Abstract
The mechanistic basis of sensory adaptation and gradient sensing in bacterial chemotaxis is reversible covalent modification of transmembrane chemoreceptors, methylation, and demethylation at specific glutamyl residues in their cytoplasmic domains. These reactions are catalyzed by a dedicated methyltransferase CheR and a dedicated methylesterase CheB. The esterase is also a deamidase that creates certain methyl-accepting glutamyls by hydrolysis of glutamine side chains. We investigated the action of CheB and its activated form, phospho-CheB, on a truncated form of the aspartate receptor of Escherichia coli that was missing the last 5 aa of the intact receptor. The deleted pentapeptide is conserved in several chemoreceptors in enteric and related bacteria. The truncated receptor was much less efficiently demethylated and deamidated than intact receptor, but essentially was unperturbed for kinase activation or transmembrane signaling. CheB bound specifically to an affinity column carrying the isolated pentapeptide, implying that in the intact receptor the pentapeptide serves as a docking site for the methylesterase/deamidase and that the truncated receptor was inefficiently modified because the enzyme could not dock. It is striking that the same pentapeptide serves as an activity-enhancing docking site for the methyltransferase CheR, the other enzyme involved in adaptational covalent modification of chemoreceptors. A shared docking site raises the tantalizing possibility that relative rates of methylation and demethylation could be influenced by competition between the two enzymes at that site.
Reversible covalent modification of transmembrane receptors is the mechanistic basis of sensory adaptation and gradient sensing in bacterial chemotaxis (see refs. 1–3 for reviews of our understanding of these processes). Chemoreceptors are methylated and demethylated at specific glutamyl residues by the dedicated methyltransferase CheR and the dedicated methylesterase CheB. An increase in occupancy at an attractant binding site causes the receptor to signal across the membrane, altering the activity of a protein kinase and ultimately changing the swimming behavior of the cell. However, that change is transient because a compensatory increase in methylation resets the receptor, and thus cellular behavior, back to a null, adapted state. Conversely, a reduction in receptor occupancy is balanced by compensatory demethylation. Swimming bacteria sense spatial gradients of attractants and repellents by detecting concentration changes over time as they move. The chemosensory system compares a measure of current attractant concentration to a record of the just-previous concentration (4). Current concentration is measured by receptor occupancy. The record of the past concentration is the level of methylation. This level reflects the just-previous extent of receptor occupancy because rates of compensatory covalent modification are slow relative to changes in receptor occupancy. Thus dynamic methylation and demethylation are crucial to chemosensory function.
The most extensive body of information about adaptational modifications is for the well-characterized chemoreceptors of Escherichia coli and Salmonella typhimurium (see refs. 1–3 for reviews). These receptors exist as stable homodimers organized in three domains, periplasmic, transmembrane, and cytoplasmic. The periplasmic domain carries ligand-binding sites, the transmembrane domain conveys conformational signals across the membrane, and the cytoplasmic domain contains the adaptational, methyl-accepting sites as well as controlling the activity of a noncovalently associated histidine kinase, CheA. This kinase phosphorylates one of its own histidines and then donates the phosphoryl group to the response regulator CheY. Cellular levels of phospho-CheY determine the rotational bias of the flagellar motors because the motors rotate only counterclockwise in the absence of interaction with the phosphorylated response regulator. Modulation of rotational bias by receptor-mediated control of phosphorylation directs cells to favorable environments. The sequences of the cytoplasmic domains of chemoreceptors are closely related, and four methyl-accepting sites occur at conserved positions (5–10), spaced in a helical periodicity that would place them on solvent-exposed faces of amphipathic helices (11). For each receptor, two methyl-accepting glutamates are created by deamidation of a glutamine (5). This modification is catalyzed by the same protein, CheB, that catalyzes demethylation. Thus CheB is both a methylesterase and a deamidase. Amides at methyl-accepting sites are in large part the functional equivalents of methylesters, and deamidation has the same functional consequences as demethylation, but is not reversible (12–14).
