Abstract
Previous studies indicated that the vaccinia virus D10 protein, which is conserved in all sequenced poxviruses, participates in the rapid turnover of host and viral mRNAs. D10 contains a motif present in the family of Nudix/MutT enzymes, a subset of which has been shown to enhance mRNA turnover in eukaryotic cells through cleavage of the 5′ cap (m7GpppNm-). Here, we demonstrate that a purified recombinant D10 fusion protein possesses an intrinsic activity that liberates m7GDP from capped RNA substrates. Furthermore, point mutations in the Nudix/MutT motif abolished decapping activity. D10 has a strong affinity for capped RNA substrates (Km ≈ 3 nm). RNAs of 24–309 nt were decapped to comparable extents, whereas the cap of a 12-nt RNA was uncleaved. At large molar ratios relative to capped RNA substrate, competitor m7GpppG, m7GTP, or m7GDP inhibited decapping, whereas even higher concentrations of unmethylated analogs did not. High concentrations of uncapped RNA were also inhibitory, suggesting that D10 recognizes its substrate through interaction with both cap and RNA moieties. Thus far, poxviruses represent the only virus family shown to encode a Nudix hydrolase-decapping enzyme. Although it may seem self-destructive for a virus to encode a decapping and a capping enzyme, accelerated mRNA turnover helps eliminate competing host mRNAs and allows stage-specific synthesis of viral proteins.
Keywords: mRNA metabolism, MutT motif, Nudix hydrolase, poxvirus, mRNA turnover
The steady-state concentration of an mRNA is determined by synthesis and decay, allowing cells to rapidly adapt their pattern of gene expression (1). Similarly, some viruses accelerate mRNA turnover to suppress synthesis of cellular proteins and regulate expression of their own genes (2). Studies of vaccinia virus (VACV), the laboratory prototype poxvirus, indicated that viral mRNAs are relatively unstable compared with those of uninfected cells (3, 4). Thus, rapid mRNA turnover coupled with robust and sequential transcription of viral early, intermediate, and late genes allow stage-specific protein synthesis (5). Likewise, VACV infection induces the destabilization of cellular transcripts, a process thought to contribute to the shutdown of host protein synthesis (6–9). The latter may enhance viral replication by alleviating competition from cellular mRNAs for the protein synthetic machinery and by diminishing host antiviral responses. Nevertheless, the mechanisms used by VACV to regulate mRNA turnover have not been elucidated.
The VACV D9 and D10 proteins were identified as putative negative regulators of gene expression during a transfection-based DNA library screen used to isolate activators of late VACV transcription (10, 11). Both proteins are highly conserved: D10 homologs are present in all sequenced poxviruses, and D9 homologs are in all members of the chordopoxvirus subfamily. Overexpression of D10 during infection significantly reduced the amount of VACV transcripts, and overexpression of D9 had a similar but lesser effect (11). Remarkably, the inhibitory effect of D10 was only manifested on mRNAs that were capped (11), a structural feature of both VACV and host mRNA transcripts (12, 13). Deletion of the gene encoding D9 had little effect on virus replication, whereas a D10-deletion mutant virus (vΔD10) exhibited a 10-fold reduction in the formation of infectious virus (14). When equivalent numbers of virus particles were used for infection, viral and cellular transcripts persisted significantly longer in cells infected with vΔD10 than with wild-type virus. Furthermore, the shutoff of host protein synthesis was delayed in cells infected with vΔD10. These data are in accord with the hypothesis that D10 negatively regulates viral and cellular gene expression through a cap-dependent mRNA turnover mechanism.
Both VACV D9 and D10 contain a Nudix/MutT motif, a signature sequence characteristic of a group of small phosphohydrolases with varied functions that cleave nucleoside diphosphates linked to another moiety (15, 16). Because of the presence of this motif, we had suggested that D9 and D10 might cleave the m7GpppNm- cap, thereby exposing the RNA to exonucleases (11). Subsequently, the Nudix/MutT protein Dcp2 of yeast, humans, and nematodes was shown to possess intrinsic RNA decapping activity (17–20). To determine whether D10 regulates mRNA turnover through a decapping mechanism, we expressed VACV D10 as a fusion protein in bacteria and analyzed the enzymatic properties of the purified recombinant protein. D10 hydrolyzed the RNA cap, generating m7GDP as a product, and this activity was abolished by mutation of the Nudix/MutT motif. Both excess uncapped RNA and methylated nucleotides, including m7GpppG, m7GTP, and m7GDP, reduced decapping activity of D10, suggesting that substrate recognition involves both the cap and the RNA body.
