Abstract
Accumulating evidence has shown that many molecules, including some cyclin-dependent kinases (Cdks) and cyclins, as well as the death-effector domain (DED)-containing FADD, function for both apoptosis and cell cycle. Here we identified that DEDD, which also possesses the DED domain, acts as a novel inhibitor of the mitotic Cdk1/cyclin B1 complex. DEDD associates with mitotic Cdk1/cyclin B1 complexes via direct binding to cyclin B1 and reduces their function. In agreement, kinase activity of nuclear Cdk1/cyclin B1 in DEDD-null (DEDD−/−) embryonic fibroblasts is increased compared with that in DEDD+/+ cells, which results in accelerated mitotic progression, thus exhibiting a shortened G2/M stage. Interestingly, DEDD−/− cells also demonstrated decreased G1 duration, which perhaps enhanced the overall reduction in rRNA amounts and cell volume, primarily caused by the rapid termination of rRNA synthesis before cell division. Likewise, DEDD−/− mice show decreased body and organ weights relative to DEDD+/+ mice. Thus, DEDD is an impeder of cell mitosis, and its absence critically influences cell and body size via modulation of rRNA synthesis.
Keywords: apoptosis, cell cycle, cell size, cyclin-dependent kinase, mitosis
Cell size control is tightly associated with cell cycle regulation. The consensus is that cells predominantly increase their volume during the G1 cell cycle phase (1–3). However, evidence has demonstrated that a proper cell growth also undergoes before cell division (4, 5), where rRNA and protein synthesis achieve maximal activity. The duration of this period is defined by the activation of cyclin-dependent kinase 1 (Cdk1)/cyclin B1 complexes, which is proceeded through dephosphorylation of the inhibitory residues of Cdk1 and Tyr-15, and also Thr-14 in higher eukaryotes, by the Cdc25 phosphatase family (4–10), followed by translocation of Cdk1/cyclin B1 into the nucleus. Thus, defects in Cdc25 genes delay mitotic entry, resulting in increased cell size (11–13). In contrast, an inadequate duration before division, due to aberrant phosphorylation of Tyr-15, causes cells to enter mitosis before sufficient growth, resulting in decreased size of the daughter cells (14–17). Interestingly, the loss of Wee1-related kinases, responsible for phosphorylation of Thr-14/Tyr-15, causes premature cell division in yeast, Xenopus, or Drosophila cells, but not in higher mammalian cells (4, 5, 9, 10, 15–18). This suggests the presence of alternative mechanism(s), which may also influence cell size, particularly in mammalian cells. However, the responsible mechanisms have remained unclear.
Linkage of cell cycle and apoptosis has been recognized for many molecules, including some Cdks and cyclins (19, 20). Recently, it was also demonstrated that the death-effector domain (DED)-containing molecule FADD regulates mitosis (21). The DED domain of ≈80 amino acid residues is well conserved in various death-inducing proteins (22–24). The DED of FADD recruits two DED-containing caspases, caspase-8 and caspase-10, to form the death-inducing signal complex, thereby initiating apoptosis (23–30). Although the DEDs have no enzymatic function, they link participants in a signaling chain through homotypic interactions (31). A DED domain is also present in the N terminus of the DEDD molecule (32). DEDD is localized to the nucleus, with a strong accumulation at the nucleoli, consistent with the presence of multiple nuclear localizing sequences (32). In vitro binding analysis showed DEDD can also associate with FADD or caspase-8/10 via DEDs. Overexpression studies have suggested a weak proapoptotic effect for DEDD; however, the physiological role of DEDD has remained unelucidated until now (32, 33).
In this study we found that DEDD in fact inhibits the activity of Cdk1/cyclin B1 complexes subsequent to their translocation into the nucleus. This finding proposes a novel impeditive mechanism of Cdk1/cyclin B1 activity within the nucleus, independent of its activation through phosphorylation and dephosphorylation of the inhibitory residues in the cytoplasm. We also suggest that this DEDD-mediated decrease of the Cdk1/cyclin B1 activity extends the progression of mitosis and thus appears to play a role in cell size regulation before cell division.
Results
Normal Apoptosis Responses but Shortened Mitotic Progression in DEDD−/− Cells.
To investigate physiological roles of DEDD, we created DEDD−/− mice [supporting information (SI) Fig. 5A] and first compared the induction of apoptosis in primary and 3T3 mouse embryonic fibroblast (MEF) cells generated from mutant and control wild-type mice. Complete lack of DEDD expression was confirmed by Northern blot analysis of RNA from those cells (SI Fig. 5B). In contrast to previous overexpression studies, apoptosis induction was unaffected in DEDD−/− cells: activation of caspase-3 and caspase-8 in response to CD95 ligation, TNFα, or staurosporine was comparable in mutant and wild-type cells (SI Fig. 6 A and B). Also, both types of cells harbored similar proportions of TUNEL-positive apoptotic or propidium iodide-stained dead cells after induction of apoptosis (SI Fig. 6 C and D).
