATGTTGCGGATCCCCGTTGATCCTGTAGGTGGA-3′), SMC2 (5′-GTTGAATTCGACACCAAGGCGCTGGAC-3′), SMC3 (5′-AAAACTGCAGGTGCTGTTCACGCTGAAGG-3′), and SMC4 (5′-TCAACGGGGATCCGCAACATGCTGGACGAGAT-3′), and chromosomal DNA as template. The fragment was cloned into the integration plasmid pNPTS138 (M. R. K. Alley, personal communication), resulting in pRBJ560. A gentamycin resistance (GentR) cassette from pUCGM (28) was inserted in the BamHI site between the upstream and downstream sequences, resulting in plasmid pRBJ561. This plasmid was mobilized by bacterial conjugation (27) into CB15N, and clones containing tandem copies of the wild-type and the disrupted smc genes were selected. Excision of the integrated plasmid was selected for by growing on PYE plates containing 3% sucrose at 20°C. The Δsmc∷GentR allele was transduced into clean background by using ΦCR30 (27), resulting in the strain CB15NΔsmc.
Identification of the Caulobacter smc Gene.
Parts of the Caulobacter smc gene were identified by searching the partial genome sequence obtained from the TIGR database (Institute for Genomic Research). Sequencing of the entire gene was performed at the Protein and Nucleic Acid (PAN) facility at Stanford University. The DNA sequence has been submitted to GenBank and assigned the accession number AF172724.
Synchronization, Immunoblotting, and Flow Cytometry.
Pure populations of swarmer cells were isolated by Ludox centrifugation (26). Immunoblotting was performed as described (29) by using antibodies to CtrA (29), McpA (30), and the major 25 kDa Flagellin (U. Jenal, personal communication). Samples for flow cytometry were prepared and analyzed as described (31).
FISH. Probes for FISH were synthesized as described (32). Plasmid pGM1342 (G. Marczynski, personal communication) that contains a 13-kb EcoRI fragment overlapping the Caulobacter origin of replication was used for the origin probe. Plasmid pKH1.1 (33), which contains a 15-kb EcoRI fragment with the flbN flaDBC flbO motC genes, was used for the terminus probe. These genes are expected to be close to the terminus, because they are located opposite the origin on the Caulobacter genetic map (B. Ely, personal communication). Cy3-dCTP and FluorX-dCTP (Amersham Pharmacia Biotech) were used for labeling.
Cells in PYE were fixed with 2.5% formaldehyde in 30 mM sodium phosphate (pH 7.5) for 15 min at room temperature and 45 min on ice. The fixed cells were washed three times in PBS (140 mM NaCl/3 mM KCl/8 mM Na2HPO4/1.5 mM KH2PO4) and resuspended in GTE (50 mM glucose/20 mM Tris⋅HCl, pH 7.5/10 mM EDTA). Lysozyme was added to 2.5 μg/ml, and 12 μl cells were placed on a poly-l-lysine-treated slide and kept for 10 min at room temperature. The cells were dried, placed in methanol (−20°C) for 10 min, in acetone (−20°C) for 30 s, and allowed to dry. The slides were washed two times in 2 × SSCT (300 mM NaCl/30 mM NaCitrate/0.1% Tween-20, pH 7.0) for 5 min and incubated in 2 × SSCT + 50% formamide at 37° for 30–60 min. Then 12 μl hybridization solution (3 × SSC/50% formamide/10% dextran sulfate/4–8 ng/μl of each probe) was added and covered with a 22 × 22 mm coverslip. The DNA was denatured by placing the slide on a 94°C heat block for 2 min, and the probes were hybridized overnight at 42°C. The slides were washed two times in 2× SSCT + 50% formamide for 30 min at 37°, one time in 2× SSCT + 25% formamide for 10 min, three times in 2× SSCT for 10 min, and briefly with PBS. When relevant, immunofluorescence localization of a protein was performed as described (29), starting with the blocking step. The slides were mounted with Fluoroguard (Bio-Rad) + 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI).
