Abstract
FOX (forkhead box) transcription factors have diverse regulatory roles in development, signaling, and longevity, as well as being able to bind stably to target sites in silent chromatin. Crystal structure analysis showed that the FOXA DNA binding domain folds into a helix-turn-helix (HTH) motif flanked on either side by “wings” of polypeptide chain. The wings have the potential to interact with the DNA minor groove along the long axis of the DNA helix, flanking the HTH interactions with the major groove. Diverse FOX family homologs exist, and structural studies with certain DNA target sites suggest that neither of the wing regions are well ordered or provide a stable contribution to DNA target site binding. However, FOXA1 binds certain DNA target sites with high affinity, and as a monomer. To determine whether the wing domains contribute to stable DNA binding, we assessed complexes of FOXA with high and lower affinity DNA target sites by hydroxyl radical footprinting and site-directed mutagenesis. The data revealed clear protections predicted for wing interactions at the high affinity target, but less so at the lower affinity target, indicating that the wing domains stably interact with high affinity DNA sites for FOXA proteins.
Keywords: FOX transcription factors, wing domains, DNA binding, hydroxyl radical footprinting
The FOXA (forkhead box A) transcription factors, FOXA1 (HNF3α), FOXA2 (HNF3β), and FOXA3 (HNF3γ), were initially discovered to regulate liver- and other gut organ-specific genes 1, and then to be necessary for the development of the endoderm and liver 2; 3; 4. FOXA target sites in chromatin are occupied in gut progenitor cells in embryos 5; 6, purified FOXA1 transcription factor can bind stably to its target sites on nucleosomes 7; 8, and FOXA factors help other proteins engage nearby sites in chromatin 9; 10. Thus, FOXA1 qualifies as a “pioneer factor,” capable of dominantly entering silent chromatin and initiating regulatory events 11. Recent genome-wide binding studies of FOXA1 emphasize the need to better predict where the factor binds DNA with high affinity 12. The goal of this study is to better understand the mechanism of FOX factor interactions with DNA.
The FOXA homologs govern gut development in flies 13, worms 14; 15, frogs 16, and fish 17; all are related by a highly conserved, ∼110 amino acid DNA binding domain. Sequence variants of this domain define the classes FOXA-FOXQ 18; 19. Humans, for example, contain 43 FOX factor genes. FOX factors of different classes function in diverse gene regulatory contexts, including the development of non-gut tissues, signal transduction, and longevity 19.
In a co-crystal with DNA, part of the FOXA3 DNA binding domain folds into a helix-turn-helix (HTH) structure that protrudes into the DNA major groove and makes base-specific contacts for target site recognition (20; Fig. 1A, “HTH”). Two loops, or “wings,” of polypeptide chain flank the HTH region, allowing the protein to bind DNA along the long axis. Wing 2 makes DNA minor groove contacts in the crystal (Fig. 1A). Wing 1 extends from the other side of the HTH along the long axis of DNA. The DNA used for FOXA3 crystallization contained an incomplete target sequence (Fig. 1A, segment with base pairs shown; Fig. 1B) and was too short to determine if there were direct DNA contacts for wing 1. Recent studies of FOXK1 (formerly called ILF-1) co-crystals with a longer DNA substrate showed that indeed wing 1 makes direct base and phosphate contacts in the DNA minor groove 21. However, two molecules of FOXK1 were unexpectedly bound on opposite sides of the DNA in the crystal asymmetric unit, and this FOX factor lacks wing 2, having another alpha-helix in its place. NMR studies of FOXD3 (formerly called Genesis) bound to a long DNA segment concluded that wing 1 interacts with the DNA minor groove, but the interactions are relatively weak and unstable, and that wing 2 is not well ordered 22. Furthermore, wing 2 was shown to be dispensable for target site binding by FOXJ2 protein23.
Figure 1.