Receptor modification is a dynamic and finely balanced phenomenon. In the steady state, a population of receptors, either unoccupied or occupied but adapted, is constantly undergoing methylation and demethylation, yet maintains a net level of modification that balances occupancy and generates an intermediate (null) level of kinase activation. What controls the rates and extents of these reactions? The methylesterase is activated by phosphorylation (15). The activity of its catalytic domain is controlled by a regulatory domain (16) that is a homolog of the response regulator CheY and is phosphorylated by the same, chemoreceptor-controlled kinase (17). Phosphorylated CheB has at least a 70-fold higher methylesterase activity than the unmodified enzyme (18). In contrast, methyltransferase activity does not appear to be modulated by alterations of the enzyme but rather by an apparent conformational change in the receptor substrate upon changes in ligand occupancy (19). In addition, recent work has shown that a pentapeptide sequence Asn-Trp-Glu-Thr-Phe (NWETF in the one-letter code), present at the extreme carboxyl terminus of some chemoreceptors (the “high-abundance” receptors) provides a docking site for the methyltransferase (20) and greatly enhances methylation of receptors on which it is present (21–24). Receptors naturally lacking the methyltransferase-docking site (low-abundance receptors) are poorly methylated and thus are ineffective, in the absence of high-abundance receptors, in both adaptation and the ability to mediate chemotaxis (23–27). In the course of our ongoing investigations of the low-abundance receptor Trg, we observed an indirect indication that the presence of the carboxyl-terminal pentapeptide also influenced methylesterase activity. We pursued that observation and found that the pentapeptide is a major determinant of esterase action, serving as a docking site for this adaptional enzyme just as it does for its complementary partner, the methyltransferase.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
RP3098 (28) is a strain of E. coli K12, provided by J. S. Parkinson (University of Utah, Salt Lake City), that carries a deletion from flhA through flhD and thus lacks the genes for all Che proteins. pNT201 (29) carries tar under the control of a tac promoter. pAL61, in which a termination codon was introduced at the fifth-to-last codon of tar, was constructed by PCR-based mutagenesis of pNT201 and confirmed by sequencing. pCW/cheB, which carries cheB under the control of tandem tac promoters (30, 31), was obtained from F. W. Dahlquist (University of Oregon, Eugene).
Protein Preparation, Purification, and Quantification.
CheA, CheW, and CheY were produced and purified, membranes containing chemoreceptors were prepared, and all proteins were quantified as described (23), except that EDTA was 10 mM, membranes were prepared from larger cultures, and thus all volumes were proportionately larger, cells in a buffer containing 250 mM sucrose were disrupted by two passages through a French Press cell (Aminco) at 8,000 psi, and low-speed and high-speed centrifugations were at 10,000 rpm, 4°C in a Sorvall SS-34 rotor, and for 40 min at 45,000 rpm in Beckman Ti-60 rotor (140,000 × g), respectively. Cell lysates containing CheB were prepared as for the other Che proteins by using RP3098 harboring pCW/cheB, except that cell disruption was at 20,000 psi.
In Vitro Assays.
Phosphorylation (23), methylation (23), demethylation (16, 32), and deamidation (31) were assayed essentially as described. To prepare substrates for demethylation, membranes containing Tar were incubated for 10 min with a cell lysate containing CheR (≈1 μM) and 50 μM S-adenosyl-l-[methyl-3H]methionine ([methyl-3H]AdoMet) (Amersham Pharmacia) in the conditions of a methylation assay. Membranes containing TarΔpp were incubated with [methyl-3H]AdoMet at a ≈17-fold higher concentration of CheR-containing lysate for 60 min. Incubated membranes were diluted in an excess of ice-cold 50 mM Tris⋅HCl, pH 7.5, 0.5 mM EDTA, 2 mM DTT, 10% glycerol plus 2 M KCl (TEDG+2 M KCl) and centrifuged in a TLA-100.4 rotor at 70,000 rpm for 20 min. Pelleted membrane was suspended in TEDG-2M KCl, centrifuged again, and suspended in TEDG. The extent of receptor labeling with 3H-methyl groups was determined as for methylation assays (23). 3H-methyl-labeled membranes were diluted in TEDG containing 50 mM KCl and 10 mM MgCl2 to yield a final receptor concentration of 10 μM. When present, CheA and CheW were at 5 μM, and ATP was 1 mM. For most reactions, all components except CheB and ATP were incubated at room temperature for 30 min and reactions initiated by addition of CheB (to 0.5 μM) or CheB plus ATP (to 0.5 μM and 1 mM, respectively). In some cases ATP was added to 1 mM 15 min before addition of CheB. At various times 4-μl samples were removed and mixed with 20-μl 5 N acetic acid, and radioactive methanol released was quantified by the vapor-phase equilibrium method (33).