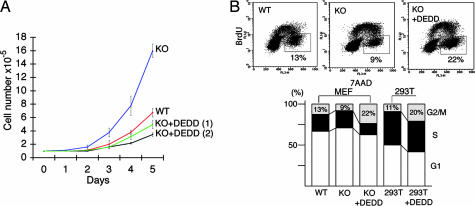
Interestingly, however, the proliferative status of DEDD−/− MEF cells was notably increased compared with DEDD+/+ cells (Fig. 1A, WT and KO). The doubling time was 45.8 ± 6.5 h in WT cells and 37.5 ± 2.5 h in knockout (KO) cells (n = 9 each). The acceleration in proliferation was solely mediated by DEDD deficiency, because reexpression of mouse DEDD in mutant cells tempered the phenotype (Fig. 1A, KO+DEDD 1 and 2). BrdU incorporation analyses revealed a specific decrease in the proportion of cells at the G2/M fraction in DEDD−/− MEF cells. Reexpression of DEDD in DEDD−/− cells specifically increased the G2/M fraction (Fig. 1B Upper and first three lanes of Lower), indicating that the change in G2/M fraction in DEDD−/− cells was solely mediated by DEDD deficiency. Similarly, in 293T cells, overexpression of DEDD enlarged the proportion of G2/M (Fig. 1B, last two lanes of Lower). We measured the actual length of each cell cycle stage based on both the doubling time and the proportion of each cell cycle stage obtained through the BrdU incorporation analysis. As displayed in Table 1, the comparative difference (% ratio) between DEDD−/− and DEDD+/+ cells or between DEDD−/− and DEDD-transfected DEDD−/− cells was most striking for G2/M, although the actual length of G1 was also shortened. In addition, the length of early and late mitosis stages (before and after the metaphase) in DEDD+/+ and DEDD−/− cells was investigated by counting mitotic index in asynchronous cells, which was estimated based on the morphology after the DNA staining with 7-amino-actinomycin D, and then multiplying the percentage of cells in early or late mitosis by the cell cycle length. As also shown in Table 1, in DEDD−/− cells the early mitosis stage (before the metaphase) was most profoundly shortened. When the mitotic index was analyzed after synchronizing MEF cells in G1 via starvation, the decrease after the peak in the percentage of cells in mitosis was accelerated in DEDD−/− cells compared with control DEDD+/+ cells (SI Fig. 7), indicating that the M phase rather than the G2 phase is shortened in the absence of DEDD. This result is consistent with the data above (presented in Table 1) observed using asynchronous cells. Again, DEDD−/− cells complemented with DEDD expression revealed a similar profile of the mitotic index with that of wild-type cells (SI Fig. 7).
Fig. 1.
Accelerated mitotic progression in DEDD−/− MEF cells. (A) Proliferation of 3T3 MEF cells. Cell numbers are plotted each day after the initial plating of 105 cells in a 10-cm-diameter plate. WT, DEDD+/+; KO, DEDD−/−; KO+DEDD 1 and 2, two clones of DEDD−/− cells transfected with DEDD. Data are averages of triplicates. Error bars indicate SEM. (B) BrdU incorporation analysis. (Upper) BrdU/7-amino-actinomycin D FACS profiles of WT, KO, and KO+DEDD cells. (Lower) First three lanes, various types of 3T3-MEF cells; last two lanes, 293T cells with or without DEDD overexpression.
Table 1.
Actual length of each cell cycle phase
| Cell type | WT, h | KO, h | KO + DEDD, h | KO/WT × 100, % | KO + DEDD/KO × 100, % |
|---|---|---|---|---|---|
| Doubling time | 45.8 ± 6.5 | 37.5 ± 2.5 | 50.8 ± 7.2 | 81.0 | 135 |
| G1 | 29.2 ± 4.6 | 25.1 ± 3.2 | 28.4 ± 4.1 | 85.9 | 111 |
| S | 9.1 ± 1.9 | 8.6 ± 1.1 | 10.8 ± 1.0 | 94.5 | 126 |
| G2/M | 7.5 ± 0.5 | 3.8 ± 0.3 | 12.1 ± 2.0 | 50.7 | 312 |
| Mitosis | |||||
| Whole | 5.9 ± 0.4 | 2.9 ± 0.3 | ND | 49.2 | ND |
| Early | 3.8 ± 0.8 | 1.6 ± 0.2 | ND | 42.1 | ND |
| Late | 2.1 ± 0.5 | 1.3 ± 0.4 | ND | 61.9 | ND |
| G2 | 1.6 ± 0.4 | 0.9 ± 0.3 | ND | 56.3 | ND |
Data are averages from nine different experiments ± SEM.
DEDD Associates with Mitotic Cdk1/Cyclin B1 Complexes.