Results
Recombinant VACV D10 Catalyzes RNA Decapping.
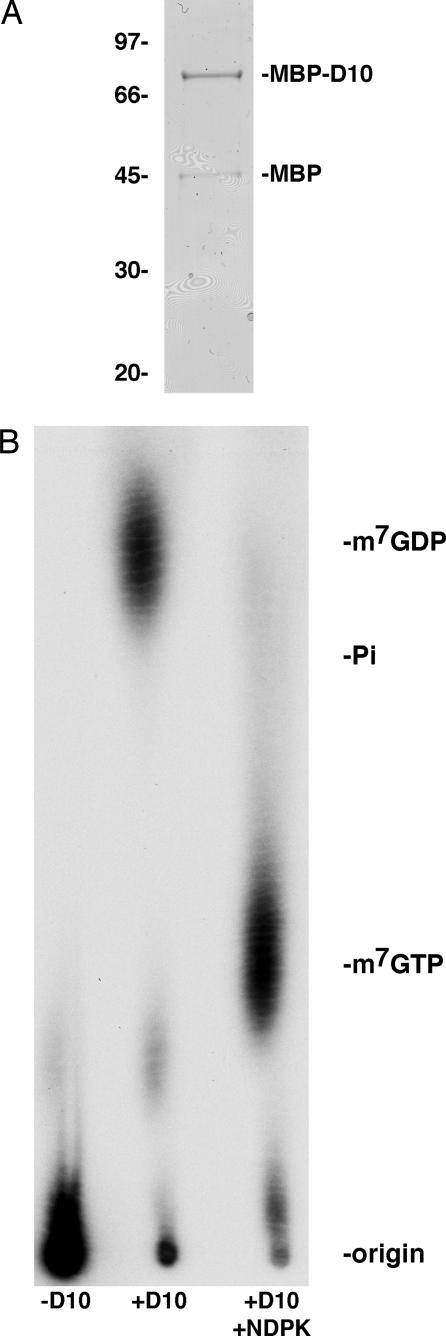
To investigate the role of D10 in mRNA turnover, a maltose-binding protein (MBP)–D10 fusion was synthesized in Escherichia coli and purified sequentially over amylose and heparin columns. A band corresponding to the full-length fusion protein and a minor one identified as MBP by mass spectrometry and Western blotting was detected upon SDS/PAGE (Fig. 1A). The free MBP presumably arose from partial degradation of MBP–D10. A 309-nt RNA was synthesized by in vitro transcription and subsequently capped and 32P-labeled by using recombinant VACV mRNA guanylyltransferase for use as a substrate in a decapping assay. After incubation of the recombinant D10 protein with the capped RNA substrate, the products of the reaction were resolved on polyethyleneimine-cellulose thin-layer chromatography (TLC) plates and detected by autoradiography. Unlabeled standards were run in parallel and visualized by UV shadowing. In the absence of D10, the RNA remained near the origin. However, the addition of recombinant D10 resulted in the release of a product that comigrated with the m7GDP standard (Fig. 1B). The identity of m7GDP was confirmed by treating the product of the reaction with nucleoside diphosphate kinase, an enzyme that specifically converts nucleoside diphosphates into nucleoside triphosphates. As expected, the kinase converted the product into a species that comigrated with the m7GTP standard (Fig. 1B).
Fig. 1.
Recombinant VACV D10 has RNA-decapping activity. (A) VACV D10 was expressed in E. coli as an MBP–D10 fusion protein and purified sequentially over amylose and heparin columns. Purified protein was resolved by SDS/PAGE and stained with Coomassie blue. Protein mass standards (in kDa) are indicated at the left. (B) MBP–D10 (30 ng) was incubated with 20 fmol of 32P-cap-labeled actin RNA (309 nt) in decapping buffer for 30 min at 37°C. A portion of the reaction was treated with two units of nucleoside diphosphate kinase in the presence of 1 mM ATP for 30 min at 37°C. Aliquots of the reaction were resolved by polyethyleneimine-cellulose TLC in 0.75 M LiCl. Unlabeled standards were run in parallel and visualized by UV shadowing. Radioactivity was detected by autoradiography. NDPK, nucleoside diphosphate kinase.
To verify that recombinant D10, and not a contaminating E. coli protein, was responsible for cap cleavage, MBP was expressed and purified in parallel with MBP–D10. Neither MBP nor any copurifying proteins added to the decapping reaction induced m7GDP formation, suggesting that D10 was solely responsible for the decapping activity (data not shown).