Peptide Affinity Columns for Analysis and Purification.
Peptides NWETF, EENWETF, DPNWETF, and FTEWNPD were synthesized by solid-phase synthesis using fluorenylmethoxycarbonyl chemistry on an Applied Biosystems 431A machine. Reverse-phase chromatography of the final products on a C8 RP-300 column (Applied Biosystems) revealed single peaks. The peptides had a single amino group, on their amino termini, and thus could be coupled specifically at that terminus to an appropriate resin by using 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide-N-hydroxysuccinimide (NHS) coupling chemistry. We did this by using 1 ml (analytical) or 5-ml (preparative) Hi-Trap NHS-activated columns (Amersham Pharmacia), 5 mg/ml peptide, and the protocol provided with the columns. A column was first equilibrated with TEDG, cell lysate (≈2 mg protein/ml) containing CheR or CheB produced from an induced, plasmid-borne gene was applied, followed by two bed volumes of TEDG and three bed volumes of TEDG containing 5 mg/ml pentapeptide or heptapeptide. For purification of preparative amounts of CheB, 5-ml columns carrying any one of the natural sequences were eluted with 2 M NaCl instead of pentapeptide, yielding a protein more than 99% pure as estimated by Coomassie blue staining of overloaded SDS polyacrylamide gels. The ≈20-kDa species corresponding to the proteolytically produced, catalytic domain of CheB was only ≈0.02% the intensity of intact CheB. CheB concentration was determined by using a molar extinction value of 22,000 cm−1 at 280 nm (34).
RESULTS
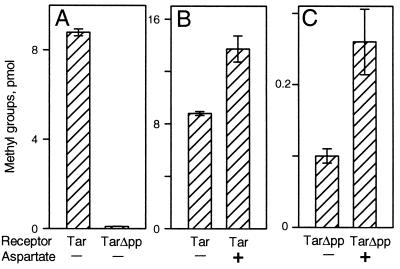
Tar Deleted for Its Carboxyl-Terminal Pentapeptide. To assess the role of the carboxyl-terminal pentapeptide sequence NWETF in methylesterase action on chemoreceptors, we created a gene coding for a form of the high-abundance receptor Tar lacking those residues. We called that truncated receptor TarΔpp, for Tar deleted of pentapeptide. Both the altered gene and the natural form of tar were contained in the same plasmid vector under the control of a modified lac promoter, allowing production as desired at a range of cellular dosages. Immunoblots of fully induced cells and of membranes isolated from such cells revealed that TarΔpp was present at approximately the same level as full-length Tar and that neither protein exhibited any significant proteolysis (data not shown). We compared the functional properties of the two forms of Tar by using in vitro assays. As observed in a previous study in which a pentapeptide-truncated form of Tar was created (22), removal of the carboxyl-terminal residues reduced methyl-accepting activity in vitro to a few percent of the activity of the full-length receptor (Fig. 1A), a result consistent with the identification of NWETF as a methyltransferase-docking site (20). Even though methyl-accepting activity of TarΔpp was low, stimulation by a saturating concentration of a Tar ligand, aspartate, increased this activity (Fig. 1C) to an extent similar to that observed for intact Tar (Fig. 1B), indicating that transmembrane signaling, as assessed by methylation in vitro, was not significantly disrupted by elimination of the carboxyl-terminal pentapeptide.
Figure 1.
Methyl-accepting activities of Tar and TarΔpp. Membranes providing ≈5 μM Tar or TarΔpp were incubated for 1 min at room temperature with 50 μM [methyl-3H]AdoMet, 1 mM aspartate (where indicated), and a cell extract providing ≈1 μM CheR. Samples containing ≈50 pmol receptor were analyzed for radiolabeled methyl groups. The data are averages of experiments using two independent membrane preparations. Error bars show SEs.