Microscopy.
To examine nucleoid morphology by using DAPI staining, the cells were collected by centrifugation, resuspended in the remaining media, added to methanol (−20°C), and incubated for at least 10 min at −20°C. Cells were placed on poly-l-lysine-treated coverslips, allowed to dry, washed three times with PBS, and mounted with Slowfade (Molecular Probes) + 1 μg/ml DAPI. Images were obtained with a MicroMAX cooled CCD camera (Princeton Instruments, Trenton, NJ) by using a Nikon Eclipse E800 microscope with a Nikon Plan APO 100×/1.40 differential interference contrast microscopy objective. The images were acquired and processed by using metamorph 3.6A (Universal Imaging, Media, PA).
RESULTS
Identification of the Caulobacter crescentus smc Homolog.
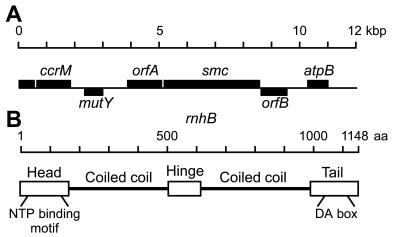
A search of the partial Caulobacter genome sequence database revealed parts of a gene that were homologous to eukaryotic and prokaryotic smc genes (25–40% identity and 45–60% similarity). The entire gene was sequenced and found to encode a predicted ORF of 1,148 amino acids with a calculated molecular mass of 123.2 kDa. The genetic organization of the chromosomal region is shown in Fig. 1A. The Caulobacter smc gene is expected to be transcribed as the second gene in an operon with a gene that shows homology to hypothetical genes from other bacteria. All the SMC signature motifs and domain organization (24) are conserved in the Caulobacter SMC protein (see Fig. 1B): an amino-terminal NTP-binding domain, two heptad repeat regions predicted to form coiled-coils separated by a hinge region, and a carboxyl-terminal region containing the DA-box signature motif (16). Thus, the gene identified is likely to encode the Caulobacter SMC protein.
Figure 1.
Organization of the Caulobacter chromosomal region containing the smc gene and SMC protein domain structure. (A) Predicted genetic organization of the smc chromosomal locus. Bars above the line represent genes transcribed from left to right; bars below the line represent genes transcribed in the opposite direction. The orfA and orfB genes are putative ORFs that show homology to genes of unknown function in other bacteria. (B) Predicted domain structure of the Caulobacter SMC protein. The domains were assigned by homology to other SMC proteins (24) and the locations of the coiled-coil regions were predicted as described (40).
Defects in Cell Growth in a smc Null Mutant.
To examine the function of smc in Caulobacter, we constructed a null mutation by replacing the majority of the coding region with a gene that confers resistance to gentamycin, resulting in the strain CB15NΔsmc. On rich medium (PYE) plates, the smc null mutant was temperature sensitive for growth, because it formed colonies at 20°C but not at higher temperatures. The mutant strain appeared to rapidly acquire compensatory mutations that partially relieved the growth defect. The growth defect was less severe on M2 glucose minimal medium (M2G) plates, because the null mutant was viable, but grew poorly at all temperatures. When grown in liquid medium, the growth defect was more severe. The smc null mutant was inviable at all temperatures in rich medium, and in minimal medium it was viable only below 25°C, although the generation time (≈6 hr in M2G at 20°C) was longer than that of the parent strain (≈4 hr). Shift of a culture grown at 20°C in minimal medium to 30°C or to rich medium resulted in rapid loss of viability, because the number of colony-forming units dropped 100-fold within 2 hr (data not shown).