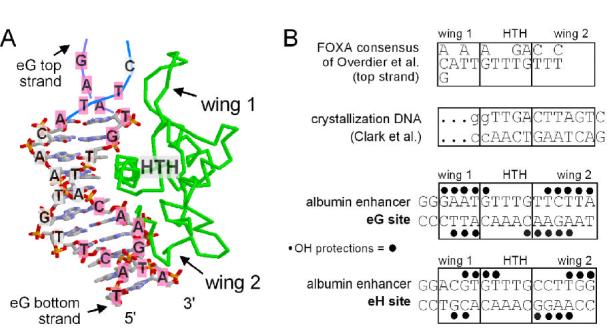
Structure of the FOXA3-DNA complex, target sequence alignments, and hydroxyl radical footprinting. A. Image of the FOXA3-DNA co-crystal 20 generated with RasMol 2.6 (courtesy of Roger Sayle). The sequence of the albumin enhancer eG site is superimposed on the DNA; bases highlighted in pink are where the phosphodiester backbone was protected from cleavage by hydroxyl radicals in our experiments. The GAAT DNA segment at the top, without base pairs, was not present in the crystal and is shown hypothetically in the image. The image also shows the polarity of the DNA strands, the helix-turn-helix (HTH) motif and the “wings” of polypeptide chain of FOXA3 (green). B. Sequence alignments of the FOXA consensus determined by SAAB 29, the oligonucleotide used for crystallization, and the eG and eH sites of the albumin enhancer, where FOXA proteins bind and activate transcription 30; 31. The wing 1 and wing 2 regions are denoted, relative to the HTH region, based on FOXA3-DNA contacts in the co-crystal, including any direct wing contact to a base or its 5′ phosphate, or an indirect contact via a water molecule. The positions of hydroxyl radical protection are shown by black dots above and below the eG and eH site sequences; see data in Figure 2.
In summary, studies of FOXD and FOXJ factors suggest that one or both of their wing regions do not make stable DNA contacts, and the crystal structure failed to resolve the issue for wing 1 of FOXA3. However, our prior studies with purified FOXA1 showed that it binds a functional site at the albumin gene enhancer eG site with high affinity (kD ∼0.15 nM)24. Also, an optimal FOXA binding consensus, derived from a SAAB analysis 25 and related to one created by sequence comparison 26, contains DNA sequences upstream of the region bound by the HTH, and thus would be predicted to be bound by wing 1 (Fig. 1A, B). To better understand how FOXA1 protein binds its targets so efficiently as a monomer, we first performed hydroxyl radical footprinting. In contrast to bulky nuclease probes, hydroxyl radicals have a solvent radius of ∼1.4 Å and thus are highly sensitive to close protein-DNA interactions 27.
Hydroxyl radicals attack each ribose moiety along the phosphodiester backbone of DNA, and thus are sensitive to both major and minor groove binding 25; 28. As seen in Fig. 2 and denoted in Fig. 1B, FOXA1 protein protects two regions at each of the albumin enhancer eG and eH sites from hydroxyl radical cleavage, on both the top and bottom strands of DNA. These data are representative of 3 independent footprinting experiments using different cleavage conditions. It was previously shown that FOXA1 binds the eG site with a 3-4x higher affinity than the eH site 24; consistent with this, the eG site footprints are stronger than that for eH (Fig. 2). Comparison of the eG site with the FOXA binding site consensus 29 and the crystallization DNA (Fig. 1B) allowed us to unambiguously orient the protein on the eG sequence (Fig. 1A), and thus determine which wing domain gives which hydroxyl radical protections (Fig. 1B).
Figure 2.