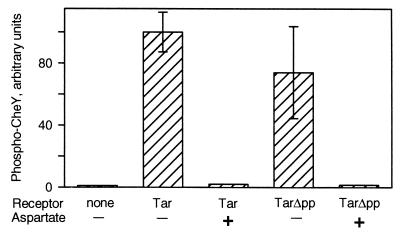
Formation of kinase-activating complexes with CheA and CheW is a crucial receptor activity. We tested the ability of TarΔpp to stimulate the kinase activity of CheA, using a standard in vitro assay in which the formation of phospho-CheY is a direct measure of CheA autophosphorylation (35) and found that the truncated receptor was almost as effective as full-length Tar (Fig. 2). This result is consistent with our previous observation that Trg, a chemoreceptor naturally lacking the carboxyl-terminal pentapeptide, activates kinase within a factor of 2 as well as Tar, a receptor that naturally has the pentapeptide (23). We used phosphorylation as a second assay for transmembrane signaling and found that aspartate binding to TarΔpp resulted in an approximately 100-fold reduction in kinase activity, an effect equivalent to that observed for intact Tar (Fig. 2). Taken together, our data indicated that deletion of the pentapeptide from Tar had the expected effect of reducing methyl-accepting activity but otherwise left the receptor functional. Thus we could use the truncated receptor to investigate the possibility of a specific role for the pentapeptide in CheB action by comparing TarΔpp and intact Tar.
Figure 2.
Kinase activation by unoccupied and ligand-occupied forms of Tar and TarΔpp. Membranes providing ≈2 μM Tar or TarΔpp were incubated with 0.25 μM CheA, 4 μM CheW, and 10 μM CheY to allow complex formation, 32P-ATP was added, samples were taken 10 s later, and 32P-labeled phospho-CheY was determined by SDS/PAGE and PhosphorImaging. Aspartate (1 mM) was present where indicated. Replicates and error bars are as for Fig. 1.
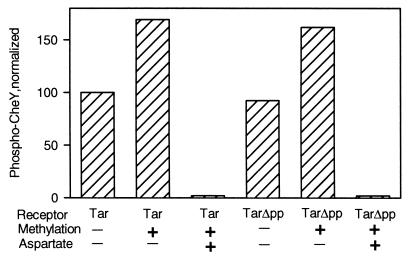
Demethylation. To compare CheB-catalyzed demethylation of TarΔpp and Tar, we prepared samples of the two receptors modified with radiolabeled methyl groups. We isolated membranes from cells lacking the general components of the chemosensory system and producing Tar or TarΔpp as the sole chemoreceptor. The two available methyl-accepting sites of those receptors (the other two were gene-encoded glutamines) were modified with radioactive methyl groups by incubation of membrane samples with S-adenosyl[methyl-3H]methionine plus a cell lysate containing the methyltransferase CheR. As noted above, in vitro methylation was much less efficient for TarΔpp than for full-length Tar, but incubation of the truncated receptor with a higher concentration of enzyme-containing lysate for a longer time produced essentially the same level of methylation for the two receptor species. Before using such methylated receptors to test CheB activity, it was important to determine that the two different preparation conditions had left the respective receptors functionally intact. This was done by testing for kinase activation and for control of kinase activity by ligand occupancy. Fig. 3 shows an example of such a test. The two membrane preparations, one containing Tar and the other TarΔpp, had the same concentration of receptor and exhibited almost the same extent of kinase activation (see Fig. 2 for the variation we observed over several preparations). After methylation to a level of approximately 0.4 3H-methyl groups per receptor, both Tar and TarΔpp mediated increased kinase activation, as expected from previous demonstrations that receptor methylation enhances kinase activation (14) and those increases were of essentially the same magnitude. Saturation of methylated Tar or TarΔpp with the chemoattractant aspartate resulted in drastic reduction of kinase activity, the pattern exhibited by the receptors prior to methylation (Fig. 2). Thus both methylated receptors preparations appeared equivalently functional in kinase activation and transmembrane signaling. This pattern was observed each time we made such preparations.