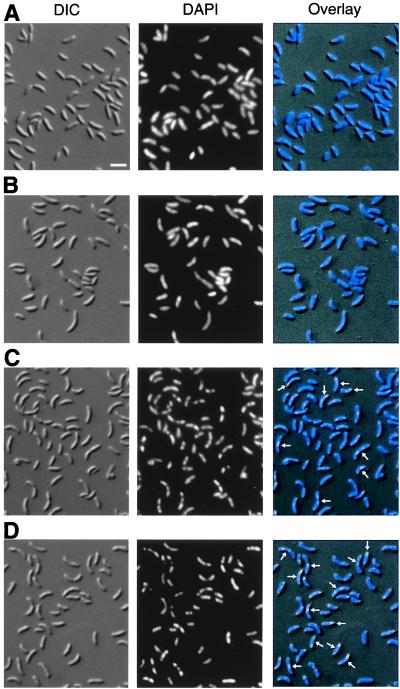
Abnormal Nucleoid Morphology and Distribution of Cell Types in the smc Null Mutant. In B. subtilis and in eukaryotes, SMC proteins are involved in different aspects of chromosome organization (17–20). We therefore examined nucleoid morphology in the smc null mutant by combined Nomarski differential interference contrast microscopy and DAPI staining (Fig. 2). In the parent strain CB15N, the nucleoids were found to be smooth, and staining was observed in the entire cell (Fig. 2A and B). Staining of live or fixed cells with the DNA dyes Hoechst33258, Propidium Iodide, or SYTO16 confirmed that no nucleoid-free regions are detectable in wild-type Caulobacter cells (data not shown). However, in the smc mutant cells, the nucleoids appeared irregular in many cells, and chromosome-free regions were observed (cells marked with arrows in Fig. 2C). When the cells were incubated at the nonpermissive temperature (30°C for 4 hr), the effect was exaggerated, and the majority of the cells had highly irregularly shaped nucleoids and chromosome-free regions (Fig. 2D). However, less than 0.1% of the smc null mutant cells were found to be anucleate when grown at both low and high temperatures.
Figure 2.
Nucleoid morphology in wild-type and smc null mutant cells. (A) The parent strain CB15N grown at 20°C. (B) CB15N grown at 20°C and shifted to 30°C for 4 hr. (C) The smc null mutant strain CB15NΔsmc grown at 20°C. (D) CB15NΔsmc grown at 20°C and shifted to 30°C for 4 hr. Nomarski differential interference contrast microscopy images of the cells are shown in the first column, DAPI-stained nucleoids are shown in the second column, and the third column is an overlay of the images. Arrows indicate cells with abnormal nucleoid morphology, and the white scale bar represents 2 μm.
In addition to the defects in nucleoid structure, we observed abnormal distribution of cell types in the smc null mutant cultures. When grown in minimal medium at 20°C, cell cultures were almost normal, but after incubation at the nonpermissive temperature for 4 hr, swarmer cells were no longer observed, and a high proportion of the culture accumulated at the predivisional cell stage (Fig. 2D). The abnormal distribution of cell types indicates that the smc null mutant has a cell division defect, in addition to a defect in nucleoid organization.