The Nudix/MutT Motif of VACV D10 Is Essential for RNA Cap Cleavage.
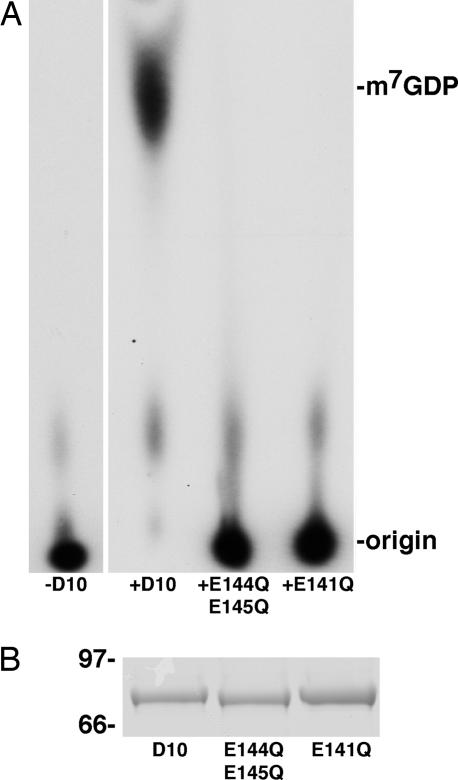
The highly conserved Nudix/MutT motif comprises the 23-aa sequence GX5EX7REUXEEXGU, where “U” represents one of the bulky hydrophobic amino acids (isoleucine, leucine, or valine) and “X” is any amino acid (15). The critical residues in the Nudix/MutT motif required for pyrophosphatase activity have been determined for the E. coli MutT protein, an enzyme responsible for conversion of mutagenic 8-oxo-dGTP into 8-oxo-dGMP (16, 21). To assess the requirement for the Nudix/MutT motif of D10 in RNA decapping, mutations were made in selected critical residues of the Nudix/MutT motif, and the resultant proteins were expressed and purified in parallel with wild-type recombinant D10 (Fig. 2B). Two mutant D10 proteins were analyzed: one in which two conserved glutamic acid residues at positions 144 and 145 were converted to glutamine (E144Q/E145Q) and a second in which a conserved glutamic acid at position 141 was changed to glutamine (E141Q). Even upon addition of excess protein (1 μg per reaction) only the unmutated recombinant D10 decapped RNA (Fig. 2A). Thus, the Nudix/MutT motif of D10 is essential for RNA decapping, conclusively demonstrating that D10 is directly responsible for this activity.
Fig. 2.
The Nudix/MutT motif of D10 is essential for RNA decapping. (A) Two mutated versions of MBP–D10 were expressed in E. coli and purified in parallel with wild-type MBP–D10 over an amylose column. One mutant, E144Q/E145Q, contains two point mutations in which glutamic acid residues at positions 144 and 145 were converted to glutamine. The second mutant, E141Q, contains a point mutation at position 141 that converts glutamic acid to glutamine. One microgram each of MBP-D10, E144Q/E145Q, and E141Q was used for decapping assays, and the products were resolved by TLC as in Fig. 1. (B) Equivalent amounts of wild-type MBP–D10 and the two mutant MBP–D10 proteins (E144Q/E145Q and E141Q) were resolved by SDS/PAGE electrophoresis and stained with Coomassie blue. Protein mass standards (in kilodaltons) are indicated at the left.
Determination of the Affinity of D10 for Capped RNA.