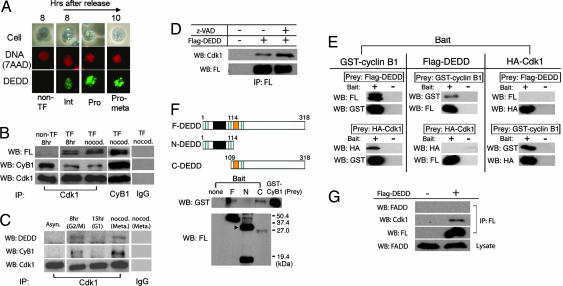
Progression of mitosis is promoted by the many protein phosphorylation events mediated by the Cdk1/cyclin B1 complex that translocates into the nucleus, after being activated through dephosphorylation at Thr-14 and Tyr-15 inhibitory residues of Cdk1 (6–8). As depicted in Fig. 2A, DEDD protein is present in the whole nucleus during the early mitosis. Therefore, we next assayed possible interactions between DEDD and Cdk1/cyclin B1. Significantly, an anti-Cdk1 antibody coprecipitated Flag-tagged DEDD and cyclin B1 together with Cdk1, from lysates of Flag-DEDD transfected 293T cells that were enriched at the G2 and M phases (8 h after release from the double-thymidine block) (Fig. 2B, second lane). This result indicates that DEDD associates with the Cdk1/cyclin B1 complex. Equivalent levels of DEDD were also coprecipitated with Cdk1/cyclin B1 in cells arrested at metaphase by a nocodazole treatment (Fig. 2B, third and fourth lanes). In addition, association of endogenous DEDD with Cdk1/cyclin B1 was also confirmed by generating a new polyclonal antibody against DEDD. As demonstrated in Fig. 2C, association of endogenous DEDD with Cdk1/cyclin B1 was observed at G2/M (second lane) or metaphase (fourth lane) after the immunoprecipitation of the complex using an anti-cyclin B1 antibody. At the postmitotic G1 phase, association of Cdk1 with either the endogenous DEDD or cyclin B1 was diminished (third lane), clearly indicating that the Cdk1/cyclin B1/DEDD complex is specifically present before cell division. Previous reports suggested that the nuclear localization of DEDD depends on caspase activation following induction of apoptosis (32, 33). Therefore, we tested whether the interaction between DEDD and the nuclear Cdk1/cyclin B1 is diminished in the presence of the pan-caspase inhibitor, z-VAD. The z-VAD, however, did not influence the association of DEDD with Cdk1/cyclin B1 within the nucleus (Fig. 2D), indicating that DEDD is constitutively located in the nucleus under nonapoptotic circumstances. Furthermore, in vitro analysis using recombinant proteins revealed that DEDD bound to cyclin B1, but not to Cdk1 (Fig. 2E). Thus, DEDD associates with the Cdk1/cyclin B1 complex via a direct binding to cyclin B1. To test whether the DED domain of DEDD is needed to couple DEDD with cyclin B1, we created DEDD variants, containing either only the DED domain (N-DEDD) or the COOH-region without the DED domain (C-DEDD), and compared the binding to cyclin B1 of these DEDD truncated mutants with that of full-length DEDD (F-DEDD). In vitro binding assays revealed that C-DEDD, but not N-DEDD, binds to cyclin B1 like F-DEDD, clearly indicating that the DED domain is not involved in the association of DEDD with Cdk1/cyclin B1 (Fig. 2F). In addition, because it was demonstrated that DEDD interacts with FADD through homophilic association via the DED domains (32), and that FADD also appears to play a role in mitosis (21), we tested whether FADD also participates in the Cdk1/cyclin B1/DEDD complex during mitosis. However, no detectable FADD was coprecipitated with Cdk1/cyclin B1/DEDD in cells arrested at metaphase by nocodazole treatment (Fig. 2G).
Fig. 2.
DEDD associates with the Cdk1/cyclin B1 complex. (A) 293T cells expressing Flag-DEDD were synchronized and cytostained for DEDD (green) and 7-amino-actinomycin D (red) at indicated cell cycle phases. Subphase during mitosis was judged by the morphology of cells and DNA (chromosome). Non-TF, nontransfected 293 T cells; Int, interphase; Pro, prophase; Pro-meta, prometaphase. (B) In vivo binding assay. Flag-DEDD protein was coimmunoprecipitated (IP) with endogenous Cdk1/cyclin B1 from Flag-DEDD transfected 293T cells synchronized at G2/M (second lane) or at metaphase (third lane). As a control, nontransfected 293 cells at the G2/M phase (first lane) were analyzed. Cells at metaphase were also used for immunoprecipitation using an anti-cyclin B1 antibody (fourth lane) or a control rabbit IgG (fifth lane). Precipitates were analyzed by Western blotting using antibodies against Flag (FL), cyclin B1 (CyB1), or Cdk1. (C) Association of endogenous DEDD with Cdk1/cyclin B1 complexes. Precipitates by an anti-Cdk1 antibody from cell lysates of wild-type (nontransfected) 293T cells synchronized at G2/M (second lane), postmitotic G1 (third lane), or metaphase (fourth lane) were tested by Western blotting using antibodies against DEDD (Top), cyclin B1 (Middle), or Cdk1 (Bottom). As a control, asynchronous 293T cells were analyzed (first lane). Precipitates with a control rabbit IgG from cells at metaphase were also Western-blotted (fifth lane). The anti-human and anti-mouse DEDD polyclonal antiserum was newly generated by immunizing rabbits with a KLH-conjugated DEDD peptide (CPDLVDKYLEETSIRYVT). (D) Pan-caspase inhibitor z-VAD does not inhibit Cdk1/cyclin B1/DEDD complex formation. 