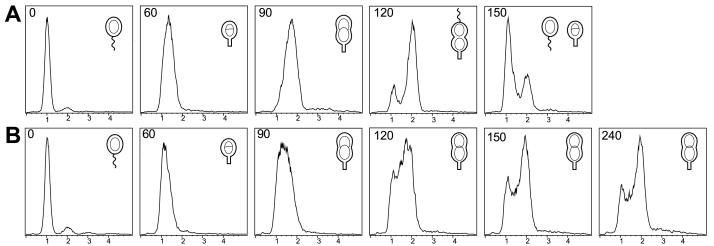
Characterization of the Cell Division Defect in the smc Null Mutant. To characterize the cell division phenotype of the smc null mutant, swarmer cells were isolated from wild-type and mutant cultures grown in minimal medium at 20°C. The swarmer cells were then incubated at 32°C and allowed to proceed synchronously through the cell cycle. Progression through the cell cycle was monitored by microscopy and by measuring the DNA content of the cells by using flow cytometry (Fig. 3). The parent strain CB15N progressed normally through the cell cycle at 32°C (Fig. 3A). The mutant strain CB15NΔsmc carried out the swarmer-to-stalked cell transition and initiated DNA replication at the same time (approximately 60 min) as the parent strain (Fig. 3B). However, approximately 80% of the mutant cells appeared to arrest as elongated cells with a slight invagination in the middle of the cell. Flow cytometry analysis showed that these cells had two completely replicated chromosomes (Fig. 3B). The remaining ≈20% of the mutant cells appeared to divide normally or were somewhat delayed. No filamentous cells or cells with more than two chromosomes were observed even after prolonged incubation at 32°C. The CB15NΔsmc culture increased in cell mass for approximately 4 hr after the shift to the nonpermissive temperature, but eventually stopped growing. A similar flow cytometry analysis of nonsynchronized populations of wild-type and smc mutant cells also showed that the majority of the mutant cells arrested as predivisional cells, confirming that the observed cell cycle arrest was not caused by the synchronization procedure (data not shown).
Figure 3.
Characterization of the cell cycle block in the smc mutant strain. Swarmer cells from the parent strain CB15N (A) and the smc null mutant strain CB15NΔsmc (B) grown at 20°C were isolated and shifted to 32°C. Progression through the cell cycle was examined by measuring the DNA content of chromomycin A3-stained cells by using flow cytometry. Chromosome equivalents are indicated on the x-axis and relative number of cells on the y-axis. The times (in minutes) when the samples were removed and fixed are indicated. Cell cycle progression is shown schematically. In the wild-type strain, swarmer cells are present at 0 min, stalked cells at 60 min, early predivisional cells at 90 min, and late predivisional cells at 120 min. Cell division occurs at 140 min.
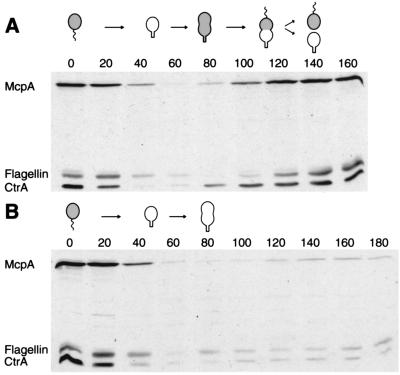
Because the smc mutant cells appeared to arrest as predivisional cells, we tested other events that normally occur in the predivisional cell. These include synthesis of the CtrA cell cycle regulator (29, 34) and the McpA chemoreceptor (35) and assembly of a flagellum at the pole opposite the stalk (25). The analysis was done by measuring the levels of the CtrA, McpA, and 25-kDa flagellin proteins by using immunoblot analysis of synchronized cultures that were allowed to progress through the cell cycle at 32°C. In the parent strain (Fig. 4A), CtrA and McpA were degraded, and the flagellum was shed at the swarmer-to-stalked cell transition. CtrA was then synthesized at the stalked-to-predivisional cell transition, and McpA and the 25-kDa flagellin reappeared in the predivisional cells. In the smc mutant cells (Fig. 4B), the proteins were turned over normally at the swarmer-to-stalked cell transition but did not reappear later in the cell cycle. Thus, it appears that the smc null mutants arrest as predivisional cells after the first sign of constriction becomes visible, but before the master cell cycle regulator CtrA is synthesized.
Figure 4.
Presence of the McpA chemoreceptor, the CtrA cell cycle regulator, and flagellin during the cell cycle in wild-type and Δsmc cells. The McpA, CtrA, and 25-kDa flagellin proteins were detected by immunoblotting samples from synchronous cultures of CB15N (A) or CB15NΔsmc (B) grown at the restrictive temperature of 32°C. Samples were taken every 20 min, and equal amounts of cells were applied to each lane. The cell cycle progression of each strain is shown schematically. Gray shading indicates the presence of CtrA in the different cell types as determined previously (29).