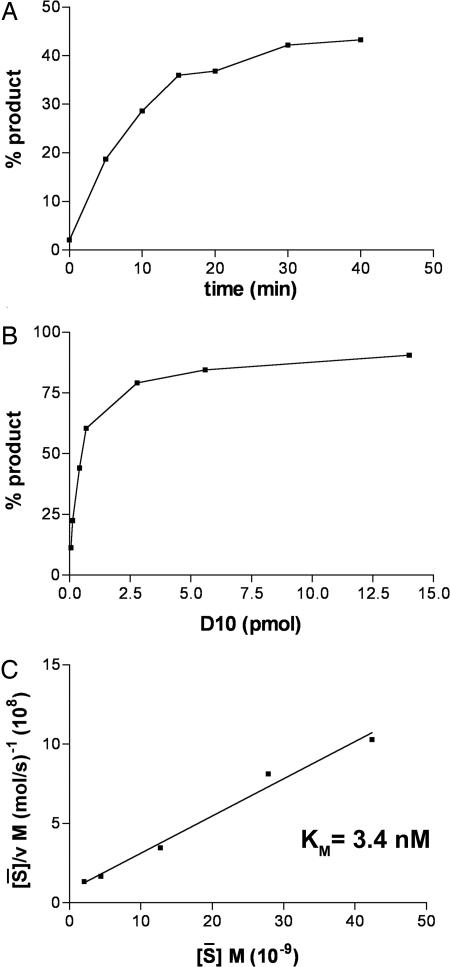
Decapping of the RNA substrate was shown to increase with time and enzyme concentration (Fig. 3 A and B). To estimate the affinity of D10 for its capped RNA substrate, the kinetic parameters of the D10 decapping reaction were determined. For technical reasons, the reactions were carried out at only moderate RNA excess, and therefore arithmetic mean substrate concentrations were used to determine the Km, defined as the substrate concentration at half maximum velocity, as described in refs. 22 and 23. The Km was determined to be 3.4 nM (Fig. 3C), indicating that D10 has a strong affinity for a capped RNA substrate.
Fig. 3.
Determination of kinetic parameters. (A) Time course of RNA decapping. In this reaction, 40 fmol of 32P-cap-labeled RNA was incubated with 10 ng of MPB–D10; at indicated times, the percent product released as m7GDP was determined by TLC as in Fig. 1 and quantified with a PhosphorImager (GE Healthcare, Piscataway, NJ). (B) Effect of D10 concentration on decapping. The percent product released in 30 min was determined with the indicated amounts of D10 as in A. (C) Determination of Km. Decapping reactions were performed in triplicate with 10 ng of MBP–D10 and 45, 90, 224, 449, or 673 fmol of 32P-cap-labeled RNA for 30 min and quantified as in A. A Km of 3.4 nM was determined with a Hanes plot by using arithmetic mean substrate concentrations (S̄) as described in ref. 22.
Effect of RNA Length on Decapping by D10.
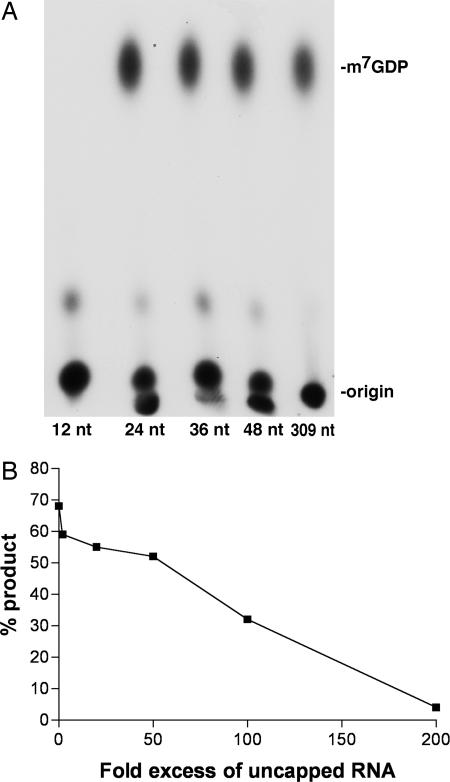
The effect of RNA length on D10 decapping activity was determined by using 32P cap-labeled RNAs with overlapping sequence. After the addition of 30 ng of recombinant D10, the 24-, 36-, 48-, and 309-nt RNAs were decapped to comparable extents, whereas the cap of the 12-nt RNA was uncleaved (Fig. 4A), suggesting that D10 prefers RNA substrates >12 nt in length.
Fig. 4.
Effect of RNA length on decapping and competition with uncapped RNA. (A) Decapping of defined-length capped RNAs. 32P cap-labeled RNAs of 12-, 24-, 36-, 48-, or 309-nt and 30 ng of MBP–D10 were mixed in the decapping assay, and the products were resolved by TLC as in Fig. 1. (B) Uncapped RNA inhibits decapping. Uncapped, unlabeled 309-nt actin RNA was added in increasing amounts to the decapping reaction, and the products were resolved by TLC. The percentage of product released was calculated with a PhosphorImager.
Uncapped RNA Inhibits Decapping.