Figure 3.
In vitro treatment to methylate Tar and TarΔpp increased kinase activation and preserved control by ligand occupancy. Membranes containing Tar or TarΔpp were incubated in the absence (methylation −) or presence (methylation +) of [methyl-3H]AdoMet and a CheR-containing extract. The latter condition resulted in ≈0.4 methyl groups per receptor. Kinase activation by washed, reisolated, receptor-containing membranes in the absence (−) or presence (+) of 1 mM aspartate was determined as in Fig. 2. Levels of phospho-CheY were normalized to the value for Tar in the absence of methylation and aspartate.
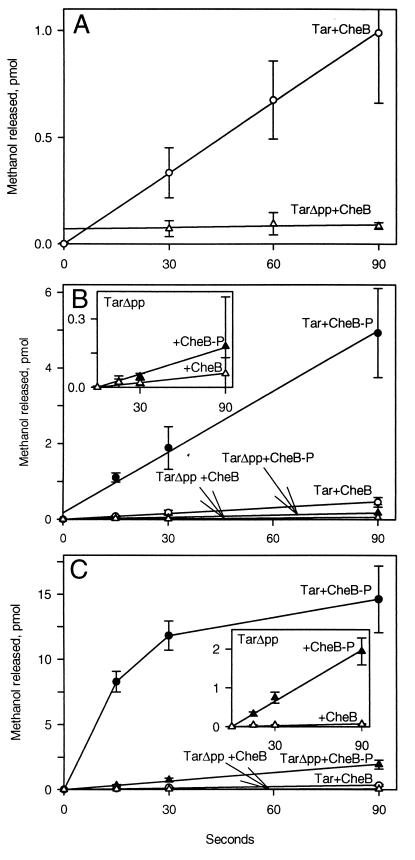
We compared the action of purified CheB on Tar and TarΔpp by using the 3H-methyl-labeled preparations described in the preceding paragraph, measuring demethylation by detecting release of radiolabeled methanol. Demethylation of full-length Tar proceeded significantly more rapidly than demethylation of TarΔpp (Fig. 4A). Phosphorylation greatly enhances the activity of CheB (15, 18), so we designed experiments to expose 3H-methyl-labeled receptors to phospho-CheB. Unfortunately it is not possible to maintain CheB in a stoichiometrically phosphorylated state because the phosphate is rapidly hydrolyzed (18), but some proportion of the enzyme is phosphorylated in the presence of the kinase CheA plus ATP (15). In such conditions (Fig. 4B) rates of demethylation increased for both Tar and TarΔpp, indicating that the more active, phosphorylated form of CheB was present. As observed for unphosphorylated CheB, demethylation catalyzed by phospho-CheB was substantially more rapid for intact Tar than for Tar lacking the pentapeptide (Fig. 4B). In the cell, chemoreceptors are thought to exist in relatively stable ternary complexes with the kinase CheA and the accessory protein CheW (31, 36). Thus it was important to compare demethylation of Tar and TarΔpp assembled into such complexes. We incubated membranes containing 3H-methyl-labeled receptors with CheA and CheW in conditions that resulted in formation of ternary complexes. In these complexes, receptor-activated CheA could efficiently phosphorylate CheB. We provided ATP for phosphorylation either at the time of CheB addition or before addition to allow creation of a pool of autophosphorylated CheA. Both conditions resulted in faster demethylation than the situation in which CheB was phosphorylated by free CheA, indicating higher levels of phospho-CheB. Simultaneous addition of ATP and CheB provided the most rapid demethylation, and thus the highest apparent concentration of phospho-CheB. Data from such experiments are shown in Fig. 4C. As for receptor alone, Tar in complex with CheA and CheW was demethylated much more rapidly as the full-length receptor than in the form lacking the pentapeptide.
Figure 4.