Use of FISH to Examine the Intracellular Localization of Specific Regions of the Caulobacter Chromosome. Because the smc mutation causes defects in chromosome organization as visualized by using DAPI staining, we examined the intracellular localization of specific regions of the chromosome by using FISH during the cell cycle of wild-type and smc mutant cells. Swarmer cells from wild-type and smc mutant cultures grown in minimal medium at 20°C were isolated and then incubated at 32°C. Aliquots of synchronized cells at different stages of the cell cycle were fixed and hybridized with origin and terminus probes simultaneously.
The intracellular localization of the origin and the terminus proximal regions of the Caulobacter chromosome during the cell cycle in the wild-type strain CB15N is shown in Fig. 5A. In swarmer cells (Fig. 5A, 0 min), an origin focus was observed at one pole of the cell, and a terminus focus was observed at the opposite end of the cell. The origin was located at the extreme end of the cell, but the localization of the terminus focus varied from cell to cell. In some cells, the terminus focus was located at the opposite pole and in other cells close to midcell. To examine which pole the origin localized to in swarmer cells, we simultaneously visualized the location of the origin by using FISH and the location of the McpA chemoreceptor protein by using immunofluorescence microscopy (Fig. 5B). The origin of replication and the McpA protein were found to colocalize. Because McpA localizes to the flagellated pole (36), the colocalization shows that the origin localizes to the flagellated pole. After initiation of DNA replication in stalked cells (Fig. 5A, 60 min), a second origin focus was observed at the opposite pole. Only one terminus focus was observed, and it was located at midcell in most cells, but in some cells the terminus focus was located close to one pole. Very few cells with two origin foci at one end of the cell were observed (approximately 1% of the two-foci cells). These results indicates that one of the origins is rapidly moved to the opposite pole shortly after initiation of chromosome replication. In early predivisional cells (Fig. 5A, 90 min), we observed bipolar localization of the origin foci and localization of the terminus focus at midcell in the majority of the cells. In late predivisional cells (Fig. 5A, 120 min), in addition to bipolar localization of the origin foci, we observed two terminus foci in many cells. The two terminus foci were generally located close to midcell, but the distance between the foci varied from cell to cell.
Mislocalization of the Origin of Replication and the Terminus in smc Mutant Cells.
Fig. 5C shows the localization of the origin and the terminus proximal regions of the chromosomes in the smc mutant. In the majority of the mutant cells, the origin and terminus foci were observed at the same sites as in wild-type cells (Fig. 5A). But 10–15% of the cells had mislocalized origin or terminus foci (cells marked with arrows in Fig. 5C). Cells with mislocalized origins were rare in the wild-type strain (<1% of the cells). We also observed that the FISH foci in the smc mutant cells were less condensed and less regular in size than in wild-type cells when samples were prepared in parallel. Additionally, more cells that did not have detectable foci were observed in the smc mutant. These results indicate that the smc mutant has a defect in chromosome condensation or organization, which could interfere with the process that localizes the origin, resulting in the increased frequency of cells with mislocalized origins or termini.
DISCUSSION
The Caulobacter genome contains a gene that belongs to the prokaryotic and eukaryotic smc gene family. To characterize the functions of Caulobacter smc, we constructed a null mutation by exchanging the majority of the coding region with a gene that confers resistance to the antibiotic gentamycin. The null mutant was found to have a conditionally lethal phenotype; it was viable only in minimal media at 25°C and below. DAPI staining showed that the smc null mutant had abnormal nucleoid structure, because the nucleoids appeared to be irregular and chromosome-free regions in the cells were observed. This effect was more severe when the cells were incubated at nonpermissive conditions. The observation of irregular nucleoids indicates that the smc mutant has a defect in nucleoid organization at a global level.