The observation that decapping is influenced by RNA length raised the possibility that D10 may physically interact with RNA. To evaluate whether uncapped RNA was capable of inhibiting cap cleavage by D10, increasing amounts of unlabeled RNA were added to the reaction and the percentage of product formed was calculated. A 100-fold molar excess of competitor RNA reduced decapping by 50%, and a 200-fold excess virtually abolished decapping (Fig. 4B). The IC50 of uncapped RNA was determined to be 3 μM. The RNA length requirement for efficient decapping and the competition by uncapped RNA suggest that D10 recognizes RNA in addition to the cap structure.
Methylated Nucleotides Inhibit Decapping.
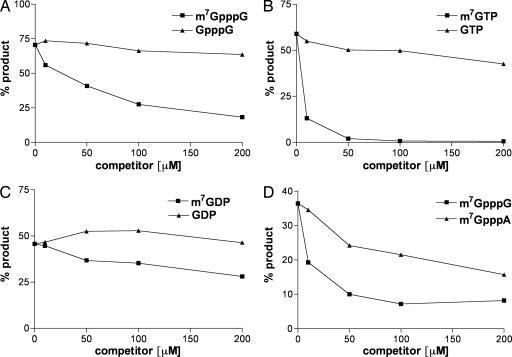
Competition experiments with methylated and unmethylated cap derivatives were carried out to further characterize the properties of D10. A substantial decrease in decapping was seen with 50 μM m7GpppG, which represents a 37,500 molar excess over capped RNA (Fig. 5A). However, the corresponding unmethylated dinucleotide GpppG was not inhibitory at four times that concentration (Fig. 5A). Interestingly, m7GTP appeared more inhibitory than m7GpppG, whereas m7GDP was less inhibitory (Fig. 5 B and C, respectively). At the same capped RNA substrate concentration, the IC50 values for m7GTP, m7GpppG, and m7GDP were ≈3 μM, 63 μM, and 300 μM, respectively. Neither unmethylated GTP nor GDP inhibited decapping, consistent with specificity for a methylated cap.
Fig. 5.
Methylated nucleotide derivatives inhibit D10 decapping. (A) m7GpppG or GpppG at the inhibitor concentrations indicated was added to the decapping reaction containing 20 fmol of capped RNA and 30 ng of MBP–D10 protein, and the products were resolved by TLC. The percent of product released was quantified with a PhosphorImager. (B) m7GTP or GTP was added to the decapping reaction and analyzed as in A. (C) m7GDP or GDP was included in the decapping reaction and analyzed as in A. (D) m7GpppG and m7GpppA were compared for their ability to inhibit D10 cap cleavage. The points are the averages of two experiments.
Cellular and VACV early mRNAs have m7GpppGm or m7GpppAm caps. However, VACV intermediate and late transcripts only contain m7GpppAm caps resulting from highly conserved transcription initiation signals (5, 24). To determine whether D10 exhibits cap sequence specificity, we compared the abilities of m7GpppG and m7GpppA to inhibit D10 decapping activity. As shown in Fig. 5D, m7GpppG inhibited RNA decapping more efficiently than m7GpppA, suggesting that D10 may exhibit a preference for m7GpppG-capped cellular and viral early transcripts.
Discussion
The present demonstration of RNA decapping activity by a recombinant VACV D10 fusion protein provides a mechanism to explain in vivo studies that identified VACV D10 as a negative regulator of cellular and viral gene expression (11, 14). VACV D10 was predicted to have decapping activity (11) because it contains a Nudix/MutT motif, a sequence characteristic of enzymes that act as nucleotide phosphohydrolases (15). Two lines of evidence confirmed that D10 and not a contaminating E. coli protein was responsible for the observed RNA decapping. First, MBP purified in parallel with the MBP–D10 fusion protein had no detectable mRNA decapping activity. Second, two versions of D10 containing point mutations in the Nudix/MutT motif were incapable of mRNA cap cleavage, even when excess protein was added. The latter result further defined the Nudix/MutT motif of D10 as essential for decapping.
The Km of the decapping reaction catalyzed by recombinant D10 was determined to be ≈3 nM, suggesting that D10 has a strong affinity for capped RNA. By comparison, the E. coli MutT protein has a 100-fold higher Km for its 8-oxo-dGTP substrate (25). We were unable to find a published Km value for the cellular decapping enzyme Dcp2. At the concentration of D10 tested, only RNAs longer than 12 nt were decapped, suggesting the RNA moiety enhanced substrate recognition and cleavage. This preference, as well as the m7GDP product, distinguishes D10 from the cellular scavenger enzyme DcpS, which hydrolyzes free cap structures to release m7GMP (reviewed in ref. 26).