Demethylation of Tar and TarΔpp catalyzed by CheB and phospho-CheB. (A) Action of CheB on receptors alone. Membranes providing ≈10 μM Tar or TarΔpp modified with 3H-methyl groups as described for Fig. 3 (≈0.4 methyls per receptor) were mixed with 0.5 μM purified CheB. Samples containing ≈40 pmol receptor were removed at the indicated times and analyzed for released 3H-methanol. (B) Action of CheB and phospho-CheB on receptors alone. Receptor-containing membranes as in A were incubated 15 min with 5 μM CheA, 1 mM ATP was added to some incubations (plots labeled CheB-P), the incubation continued for 15 min, purified CheB was added to 0.5 μM, and samples were taken and analyzed as for A. (C) Action of CheB and phospho-CheB on receptors complexed with CheA and CheW. Receptor-containing membranes as in A were incubated 30 min with 5 μM CheA and 5 μM CheW. CheB or CheB plus ATP (plots labeled CheB-P) were added to 0.5 μM CheB and 1 mM ATP, and samples were taken and analyzed as for A. For all panels, data are averages of experiments on at least three separate membrane preparations. Error bars show SEs.
Deamidation.
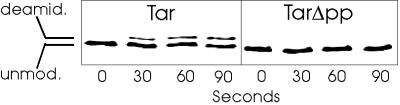
In addition to demethylation, CheB also catalyzes deamidation of glutamines at receptor modification sites. We investigated this action on full-length and truncated Tar by adding purified CheB to membrane preparations containing Tar or TarΔpp carrying gene-encoded glutamines at the two sites of potential deamidation, and assaying for deamidation by using SDS/PAGE and immunoblotting with antireceptor serum. A deamidated receptor migrates slightly more slowly in an SDS gel than the unmodified protein from which it is derived (37, 38) and thus the two forms are resolved as separate bands on an immunoblot. Fig. 5 shows results from a representative experiment. In the conditions used, there was substantial deamidation of Tar but no detectable modification of TarΔpp.
Figure 5.
Deamidation of Tar and TarΔpp catalyzed by CheB. Membranes providing ≈10 μM Tar or TarΔpp were incubated in TEDG plus 10 mM MgCl2 with 0.5 μM purified CheB. Samples containing ≈3 pmol receptor were removed at the indicated times and analyzed by SDS/PAGE and immunoblotting. Shown is the relevant portion of the blot, including the positions of unmodified (unmod.) and deamidated (deamid.) receptor. Unmodified TarΔpp migrates slightly more rapidly than unmodified Tar, consistent with a difference of five residues.
CheB Interaction with the Pentapeptide.
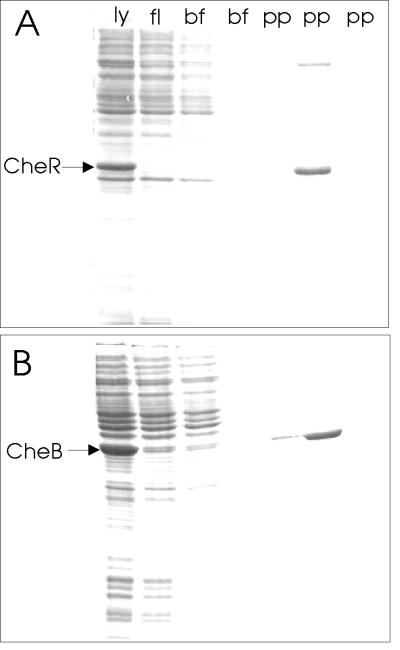
The strong dependence of CheB-catalyzed receptor modifications on the presence of the carboxyl-terminal pentapeptide implied that the enzyme interacted with that sequence. We tested this notion by making an affinity column containing the NWETF pentapeptide and testing for specific retention and elution of CheB. The pentapeptide was coupled via its amino terminus to a linker molecule attached to a resin packed in a small column. We demonstrated the efficacy of the column by showing that it retained CheR selectively and released that protein upon elution with pentapeptide (Fig. 6A). A representative experiment with CheB is shown in Fig. 6B. A soluble extract from cells containing CheB was applied to the column and the column washed with buffer and then with buffer containing 2 mM pentapeptide. CheB was retained by the pentapeptide column and eluted by free pentapeptide. In contrast, CheB was not retained by a column in which the natural sequence was reversed from NWETF to FTEWN (data not shown). We conclude that CheB interacts specifically with the pentapeptide NWETF.
Figure 6.