293T cells with (second and third lanes) or without (first lane) Flag-DEDD transfection were arrested at metaphase. A proportion of transfected cells were also incubated with 50 μM of z-VAD-fmk (Biovision) for the last hour (third lane). Cdk1/cyclin B1/DEDD complexes were immunoprecipitated by using an anti-Flag antibody. Precipitates were analyzed by Western blotting for Cdk1 (Upper) or Flag-DEDD (Lower). (E) In vitro binding assay using purified Flag-tagged DEDD, HA-tagged Cdk1, and GST-cyclin B1 fusion protein. A prey protein was pulled down by either type of bait protein. As controls, prey proteins were also incubated in the absence of the respective bait protein, followed by precipitation (Bait: −). Precipitates were analyzed by Western blotting (WB) using antibodies against Flag (FL; for DEDD), HA (for Cdk1), or GST (for cyclin B1). (F Top) Schematic showing of the structure of F-DEDD, N-DEDD, and C-DEDD. Light blue region, nuclear localizing sequences; black region, DED domain; orange region, proline-rich region. Numbers indicate amino acids starting from the first methionine. (F Middle and Bottom) Binding assay of the F, N, and C-DEDD to GST-cyclin B1. Flag-tagged DEDD variant proteins (baits) were purified from 293 T cells overexpressing the respective proteins and incubated with GST-cyclin B1 (prey), followed by a pull-down. Precipitates were analyzed by Western blotting, either for binding using an anti-GST antibody (Middle) or for bait proteins using an anti-Flag (FL) antibody (Bottom). As a control, the prey protein, GST-cyclin B1, was also loaded on the gel and Western-blotted with the anti-GST antibody (WB: GST, fifth lane). The N-DEDD protein appeared to yield dimers due to homotypic interactions (31) (indicated by an arrowhead in Bottom). (G) FADD does not associate with the Cdk1/cyclin B1/DEDD complex. The Cdk1/cyclin B1/DEDD complex was immunoprecipitated from nocodazole-treated 293T cells with (second lane) or without (first lane) Flag-DEDD transfection by using an anti-Flag antibody. Precipitates were analyzed by Western blotting for FADD (first panel), Cdk1 (second panel), or Flag-DEDD (third panel).
Association of DEDD Decreases the Kinase Activity of Cdk1/Cyclin B1 Complexes and Thereby Controls rRNA Synthesis.
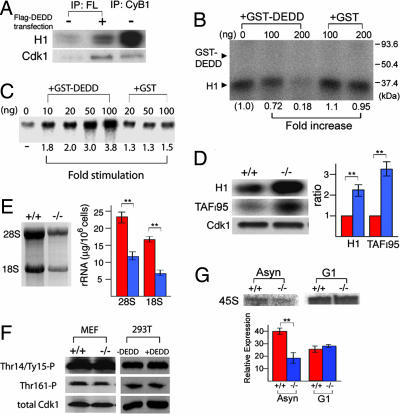
We then asked how the association of DEDD influences the function of Cdk1/cyclin B1. We first assessed the kinase activity of DEDD-bound Cdk1/cyclin B1 on histone H1. DEDD-bound Cdk1/cyclin B1 was precipitated from the nuclei of 293T cells overexpressing Flag-tagged DEDD, using an anti-Flag antibody (Fig. 3A, second lane). Cdk1/cyclin B1, immunopurified from the nuclei of nontransfected 293T cells with antibodies against either Flag (Fig. 3A, first lane), or cyclin B1 (Fig. 3A, third lane), were used, as negative and positive controls, respectively. DEDD-bound Cdk1/cyclin B1 apparently harbored much less kinase activity. More directly, the kinase activity of Cdk1/cyclin B1 isolated from DEDD−/− cells (DEDD-free Cdk1/cyclin B1) was decreased by the presence of recombinant DEDD (GST-DEDD) protein in a dose-dependent manner when tested using histone H1 (32 kDa) as a substrate (Fig. 3B). Note that GST-DEDD itself (60 kDa; indicated in Fig. 3B) was not phosphorylated. One of the prominent mitotic events mediated by Cdk1/cyclin B1 is the phosphorylation of TAFI95, which represses RNA polymerase I (Pol.I) transcription, and thus silences the rRNA synthesis, leading to termination of protein production before cell division (34, 35). Because this event undergoes at nucleoli, where DEDD protein is also strongly present (refs. 32 and 33, and also apparent in Fig. 2A), we monitored whether recombinant DEDD can prevent the Cdk1/cyclin B1-mediated Pol.I transcription repression in vitro. Consistent with the decrease in kinase activity of Cdk1 in the presence of DEDD (shown in Fig. 3 A and B), recombinant GST-DEDD, but not GST, significantly increased transcription of Pol.I in the presence of Cdk1/cyclin B1 (Fig. 3C). Comparative analysis of DEDD+/+ and DEDD−/− cells further supported these data. Cdk1/cyclin B1 isolated from DEDD−/− MEF cell nuclei phosphorylated histone H1 and the TAFI95 more efficiently than that obtained from DEDD+/+ cell nuclei (Fig. 3D). Consistent with the increased kinase activity on TAFI95 in DEDD−/− cells, levels of both 28S and 18S ribosomal RNA (rRNA) were decreased in DEDD−/− compared with control DEDD+/+ cells, reflecting an accelerated termination of rRNA synthesis (Fig. 3E). In line with this, DEDD−/− cells contained less protein per cell (0.8 ± 0.1 mg/106 WT cells vs. 0.3 ± 0.05 mg/106 KO cells; n = 8). Interestingly, evaluation of Cdk1 in DEDD+/+ and DEDD−/− cells revealed comparable levels of Cdk1 phosphorylation at Thr-14 and Tyr-15 inhibitory residues (Fig. 3G). Phosphorylation of Thr-161, a hallmark of active Cdk1 (6, 7), was also equivalent in mutant and wild-type cells (Fig. 3F). Likewise, overexpression of DEDD in 293T cells did not change phosphorylation status at any of these three amino acids (Fig. 3F Right). Thus, the decrease in Cdk1/cyclin B1 activity mediated by DEDD is not through modulation of phosphorylation status of Cdk1.