An attempt was made to determine whether D10 activity was specific for a methylated cap structure. Although an unmethylated cap on RNA was cleaved by D10, we found that similarly purified D10 with active site mutations that completely prevented cleavage of methylated caps still cleaved unmethylated ones (data not shown), indicating contamination with an E. coli pyrophosphatase. Therefore, competition studies were used to assess the interaction of D10 with cap structures and with RNA. Although relatively high concentrations of the methylated dinucleotide m7GpppG were able to reduce decapping of the RNA substrate, unmethylated GpppG was not. Similarly, m7GTP and, to a lesser extent, m7GDP inhibited decapping, whereas the corresponding unmethylated nucleotides did not. These results are consistent with the role of D10 in specifically cleaving methylated cap structures. Excess uncapped RNA also inhibited decapping, suggesting that D10 recognizes the RNA moiety, although direct binding has not yet been demonstrated.
It is of interest to compare D10 with Dcp2, the cellular decapping enzyme (17–20, 27, 28). Except for the MutT motif, there is little sequence similarity between the viral and cellular enzymes. Both liberate m7GDP and are inhibited by uncapped RNA. However, unlike D10, Dcp2 is not inhibited by m7GpppG. For human Dcp2, an RNA substrate >23 nt in length was required for cap cleavage, with increasing length further enhancing decapping activity. In addition, Dcp2 forms a heterodimer with Dcp1, which stimulates decapping by an unknown mechanism, whereas there is no information yet regarding an association of D10 with another protein. Taken together, these data suggest subtle differences in substrate recognition by D10 and Dcp2, although the catalytic mechanism is presumably the same.
Because both cellular and VACV mRNAs are capped, the intriguing question arises about whether D10 exhibits mRNA specificity. Previous studies demonstrated that both viral and cellular transcripts decayed more rapidly when D10 was overexpressed (11) and decayed less rapidly when D10 was not expressed (14). However, this finding does not exclude quantitative differences between the decapping of cellular and viral mRNAs. Because the RNA substrates for the present study were made with T7 RNA polymerase, all transcripts started with pppG- and therefore the caps were m7GpppG-. However, cellular and early VACV mRNAs have both m7GpppGm- and m7GpppAm- caps at their 5′ ends, whereas intermediate and late VACV mRNAs have exclusively m7GpppAm- caps because of the conserved sequence at the RNA start sites (5, 24, 29). (Note that the penultimate nucleotide of cellular and viral mRNAs is methylated at the 2′-O position, whereas the in vitro synthesized RNAs used here were not 2′-O-methylated.) Intriguingly, the m7GpppG cap analog reduced D10 decapping activity more effectively than m7GpppA, suggesting that D10 may preferentially recognize the m7GpppG cap of host and VACV early transcripts. However, a more direct analysis is required to confirm a decapping preference. VACV intermediate and late transcripts have a unique 5′ poly(A) leader, further differentiating them from host and VACV early transcripts (5, 30, 31). It is known that decapping of cellular mRNAs depends on shortening of the 3′ poly(A) tail and dissociation of the poly(A) binding protein, although the mechanism is not completely understood (26, 32). Although speculative, poly(A) binding protein might associate with the 5′ poly(A) leader of VACV intermediate and late mRNAs, protecting the cap to some extent. Transcript selection might also be influenced by the intracellular localization of the D10. D10 does not localize in viral factory areas (14) where intermediate and late viral transcripts are transcribed and translated (G. Katsafanas and B.M., unpublished data). Transport of D10 from the viral factory could help shield nascent viral transcripts from D10 decapping activity.
After decapping by Dcp2, the mRNA is degraded by cellular exonucleases, such as the Xrn1 protein of yeast (33, 34). Dcp2 localizes in discrete cytoplasmic sites of mRNA degradation in eukaryotic cells called processing or P bodies (reviewed in ref. 35). Whether viral or cellular nucleases are involved in degradation of decapped viral transcripts remains to be determined.
Although all poxviruses have a D10 homolog, a Nudix hydrolase decapping enzyme has not been described for members of other virus families. African swine fever virus, which is distantly related to poxviruses, does encode a Nudix hydrolase (36). However, this enzyme preferentially degrades diphosphoinositol polyphosphates and does not hydrolyze methylated dinucleotide cap structures. The double-stranded L-A virus, which infects yeast, employs a novel mechanism to decap cellular messages. The GAG capsid protein forms a covalent linkage to the RNA cap, promoting cap removal (37, 38). Because L-A transcripts are not capped, the destabilized cellular RNA transcripts are thought to saturate the degradation machinery, allowing a portion of the L-A transcripts to escape degradation by cellular factors. Herpes simplex virus type 1 regulates mRNA turnover of host and viral messages through a mechanism that does not rely on mRNA decapping. Recent reports (39) indicate that the herpes simplex virion host shutoff protein cleaves mRNA internally at single-stranded cytidine and uridine residues, similar to the cleavage pattern of RNase A.