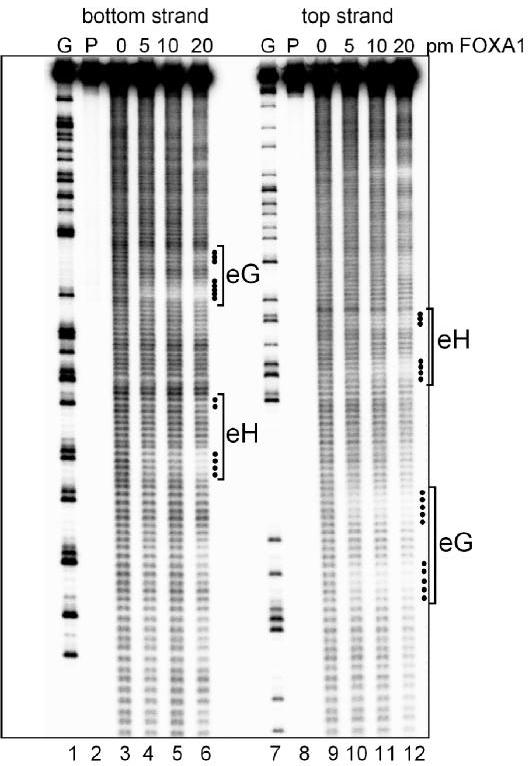
Hydroxyl radical footprinting of FOXA1 bound to DNA. Autoradiograph of a footprinting gel used to map hydroxyl radical protections by mouse FOXA1 protein, which was purified as described 24; 34. The gel depicts a fragment of the albumin enhancer from 472-651 31, end-labeled on the bottom strand (lanes 1-6) or top strand (lanes 7-12). Lanes 1 and 7 represents G sequence ladders generated by cleavage with dimethyl sulfate. Lanes 2 and 8 contained no hydroxyl cleavage reagents or FOXA1. The DNA sequence and FOXA1 binding conditions were as described 34, except that after 30 min binding in 17 μl reactions with 5 nM FOXA1, 1 μl of 200 μM Fe(II) EDTA, 3% H2O2 was added and the reactions were brought to 100 mM ascorbate. The reactions went for 2 min at room temperature and then 15 microliters of 5% glycerol were added. The reactions were diluted with 70 μl TE buffer, 2 μl of 10 mg/ml tRNA were added, and the DNAs were precipitated and run on a 6% polyacrylamide DNA sequencing gel. Gels were exposed to phosphorimager plates (Fuji BAS 2500) and X-ray film.
We find that about 5 contiguous bp of DNA are protected by FOXA1 on each of the top and bottom strands in the downstream “half” of the eG site (Fig. 1B, 2). Based on modeling, at least half of these protections appear to be due to wing 2 interactions (Figs. 1A, and 2). The eH site also exhibited clear protections in the domain that would be bound by wing 2 in the crystal (Fig 1B). Thus, FOXA1 wing 2 exhibits strong and stable contacts with DNA in solution, in contrast to that reported for FoxD3 and FoxJ2.
After a ∼5 nt gap up the gel, half a helical turn, additional nt are protected by FOXA1 at the upstream “half” of the eG site (Fig. 1B, 2). This corresponds to where wing 1 of the protein would make minor groove contacts, if the DNA structure in the original crystal was extended (Fig. 1A, pink bases denoted at top). Notably, the eH site exhibited weak protections at the wing 1 position (Fig. 1B, 2), likely reflecting the lower affinity of FOXA1 for this sequence, as seen in DNase footprinting and electromobility shift assays 24; 30. We conclude that at the eG site, both the wing 1 and wing 2 domains of FOXA1 interact closely and stably with DNA.
To directly assess the ability of wing 1 interactions to help confer high-affinity binding to the eG site by FOXA1, we mutated the eG sequence in two ways. First, we substituted wing 1 interactions from the weaker eH site into the eG site, to create the sequence designated eG-W1eH (Fig. 3A). This involved two transitions and one transversion of the sequence. Second, we introduced three transversions to create the sequence designated eG-W1mut (Fig. 3A); the first two transversions are in the relative positions of bases that are contacted by FOXK1 in its co-crystal with DNA 21. Variant and wild type double-stranded oligonucleotides were labeled and tested in quantitative electromobility shift assays for binding to a dilution series of purified FOXA1 protein (Fig. 3B). Quantitation of the data showed that at the 2 femtomole level of FOXA1, both the eG-W1eH and eG-W1mut DNAs bound about 30% poorer than the wild type DNA (p=0.01 and p=0.03, respectively), and at the 0.5 femtomole level of FOXA1, they bound about 50% poorer than wild type (p=0.05 for both). These data extend those of a prior study which showed that a single base pair change in the wing 1 binding region affected DNA binding 26. We conclude that wing 1 interactions are crucial for optimal and specific binding of FOXA1 to a target DNA site.