Fig. 3.
Association of DEDD reduces kinase activity of the Cdk1/cyclin B1 complex. (A) Kinase activity on the histone H1 substrate. Activities were normalized by the amount of precipitated Cdk1 (Western blotting; Lower). The negative control (first lane) revealed an invisible level of Cdk1 but a background level of activity. (B) DEDD-free Cdk1/cyclin B1 was immunopurified from DEDD−/− MEF cells by using an anti-cyclin B1 antibody. The kinase activity of the precipitate on the histone H1 substrate was assayed in the presence of different amounts of GST-DEDD (0, 100, and 200 ng) or GST (100 and 200 ng) protein. Fold increase indexes compared with the signal that was obtained without GST-DEDD or GST protein are displayed. The intensities of the signals were quantified by using image analysis software (NIH Image 1.63). No phosphorylation of GST-DEDD (60 kDa, size indicated) was detected. (C) Addition of recombinant GST-DEDD stimulates Pol.I transcription in vitro. Comparable amounts of GST-DEDD (second to fifth lanes) or GST alone (sixth to eighth lanes) were added to a fractionated murine nuclear extract (DEAE-280) from DEDD−/− MEF cells containing Cdk1/cyclin B1 complexes, and Pol.I transcription was initiated in the presence of nucleotides and linearized template DNA (34, 35). Radiolabeled runoff transcripts were separated on polyacrylamide gels and quantified by using a PhosphorImager. Fold stimulation indexes compared with the signal that was obtained without GST-DEDD or GST protein are displayed. Three independent experiments were performed and revealed similar results. (D Left) Kinase activity assay of Cdk1/cyclin B1 precipitated from DEDD+/+ (+/+) or DEDD−/− (−/−) 3T3 MEF cell nuclei using histone H1 (Top) or TAFI95 as substrate (Middle). Western blotting for the precipitated Cdk1 is shown in Bottom. (D Right) The relative activities in −/− cells (blue bars) to those in +/+ cells (red bars) are presented. Data are averages ± SEM of three independent assays. (E) The rRNA levels. (Left) Total RNA from 5 × 105 DEDD+/+ (+/+) and DEDD−/− (−/−) 3T3 MEF cells was separated on an agarose gel. (E Right) Absolute amounts of 28S and 18S rRNA per 106 cells. Means ± SEM are presented. (F) Phosphorylation status of Cdk1. Cdk1 was immunoprecipitated either from DEDD+/+ (+/+) or DEDD−/− (−/−) 3T3 MEF cells (first two lanes), 293T or 293T cells with DEDD transfection (293+DEDD) (third and fourth lanes), and the phosphorylation of the Thr-14/Tyr-15 (Top) and the Thr-161 (Middle) residues were analyzed by Western blotting. Data were normalized to the precipitated Cdk1 amounts (Bottom). (G) The pre-rRNA synthesis. DEDD+/+ (+/+) and DEDD−/− (−/−) 3T3 MEF cells were enriched at G1 by serum starvation. Total RNA isolated from 5 × 105 G1-enriched cells, as well as from asynchronous cells as a control, was analyzed for 45S pre-rRNA by Northern blotting by using a mouse rDNA (from −168 to + 158) fragment as a probe (Upper). The amount of 45S pre-rRNA was also analyzed by quantitative RT-PCR. Data are described as relative amounts of 45S pre-rRNA normalized to 36B4 mRNA (Lower). Means ± SEM of three independent analyses are presented.
Lack of DEDD Did Not Influence rRNA Synthesis Rates During the G1 Phase.