To examine further the defects in chromosome organization in the smc mutant, we used the FISH technique to visualize the localization of specific regions of the chromosome. We found that in wild-type cells, the origin and terminus proximal regions localized in a cell cycle-specific manner (Fig. 5A). In swarmer cells, the origin was found to localize to the flagellated pole, and the terminus was found in the opposite end of the cell. Shortly after initiation of chromosome replication in the stalked cell, one of the newly replicated origins migrated to the opposite pole of the cell, giving a bipolar organization of the origins. Thus, randomization of chromosome segregation may happen as early as after replication of the first 10 kb of the chromosome. The terminus was found to gradually move to midcell, and on completion of chromosome replication two terminus foci were observed at midcell. It is likely that an active process moves the origin, because the movement appears to be rapid, whereas the movement of the terminus is probably a passive consequence of DNA replication and nucleoid rebuilding. The localization pattern is similar, but not identical, to the patterns observed in slow-growing B. subtilis and E. coli cells (6, 8). In Caulobacter, the origins were located at the extreme ends of the cells and not at a distance from the poles as observed in B. subtilis and E. coli cells (6–8).
Examination of the intracellular localization of the origin and terminus in smc null mutants showed that in 10–15% of the cells, the origin and/or the terminus are mislocalized. Because less than 1% of wild-type cells had mislocalized origins, we conclude that the smc mutation results in DNA segregation defects. Additionally, we observed that in the smc mutant the FISH foci were less condensed and had a less regular size, suggesting that SMC is involved in organizing and/or condensing the chromosome at a local level. Defects in these functions could lead to interference with organization of the nucleoid at the global level as observed by DAPI staining. Defects in chromosome organization could interfere with the processes that localize the origins, and this may cause the high frequency of cells with abnormal origin localization. The Caulobacter ParA and ParB proteins localize to polar positions in predivisional cells, and it has been suggested that the proteins are involved in positioning the origins (3). The functions of the ParA and ParB proteins may depend on SMC.
In addition to the defects in chromosome organization, we observed a cell cycle block in the smc null mutant. At nonpermissive conditions, the cells appeared to differentiate from swarmer-to-stalked cells normally but then arrest as predivisional cells. The arrested smc mutant cells had slight invaginations at the middle of the cells and two completely replicated chromosomes, but failed to synthesize the CtrA cell cycle regulator, the McpA chemoreceptor, and to assemble a flagellum. The cell cycle arrest at a specific stage suggests that a cell cycle checkpoint senses perturbations in chromosome organization or segregation. How the putative checkpoint senses the perturbations in chromosome dynamics is unknown. Previously, it was shown that inhibition of DNA replication prevented synthesis of flagellin proteins, and the existence of a cell cycle checkpoint responsible for the inhibition of flagellum formation was suggested (37, 38). It is possible that the same mechanism senses inhibition of DNA replication and nucleoid organization or segregation defects.
The SMC protein from B. subtilis has been characterized recently. A null mutant has a conditional lethal phenotype and is deficient in chromosome organization and partitioning including misassembly of Spo0J partitioning protein foci (19, 20). The defects in chromosome organization in B. subtilis are similar to the defects we here observe in a Caulobacter smc null mutant. However, in B. subtilis, formation of anucleate cells was observed, whereas we did not observe a significant amount of anucleate cells in Caulobacter. The lack of anucleate cells is a logical consequence of the cell cycle arrest in the Caulobacter smc mutant because division fails to occur. The B. subtilis SMC protein localizes to discrete foci near the cell poles and to the nucleoid (19, 21). It was proposed that the polar foci are storage depots for SMC protein not bound to the chromosome. Purified B. subtilis SMC protein binds preferentially to single-stranded DNA (ssDNA), has a ssDNA-activated ATPase activity, and forms large nucleoprotein aggregates in a ssDNA- and ATP-dependent matter (39). The relevance of these in vitro activities for the in vivo function of SMC is unknown.