Retention of modification enzymes by immobilized pentapeptide and elution by free pentapeptide. Soluble lysate (ly) from cells containing CheR (A) or CheB (B) produced from an induced gene located on a multicopy plasmid was applied to a 1-ml column of resin carrying the immobilized pentapeptide, NWETF. Fractions collected during application of the sample (fl), two column volumes of buffer (bf), and three column volumes of buffer containing 5 mg/ml free pentapeptide (pp) were analyzed by SDS/PAGE. The positions of CheR (≈33 kDa) and CheB (≈37 kDa) are indicated on the respective gels. Analysis of the 37-kDa band eluted by pentapeptide in the experiment of B revealed an amino-terminal sequence (seven residues) identical to authentic CheB. Essentially identical results were obtained by using heptapeptides representing the carboxyl-terminal residues of Tsr (EENWETF) or Tar (DPNWETF) as the immobilized or eluting peptide.
DISCUSSION
We found that removal of the pentapeptide NWETF, naturally located at the extreme carboxyl terminus of Tar and other “high-abundance” chemoreceptors of E. coli and S. typhimurium, profoundly decreased the efficiency with which the methylesterase/deamidase CheB catalyzed receptor modification. In contrast, the truncated receptor appeared essentially unperturbed in the central functions of kinase activation and transmembrane signaling. We identified a basis for inefficient CheB-mediated modification of the truncated form of Tar by demonstrating that CheB bound to the pentapeptide. This carboxyl-terminal sequence is distinct from the positions of glutamines and glutamyl methyl esters that CheB hydrolyzes and thus the pentapeptide sequence appears to be a “docking site” for the enzyme. It is striking that this same pentapeptide also serves as an activity-enhancing docking site for the CheR methyltransferase (20), the other member of the pair of enzymes that catalyzes adaptational covalent modification of chemoreceptors. A shared docking site raises the tantalizing possibility that relative rates of methylation and demethylation could be influenced by competition between the two enzymes at that site.
Adaptation to negative stimuli is mediated by CheB-catalyzed demethylation. Thus the dependence of rapid demethylation in vitro on the pentapeptide implies that adaptation in vivo to negative stimuli should be slowed by disruption of the docking site. In fact, this has been documented. Okumura et al. (39) observed that mutational alterations in the carboxyl-terminal pentapeptide of the high-abundance receptor Tcp drastically slowed adaptation to a repellent stimulus.
A Docking Site for Enzymes of Adaptational Modification. How might interaction with a carboxyl-terminal docking site enhance enzyme action at separate sites of modification? The presence of a docking site on a receptor could increase enzyme concentration in the vicinity of substrate side chains, or pentapeptide binding could cause allosteric activation of the enzyme. The two possibilities are not mutually exclusive, so both might contribute. Increasing local concentration would not require that the enzyme remain bound to the pentapeptide as it carried out catalysis whereas allosteric activation would. The combination of these two possible modes of pentapeptide-mediated enhancements for the activity of two competing enzymes would provide opportunities for sophisticated control of rates of the opposing modifications. No matter how the docking site enhances CheB activity, the central role of the site in determining the rate of demethylation offers an explanation for the puzzling observation that cellular populations of receptors at different steady-state levels of methylation exhibit the same rate of demethylation (40). If steady-state demethylation is determined primarily by the probability of interaction of enzyme with docking site, rather than by the probability of direct interaction with the methyl esters to be hydrolyzed, then the rate would not vary as a function of receptor methylation.
Interaction of CheB and pentapeptide can be used for enzyme purification. A higher capacity version of the affinity column used to demonstrate the interaction (Fig. 6B) provided a rapid, one-step isolation that produced high purity enzyme minimally contaminated by the proteolytically produced catalytic domain (16, 18). All characterizations of CheB activity reported here were performed with such highly purified enzyme. The same column also could serve to purify CheR.
Multiple Interactions of CheB with Receptor Complexes.