Because the length of G1 was also decreased in DEDD−/− cells, we asked whether DEDD might control rRNA synthesis rates during the G1 phase, in a manner independent of the Cdk1-TAFI95 pathway undergoing at the G2/M. To this end, we analyzed the 45S pre-rRNA synthesis (34, 35) in mutant and wild-type cells that were enriched in G1, by both Northern blotting and quantitative RT-PCR. As shown in Fig. 3G, the amount of pre-rRNA was not significantly different in DEDD−/− and DEDD+/+ cells enriched in G1. This result suggests that the rates of rRNA synthesis appeared to be comparable in DEDD−/− and DEDD+/+ cells. In addition, we investigated the interaction of DEDD with the Cdk2/cyclin complexes that control G1 progression (36, 37). However, an anti-Flag antibody did not coprecipitate Cdk2 from lysates of asynchronous Flag-DEDD transfected 293T cells (containing a >50% proportion of G1 cells), excluding the possibility of an association of DEDD with the Cdk2/cyclin complexes (SI Fig. 8A). Furthermore, phosphorylation of Thr-160 of Cdk2 was at similar levels in DEDD−/− and DEDD+/+ cells, indicating the activity of Cdk2 was not altered by the lack of DEDD (SI Fig. 8B). In addition, the expression levels of Cdk inhibitors, p21CIP1 and p27KIP1, which are mainly involved in the regulation of G1/S transition via inhibiting Cdk2 activity (38), were similar in DEDD−/− and DEDD+/+ cells when tested by RT-PCR (SI Fig. 8C).
Lack of DEDD Reduced Size in Both Cells and Mice.
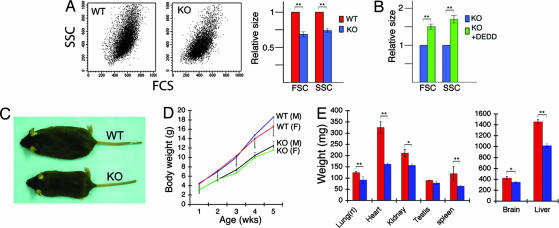
Consistent with the less rRNA and protein (39, 40), DEDD−/− cells were smaller in size than DEDD+/+ cells by FACS analysis (a 20–25% reduction in the average size in both forward- and side-scatter parameters; Fig. 4A). Forced expression of DEDD increased DEDD−/− cell volume in the FSC/SSC profile (Fig. 4B). Likewise, DEDD−/− mice revealed overall decrease in body size and weight (Fig. 4 C and D). Notably, the size and weights were decreased in all organs of DEDD−/− mice (Fig. 4E), although the cell number was not reduced, at least in organs where cell numbers were countable. Furthermore, the density of cells was increased in organs, such as liver and kidney, in DEDD−/− mice (Table 2). In consistent, the saturation density of cultured DEDD−/− MEF cells was also higher than that of DEDD+/+ cells, at a confluent stage (Table 2). All of these results may support the idea that the smaller size of DEDD−/− mice (in both organs and bodies) appears to be due to the reduction in size in DEDD−/− cells. Nevertheless, the size of single-cell in those organs was variable in different mice of the same genotype when assayed by histology or FACS, and thus its difference between DEDD−/− and DEDD+/+ mice was not significant (data not shown). Therefore, it may not mutually be excluded that this phenotype in DEDD−/− mice could derive from unknown cellular or physiological defects (e.g., certain change of metabolic status) because of loss of DEDD and are not necessarily cell autonomous. It is noteworthy, however, that levels of blood glucose, various hormones, and growth factors involved in body growth regulation were in the normal ranges in DEDD−/− mice (SI Table 3).
Fig. 4.
Lack of DEDD reduces size in cells and mice. (A Left and Center) Representative FSC/SSC profiles of DEDD+/+ (WT) and DEDD−/− (KO) MEF cells. (A Right) The FSC and SSC of DEDD−/− cells (blue bars) relative to those of DEDD+/+ (red bars) cells. (B) The FSC and SSC of DEDD−/− cells transfected with DEDD (green bars) relative to those of DEDD−/− (blue bars) cells. Data are averages from three different lines for each type of cell. Error bars indicate SEM. (C) Mouse photo showing DEDD+/+ (on the top) and DEDD−/− (on the bottom) mice. Both mice were 6 weeks of age and had been backcrossed to B6 for 10 generations. (D) Body weights. Five mice per group were analyzed, and the averages are presented. All mice used were littermates obtained from intercross-breeding of DEDD+/− mice that had been backcrossed to B6 mice for nine generations. Error bars indicate SEM. The differences between WT and KO mice were statistically significant in both males (M) and females (F) (Mann–Whitney test; P < 0.01). (E) Weights of indicated organs. Lung(rt), right lobes of the lung; red bars, DEDD+/+; blue bars, DEDD−/−. Data are averages from five male mice.
Table 2.
Cell densities in KO and WT
| Cells | WT | KO | KO:WT × 100, % |
|---|---|---|---|
| Liver | 245 ± 38 | 271 ± 51 | 111 |
| Kidney | 369 ± 30 | 479 ± 43 | 130 |
| MEF | 6.8 ± 1.2 × 105 | 2.3 ± 0.8 × 106 | 3338 |
Cell numbers per 1 × 104 μ m2 were quantified in the liver and kidney by a microscopic analysis of H&E-stained tissue sections. Data are presented as the means of six independent areas in a tissue section from three different mice for each genotype. Saturation density of MEF cells was analyzed by counting the cell numbers, when cells were at a confluent stage in a 10-cm-diameter culture dish. Data are the averages from three independent cultures for each cell type.