In conclusion, the mRNA decapping activity of D10 allows VACV to accelerate degradation of viral and host messages. Further studies are needed to determine whether D9, which also has a Nudix/MutT motif, has a similar activity. Because transcription of the D9 and D10 genes are controlled by an early and late promoter, respectively (40), the two proteins could provide continuous production of decapping enzymes. The robust synthesis and rapid decay of viral mRNAs enable the sequential transitions among early, intermediate, and late gene expression. In addition, the turnover of cellular transcripts may promote the shutoff of host protein synthesis, a process that could increase VACV access to cellular macromolecular “building blocks” and synthetic machinery. In addition, by degrading host mRNAs, VACV D10 may have an immunomodulatory role, like that of the herpes simplex virus vhs (41).
Materials and Methods
Plasmid Construction.
To construct a plasmid expressing the MBP–D10 fusion protein, the coding region of the D10 gene was amplified by PCR using VACV strain WR genomic DNA as template and oligonucleotide primers 5′-ATGAACTTTTACAGATCTAGTATAATTAGTCAGATT-ATTAAGTATAAT and 5′-GGCCGGAAGCTTTCAAT-CATC CTCAGTTAATTTTTTTAATGATTCGTAATA-ACATCCTCT. The resulting PCR product was gel purified, digested with the appropriate restriction enzyme, and ligated into pMAL-c2x (New England Biolabs, Ipswich, MA) downstream of the bacterial malE gene, to create pMAL-c2x-malE-D10. Mutations in the Nudix/MutT motif were generated by using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) or recombinant PCR using oligonucleotides containing the desired mutation.
Expression and Purification of Recombinant D10 Protein.
The pMAL-c2x-malE-D10 plasmid and derivatives were used to transform E. coli strain BL21 (EMD Biosciences, San Diego, CA), and cells were grown in LB broth containing 50 μg/ml carbenicillin and 0.2% (vol/vol) glucose. Recombinant protein expression was induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside, and cells were harvested after 4 h of growth at 30°C. The cells were lysed in B-PER detergent (Pierce, Rockford, IL), and the clarified lysate was purified sequentially over an amylose column (New England Biolabs) followed by a heparin column (Amersham Pharmacia Biotech, Piscataway, NJ). Purified protein was dialyzed against buffer containing 10 mM Tris·HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM DTT, and 2 mM Mg acetate (28).
RNA Synthesis.
A 309-nt RNA was synthesized from the pTRI-β-actin-human template (Ambion, Austin, TX) by using the MEGAshortscript in vitro transcription kit (Ambion). The 12-, 24-, 36-, and 48-nt RNAs were synthesized by in vitro transcription using annealed oligonucleotide primers containing the T7 promoter sequence and the corresponding sequence of the 309-nt actin transcript (42). The purified RNA was labeled by using recombinant VACV mRNA guanylyltransferase (Ambion) in the presence of 0.132 μM [α-32P]GTP, capping buffer [50 mM Tris·HCl (pH 8.0)/6 mM KCl/1.25 mM DTT/1.25 MgCl2/0.05 mg/ml BSA], and 1 mM S-adenosyl methionine. Unincorporated nucleotides were removed using ProbeQuant G-50 gel filtration columns (Amersham Pharmacia Biotech) or G-25 gel filtration columns (Roche Applied Science, Indianapolis, IN).
RNA Decapping Assay.
Reactions containing labeled capped RNA and purified recombinant MBP–D10 protein were carried out in 15 μl of decapping buffer [100 mM K acetate/10 mM Tris·HCl (pH 7.5)/2 mM MgCl2/0.5 mM MnCl2/2 mM DTT) at 37°C for 30 min unless indicated otherwise (28). The products of the reaction were resolved on polyethyleneimine-cellulose TLC plates (Sigma, St. Louis, MO, or Alltech Biotechnology, Columbia, MD) developed in 0.75 M LiCl. The TLC plates were dried and visualized by autoradiography and UV shadowing. Kinetic parameters were determined by using Prism (Irvine, CA) software.