Figure 3.
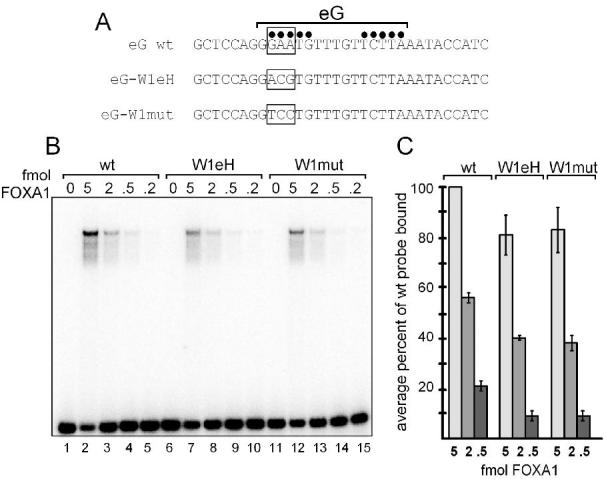
Site directed mutagenesis of the wing 1 interaction region of the high affinity eG site for FOXA1. A. Top strand of double-stranded oligonucleotide sequences of the eG site and variants are shown. Black dots indicate positions of top strand hydroxyl radical protections within the eG site. Boxed regions indicate sequences changed in the eG-W1eH and eG-W1mut variants; see text for details. B. Electromobility shift assay (EMSA) with 0.05 ng of end-labeled probes and designated femtomole (fmol) amounts of purified FOXA1 protein, as described 30 except for the addition of 5 ug of bovine serum albumin. Free probe is at the bottom of the gel; the arrowhead depicts a FOXA1-eG site complex. C. Data for two EMSA experiments quantitated on a phosphorimager and plotted as average percent of wild type probe bound at each designated amount of FOXA1 protein. The 2 and 0.5 fmol amounts of FOXA1 are bound 30% and 50% less well, respectively, to the variant eG sites compared to the wild type eG site (p=0.01 to 0.05; see text).
Our data show that FOXA1 interacts closely with one face of the DNA, along its long axis, for nearly a turn and a half of the helix. We suggest that the previous data indicating weak, dynamic interactions of the FOX protein wings with DNA could have been due to the use of non-optimal DNA target sequences in the analysis. For example, the wing 1 protections by FOXA1 at the eH site were significantly fainter than at the eG site, correlating with weaker affinity for eH 30; 31, and in vivo footprinting studies showed that in embryonic endoderm, the eG is occupied, but not the eH site 5; 6. Furthermore, conversion of the higher affinity eG sequence contacted by wing 1 to the corresponding sequence of the lower affinity eH site reduced the affinity of FOXA1 to the DNA (Fig. 3B, C). Since both the eG and eH sites are functional in adult liver cells 30,31, but the eG site is occupied earlier in development than the eH site, our findings indicate that variation in wing 1 affinity is used by FOXA1 target sequences for differential recruitment of the factor.
Although the FOXA3 crystallization DNA was short, it had a 15 degree bend in the structure 20. Subsequent studies showed that FOX factors bend DNA considerably 32, they preferentially bind pre-bent DNA 33, and they stably bind DNA that is curved around a nucleosome core particle 34. Our data showing the specificity of wing 1 interactions with DNA suggest that the interactions play an important role in allowing FOX factors to bind one side of the DNA helix, thereby allowing other transcription factors or histones9; 34 to bind the other side in a transcription factor or nucleosomal complex. This property may be crucial for the pioneering activity of FOXA proteins in chromatin and transcriptional activation 10, 11.