Discussion
Our current findings suggest that two distinct mechanisms for the modulation of Cdk1/cyclin B1 activity exist before cell division. The first checkpoint, the well established inactivation and reactivation of Cdk1 by phosphorylation at Thr-14 and Tyr-15, occurs in the cytoplasm and involves a variety of protein kinases and phosphatases (4–10). The novel subsequent inhibition occurs in the nucleus and is executed via the direct association of DEDD with Cdk1/cyclin B1. Decrease in kinase activity of mitotic Cdk1/cyclin B1 caused by binding of DEDD to cyclin B1 impedes the progression of mitosis, in turn promoting cell growth before cell division. The functional inhibition of Cdk1/cyclin B1 activity by DEDD appears to be specific for mammalian cells, because DEDD (or DEDD homologues) have not been found in databases for lower eukaryotes. This is also consistent with the fact that cell growth before cell division is not significantly affected by the loss of Wee1-related kinases, responsible for phosphorylation of Thr-14/Tyr-15, in higher mammalian cells (4, 5, 9, 10, 14–18).
It is interesting that, in DEDD−/− cells, the length was decreased not only in mitosis but also in the G1 phase (where Cdk1/cyclin B1 is not involved) during cell cycle. The lack of DEDD did not change the activity of Cdk2, or the expression levels of p21CIP1 or p27KIP1, and, thus, the precise mechanism of how the lack of DEDD influenced the length of G1 is yet unknown. Beside the fact that DEDD is a bona fide inhibitor of Cdk1/cyclin B1 during the mitosis, the effects of DEDD mutation on multiple cell cycle stages may raise the possibility of certain additional effects of DEDD, which lead indirectly to cell cycle effects. Although the rate of rRNA synthesis during the G1 was not changed (as presented in Fig. 3G), the shortened G1 phase may result in further reduction in the total amount of rRNA in DEDD−/− cells, because rRNA is also actively produced during G1 (1–3). This, in addition to the effect during early mitosis, may highlight the reduction of rRNA levels as one of the most striking phenotypes for DEDD−/− cells. In the yeast, it is indeed well known that the efficiency of ribosome synthesis is intimately connected with the cell division machinery (41–43). This possibility of alternative effect of DEDD deficiency might solve the difficulty in reconciling the short amount of time that DEDD inhibits the Cdk1/cyclin B1 activity, and the profound effects on rRNA synthesis and the cell size observed in DEDD−/− cells. Further studies on the precise mechanism that links DEDD with the control of G1 length are needed to clarify this issue. Curiously, inconsistent with our findings, Peter and colleagues (32, 33) reported that DEDD appeared to have a direct effect as a Pol.I transcriptional repressor. If so, it may be possible that there might be two (opposite) mechanisms for the regulation of rRNA synthesis by DEDD; one is a direct inhibition of Pol.I, and the other is an indirect enhancing effect via inhibiting Cdk1/cyclin B1 activity before cell division. In physiological situation in vivo, however, the latter might be dominant, because lack of DEDD did significantly decrease the rRNA amount in cells. Further studies are needed to solve this question.
Finally, it is intriguing that DEDD−/− and DEDD+/+ cells exhibited comparable responses to apoptosis induction, which is not consistent with previous reports suggesting a proapoptotic function for DEDD (31–33). One plausible explanation for this discrepancy is a functional redundancy in apoptosis-related functions by other molecule(s), presumably DEDD2, a structural homologue of DEDD (44, 45). Thus, DEDD2 may be sufficient to complement the function of DEDD in the regulation of apoptosis. Comparative functional studies of DEDD and DEDD2, are needed to address this possibility; thus, the physiological involvement of DEDD in apoptosis regulation remains an open question.
Materials and Methods
Generation of DEDD−/− Mice.
The targeting vector was constructed by replacing exons 1 and 2 and a part of the intron with a neomycin-resistance gene. Gene targeting in ES cells was performed as previously described (46). Details are described in SI Materials and Methods.
Generation of 3T3-MEF Lines.
The 3T3-MEF cell lines were generated as previously described (47). Details are described in SI Materials and Methods.
Cell Synchronization.
293T cells were synchronized either by a double thymidine block or by starvation as follows: cells were cultured in a medium containing 0.2% FBS for 24 h followed by another 24-h culture in the absence of FBS. Further details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Takashima, N. S. C. van Oers, L. DeFord, S. Subramanian (University of Texas Southwestern Medical Center), S. Ohnishi, and H. Okayama (University of Tokyo) for their assistance. This work was supported by grants from the National Institutes of Health and the Inamori Foundation (to T.M.).
Abbreviations
- Cdk
cyclin-dependent kinase
- DED
death-effector domain
- MEF
mouse embryonic fibroblast
- Pol.I
RNA polymerase I
- KO
knockout.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611167104/DC1.