It has been proposed that bacterial SMC proteins function by condensing and folding the nucleoid, and that defects in chromosome organization result in the additional phenotypes observed in the smc mutants (19, 20). Our work supports such a function for the Caulobacter SMC protein but ties the function of the SMC protein to cell cycle progression. Further studies of bacterial SMC proteins should provide new insights in basic mechanisms of chromosome dynamics that may be conserved from bacteria to higher eukaryotes.
Acknowledgments
We thank members of the Shapiro laboratory for critical reading of the manuscript and Abby Dernburg for help with FISH. R.B.J. was supported by a European Molecular Biology Organization postdoctoral fellowship. This work was supported by National Institutes of Health grants GM32506/5120MZ and GM51426 and by a grant from the Office of Naval Research (N00014-96-1-0564).
ABBREVIATIONS
- FISH
fluorescence in situ hybridization
- PYE
peptone-yeast extract medium
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF172724).
References
- 1.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ireton K, Gunther N W t, Grossman A D. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohl D A, Gober J W. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 4.Harry E J. Trends Microbiol. 1997;5:295–297. doi: 10.1016/S0966-842X(97)01091-3. [DOI] [PubMed] [Google Scholar]
- 5.Sharpe M E, Errington J. Trends Genet. 1999;15:70–74. doi: 10.1016/s0168-9525(98)01660-6. [DOI] [PubMed] [Google Scholar]
- 6.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 7.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 8.Niki H, Hiraga S. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webb C D, Graumann P L, Kahana J A, Teleman A A, Silver P A, Losick R. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 10.Teleman A A, Graumann P L, Lin D C H, Grossman A D, Losick R. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 11.Lin D C, Grossman A D. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 12.Lin D C H, Levin P A, Grossman A D. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 14.Lewis P J, Errington J. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- 15.Sharpe M E, Errington J. Mol Microbiol. 1998;28:981–990. doi: 10.1046/j.1365-2958.1998.00857.x. [DOI] [PubMed] [Google Scholar]
- 16.Koshland D, Strunnikov A V. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 17.Strunnikov A V. Trends Cell Biol. 1998;8:454–459. doi: 10.1016/s0962-8924(98)01370-1. [DOI] [PubMed] [Google Scholar]
- 18.Hirano T. Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 19.Britton R A, Lin D C, Grossman A D. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriya S, Tsujikawa E, Hassan A K, Asai K, Kodama T, Ogasawara N. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 21.Graumann P L, Losick R, Strunnikov A V. J Bacteriol. 1998;180:5749–5755. doi: 10.1128/jb.180.21.5749-5755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. EMBO J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melby T E, Ciampaglio C N, Briscoe G, Erickson H P. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenal U, Stephens C, Shapiro L. Adv Enzymol Relat Areas Mol Biol. 1995;71:1–39. doi: 10.1002/9780470123171.ch1. [DOI] [PubMed] [Google Scholar]
- 26.Evinger M, Agabian N. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ely B. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 28.Schweizer H P. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 29.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 30.Alley M R, Maddock J R, Shapiro L. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler E, Shapiro L. J Mol Biol. 1995;251:346–365. doi: 10.1006/jmbi.1995.0439. [DOI] [PubMed] [Google Scholar]
- 32.Dernburg A F, Sedat J W. Methods Cell Biol. 1998;53:187–233. doi: 10.1016/s0091-679x(08)60880-8. [DOI] [PubMed] [Google Scholar]
- 33.Hahnenberger K H, Shapiro L. J Mol Biol. 1987;194:91–103. doi: 10.1016/0022-2836(87)90718-2. [DOI] [PubMed] [Google Scholar]
- 34.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 35.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alley M R K, Maddock J R, Shapiro L. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 37.Osley M A, Sheffery M, Newton A. Cell. 1977;12:393–400. doi: 10.1016/0092-8674(77)90115-5. [DOI] [PubMed] [Google Scholar]
- 38.Sheffery M, Newton A. Cell. 1981;24:49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- 39.Hirano M, Hirano T. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]