Acknowledgments
We thank Teri Shors, Alan Townsley, Subbian Panayampalli, and George Katsafanas for helpful discussions and Stewart Shuman and Edward Niles for comments on the manuscript. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases/National Institutes of Health.
Abbreviations
- MBP
maltose-binding protein
- TLC
thin-layer chromatography
- VACV
vaccinia virus.
Footnotes
The authors declare no conflict of interest.
References
- 1.Meyer S, Temme C, Wahle E. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 2.Glaunsinger BA, Ganem DE. Adv Virus Res. 2006;66:337–394. doi: 10.1016/S0065-3527(06)66007-7. [DOI] [PubMed] [Google Scholar]
- 3.Sebring ED, Salzman NP. J Virol. 1967;1:550–558. doi: 10.1128/jvi.1.3.550-558.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oda K, Joklik WK. J Mol Biol. 1967;27:395–419. doi: 10.1016/0022-2836(67)90047-2. [DOI] [PubMed] [Google Scholar]
- 5.Baldick CJ, Jr, Moss B. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice AP, Roberts BE. J Virol. 1983;47:529–539. doi: 10.1128/jvi.47.3.529-539.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jindal S, Young RA. J Virol. 1992;66:5357–5362. doi: 10.1128/jvi.66.9.5357-5362.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. Virology. 2003;315:322–334. doi: 10.1016/s0042-6822(03)00532-4. [DOI] [PubMed] [Google Scholar]
- 9.Guerra S, Lopez-Fernandez LA, Conde R, Pascual-Montano A, Harshman K, Esteban M. J Virol. 2004;78:5820–5834. doi: 10.1128/JVI.78.11.5820-5834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keck JG, Baldick CJ, Moss B. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- 11.Shors T, Keck JG, Moss B. J Virol. 1999;73:791–796. doi: 10.1128/jvi.73.1.791-796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei CM, Moss B. Proc Natl Acad Sci USA. 1975;72:318–322. doi: 10.1073/pnas.72.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei CM, Gershowitz A, Moss B. Biochemistry. 1976;15:397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- 14.Parrish S, Moss B. J Virol. 2006;80:553–561. doi: 10.1128/JVI.80.2.553-561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bessman MJ, Frick DN, O'Handley SF. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 17.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. Proc Natl Acad Sci USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen LS, Mikhli C, Jiao X, Kiledjian M, Kunkel G, Davis RE. Mol Cell Biol. 2005;25:8779–8791. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Arch Biochem Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Lee HJ, Wilson IB. Biochim Biophys Acta. 1971;242:519–522. doi: 10.1016/0005-2744(71)90144-6. [DOI] [PubMed] [Google Scholar]
- 23.Siegel IH. Enzyme Kinetics. New York: Wiley; 1975. [Google Scholar]
- 24.Davison AJ, Moss B. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 25.Maki H, Sekiguchi M. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 26.Coller J, Parker R. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 27.Dunckley T, Parker R. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccirillo C, Khanna R, Kiledjian M. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davison AJ, Moss B. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 30.Patel DD, Pickup DJ. EMBO J. 1987;6:3787–3794. doi: 10.1002/j.1460-2075.1987.tb02714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwer B, Visca P, Vos JC, Stunnenberg HG. Cell. 1987;50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khanna R, Kiledjian M. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larimer FW, Hsu CL, Maupin MK, Stevens A. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 34.Hsu CL, Stevens A. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fillman C, Lykke-Andersen J. Curr Opin Cell Biol. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Cartwright JL, Safrany ST, Dixon LK, Darzynkiewicz E, Stepinski J, Burke R, McLennan AG. J Virol. 2002;76:1415–1421. doi: 10.1128/JVI.76.3.1415-1421.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanc A, Goyer C, Sonenberg N. Mol Cell Biol. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanc A, Ribas JC, Wickner RB, Sonenberg N. Mol Cell Biol. 1994;14:2664–2674. doi: 10.1128/mcb.14.4.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taddeo B, Zhang W, Roizman B. Proc Natl Acad Sci USA. 2006;103:2827–2832. doi: 10.1073/pnas.0510712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee-Chen GJ, Niles EG. Virology. 1988;163:52–63. doi: 10.1016/0042-6822(88)90233-4. [DOI] [PubMed] [Google Scholar]
- 41.Liang L, Roizman B. J Virol. 2006;80:7756–7759. doi: 10.1128/JVI.00587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]