Acknowledgements
This paper is dedicated to the memory of Rob Costa. L.A.C. was supported by an NRSA postdoctoral fellowship and the research was supported by grant from the NIH (R01 GM47903) to K.S.Z.
Footnotes
Corresponding author: Dr. Ken Zaret Fox Chase Cancer Center, Room W410 333 Cottman Avenue Philadelphia, PA 19111-2497 Tel.: 215-728-7066 Fax: 215-379-4305 Email: zaret@fccc.edu
References
- 1.Costa RH. In: Hepatocyte nuclear factor 3/forkhead protein family: mammalian transcription factors that possess divergent cellular expression patterns and binding specificities. Tronche F, Yaniv M, editors. R.G. Landes Company; Austin: 1994. [Google Scholar]
- 2.Ang S-L, Rossant J. HNF-3β is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr. The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 4.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–7. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 5.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 6.Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 7.Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo L, Lin FR, Cuesta I, Jarnik M, Friedman D, Zaret K. Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Molecular Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 9.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 10.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 12.Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100059. 2006 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigel D, Jürgens G, Küttner F, Seifert E, Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 14.Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-1α, β, γ homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- 16.Pohl BS, Knochel W. Of Fox and Frogs: Fox (fork head/winged helix) transcription factors in Xenopus development. Gene. 2005;344:21–32. doi: 10.1016/j.gene.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 17.Odenthal J, Nusslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–58. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- 18.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 19.Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–500. [PubMed] [Google Scholar]
- 20.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF3/fork head DNA recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 21.Tsai KL, Huang CY, Chang CH, Sun YJ, Chuang WJ, Hsiao CD. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J Biol Chem. 2006;281:17400–9. doi: 10.1074/jbc.M600478200. [DOI] [PubMed] [Google Scholar]
- 22.Jin C, Marsden I, Chen X, Liao X. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J Mol Biol. 1999;289:683–90. doi: 10.1006/jmbi.1999.2819. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Ferreria MA, Rey-Campos J. Functional domains of FOXJ2. J Mol Biol. 2003;329:631–44. doi: 10.1016/s0022-2836(03)00524-2. [DOI] [PubMed] [Google Scholar]
- 24.Zaret KS, Stevens K. Expression of a highly unstable and insoluble transcription factor in Escherichia coli: purification and characterization of the fork head homolog HNF3 alpha. Protein Expr Purif. 1995;6:821–825. doi: 10.1006/prep.1995.0014. [DOI] [PubMed] [Google Scholar]
- 25.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci U S A. 1998;95:9738–43. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux J, Pictet R, Grange T. Hepatocyte nuclear factor 3 determines the amplitude of the glucocorticoid response of the rat tyrosine aminotransferase gene. DNA Cell Biol. 1995;14:385–96. doi: 10.1089/dna.1995.14.385. [DOI] [PubMed] [Google Scholar]
- 27.Pastor N, Weinstein H, Jamison E, Brenowitz M. A detailed interpretation of OH radical footprints in a TBP-DNA complex reveals the role of dynamics in the mechanism of sequence-specific binding. J Mol Biol. 2000;304:55–68. doi: 10.1006/jmbi.2000.4173. [DOI] [PubMed] [Google Scholar]
- 28.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical “footprinting”. Curr Opin Struct Biol. 2002;12:648–53. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 29.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino acid residues adjacent to the recognition helix. Mol. Cell. Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson DA, Rowader KE, Stevens K, Jiang C, Milos P, Zaret KS. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol. Cell. Biol. 1993;13:2401–2410. doi: 10.1128/mcb.13.4.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JK, DiPersio CM, Zaret KS. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol Cell Biol. 1991;11:773–84. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. Embo J. 1994;13:5002–12. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bravieri R, Shiyanova T, Chen TH, Overdier D, Liao X. Different DNA contact schemes are used by two winged helix proteins to recognize a DNA binding sequence. Nucleic Acids Res. 1997;25:2888–96. doi: 10.1093/nar/25.14.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim E-Y, Clark EA, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]