References
- 1.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen P, Tyers M. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Kellogg DR. J Cell Sci. 2003;116:4883–4890. doi: 10.1242/jcs.00908. [DOI] [PubMed] [Google Scholar]
- 5.Rupes I. Trends Genet. 2002;18:479–485. doi: 10.1016/s0168-9525(02)02745-2. [DOI] [PubMed] [Google Scholar]
- 6.Morgan DO. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 7.Nigg EA. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 8.Smits VA, Medema RH. Biochim Biophys Acta. 2001;1519:1–12. doi: 10.1016/s0167-4781(01)00204-4. [DOI] [PubMed] [Google Scholar]
- 9.Harvey SL, Kellogg DR. Curr Biol. 2003;13:264–275. doi: 10.1016/s0960-9822(03)00049-6. [DOI] [PubMed] [Google Scholar]
- 10.Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 11.Dunphy WG, Kumagai A. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 12.Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 13.Rupes I. Trends Genet. 2002;18:479–485. doi: 10.1016/s0168-9525(02)02745-2. [DOI] [PubMed] [Google Scholar]
- 14.Gould KL, Nurse P. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 15.Parker LL, Walter SA, Young PG, Piwnica-Worms H. Nature. 1993;363:736–738. doi: 10.1038/363736a0. [DOI] [PubMed] [Google Scholar]
- 16.Stumpff J, Duncan T, Homola E, Campbell SD, Su TT. Curr Biol. 2004;14:2143–2148. doi: 10.1016/j.cub.2004.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami MS, Mood SA, Daar IO, Morrison DK. Development (Cambridge, UK) 2004;131:571–580. doi: 10.1242/dev.00971. [DOI] [PubMed] [Google Scholar]
- 18.van Vugt MA, Bras A, Medema RH. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Vermeulen K, Berneman ZN, Van Bockstaele DR. Cell Proliferation. 2003;36:165–175. doi: 10.1046/j.1365-2184.2003.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golsteyn RM. Cancer Lett. 2005;217:129–138. doi: 10.1016/j.canlet.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Alappat EC, Feig C, Boyerinas B, Volkland J, Samuels M, Murmann AE, Thorburn A, Kidd VJ, Slaughter CA, Osborn SL, et al. Mol Cell. 2005;19:321–332. doi: 10.1016/j.molcel.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Boldin MP, Varfolomeev EE, Pancer Z, Mett IL, Camonis JH, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 23.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 24.Chinnaiyan AM, O'Rourke K, Tewari M, Dixit VM. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 25.Fernandes-Alnemri T, Armstrong RC, Krebs J, Srinivasula SM, Wang L, Bullrich F, Fritz LC, Trapani JA, Tomaselli KJ, Litwack G, Alnemri ES. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin DA, Siegel RM, Zheng L, Lenardo MJ. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- 27.Muzio M, Chinnaiyan AM, Kischkel FC, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 28.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 29.Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, Gazdar A, Blenis J, Arnott D, Ashkenazi A. J Biol Chem. 2001;276:46639–46646. doi: 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ. Proc Natl Acad Sci USA. 2001;98:13884–13888. doi: 10.1073/pnas.241358198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibbetts MD, Zheng L, Lenardo MJ. Nat Immunol. 2003;4:404–409. doi: 10.1038/ni0503-404. [DOI] [PubMed] [Google Scholar]
- 32.Stegh AH, Schickling O, Ehret A, Scaffidi C, Peterhansel C, Hofmann TG, Grummt I, Krammer PH, Peter ME. EMBO J. 1998;17:5974–5986. doi: 10.1093/emboj/17.20.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JC, Schickling O, Stegh AH, Oshima RG, Dinsdale D, Cohen GM, Peter ME. J Cell Biol. 2002;158:1051–1066. doi: 10.1083/jcb.200112124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein J, Grummt I. Proc Natl Acad Sci USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dulic V, Lees E, Reed SI. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 37.Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 38.Vidal A, Koff A. Gene. 2000;247:1–15. doi: 10.1016/s0378-1119(00)00092-5. [DOI] [PubMed] [Google Scholar]
- 39.Russell J, Zomerdijk JC. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montagne J. Mol Cell Biol Res Commun. 2000;4:195–202. doi: 10.1006/mcbr.2001.0284. [DOI] [PubMed] [Google Scholar]
- 41.Dez C, Tollervey D. Curr Opin Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein KA, Baserga SJ. Mol Biol Cell. 2004;15:5038–5046. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saracino F, Bassler J, Muzzini D, Hurt E, Agostoni-Carbone ML. Cell Cycle. 2004;3:648–654. [PubMed] [Google Scholar]
- 44.Zhan Y, Hegde R, Srinivasula SM, Fernandes-Alnemri T, Alnemri ES. Cell Death Differ. 2002;9:439–447. doi: 10.1038/sj.cdd.4401038. [DOI] [PubMed] [Google Scholar]
- 45.Alcivar A, Hu S, Tang J, Yang X. Oncogene. 2003;22:291–297. doi: 10.1038/sj.onc.1206099. [DOI] [PubMed] [Google Scholar]
- 46.Miyazaki T, Hirokami Y, Matsuhashi N, Takatsuka H, Naito M. J Exp Med. 1999;189:413–422. doi: 10.1084/jem.189.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todaro GJ, Wolman SR, Green H. J Cell Physiol. 1963;62:257–265. doi: 10.1002/jcp.1030620305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.