There are now three known sites of interaction between CheB and the receptor-CheA-CheW complex: the kinase site at which CheB obtains a phosphate group, the sites at which the enzyme modifies the receptor and the pentapeptide docking site at the carboxyl terminus of the receptor. How are these sites and their consequences related? The presence of a docking site on the receptor enhanced rates of demethylation catalyzed both by unactivated CheB (Fig. 4A) and by the activated (phosphorylated) enzyme (Fig. 4 B and C). Phosphorylation of CheB by free CheA or by CheA activated in complex with receptor resulted in increased rates of demethylation for receptors lacking or carrying the docking site (Fig. 4 B and C). The experiments of Fig. 4 were not designed for detailed kinetic analysis, but estimates from that data of initial rates of demethylation catalyzed by CheB (Fig. 4A) or phospho-CheB (Fig. 4C) indicate that the enhancements resulting from either factor, phosphorylation of CheB or the presence of the docking site, were independent of each other. Thus CheB binds to the pentapeptide whether or not the enzyme is phosphorylated, and phospho-CheB is more active than unmodified CheB even without a docking site with which to interact. In the cell, CheB is thought to be phosphorylated not by free CheA but by the activated kinase in complex with receptor and CheW. Thus it is interesting to consider that the docking site on the receptor might increase the probability of CheB phosphorylation or the probability of newly phosphorylated CheB modifying the receptor complex at which it was activated.
Natural Receptors Lacking the Docking Site NWETF. The work described here used a truncated form of chemoreceptor Tar that lacked the carboxyl-terminal pentapeptide NWETF, previously characterized as a methyltransferase-docking site (20). The low-abundance receptors in E. coli naturally lack this carboxyl-terminal sequence and as a consequence are less efficiently methylated than their high-abundance counterparts that carry the methyltransferase-docking site (23, 26, 27). Our identification of the same pentapeptide as a docking site for the CheB methylesterase/deamidase implies that low-abundance receptors also should be less efficiently deamidated and demethylated. In fact, deamidation assessed in cells lacking CheR but producing large amounts of a single receptor is much less complete for the low-abundance receptor Trg (7, 9) than for the NWETF-containing receptors Tsr or Tar (37, 41). In cells lacking NWETF-containing receptors, adaptation of Trg to addition of repellents or removal of attractants is very slow (unpublished observations), a pattern consistent with slow CheB-catalyzed demethylation of receptors lacking the docking site. These observations imply that conclusions about the importance of the NWETF site for CheB-catalyzed modifications, based on comparisons in vitro of a natural and a truncated form of a high-abundance receptor, are applicable to a low-abundance receptor that naturally lacks the carboxyl-terminal docking site.
A wild-type cell contains both high-abundance and low-abundance receptors. In such cells adaptation to stimuli recognized by low-abundance receptors is effective and timely (24, 26). Inter-dimer methylation documented in vitro between forms of the high-abundance receptor Tsr (21) or Tar (22), one carrying and the other deleted of the methyltransferase-docking site NWETF, suggests that in wild-type cells NWETF-containing receptors mediate effective adaptational methylation of low-abundance receptors lacking the docking site, and thus allow timely adaptation to positive stimuli. Efficient CheB-mediated adaptation of wild-type cells to negative stimuli recognized by the low-abundance receptor Trg implies a similar cross-receptor modification for CheB-catalyzed reactions. Cross-receptor methylation or demethylation may involve receptor clustering (42). Clustering of receptors carrying docking sites near other receptors lacking docking sites could enhance modification of the latter class. This could occur by direct interaction of an enzyme tethered on one receptor dimer with a neighboring receptor or simply by providing an increased local enzyme concentration. In either case it seems likely that tethering sites contribute to the creation of a specialized local environment in which components of the chemosensory system are not uniformly distributed throughout the cell but instead are sequestered near their sites of activation and action.
Acknowledgments
We thank Angela Lilly for construction of pAL61, Gerhard Munske for synthesis of the pentapeptide and for analyses by amino-terminal sequencing, and J. S. Parkinson, F. W. Dahlquist, J. Stock, and P. Matsumura for bacterial strains and plasmids. This work was supported by Grant GM29963 from the National Institutes of Health to G.L.H.
ABBREVIATIONS
- TEDG
Tris⋅HCl/EDTA/DTT/glycerol
- TarΔpp
Tar deleted of pentapeptide
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hazelbauer G L. In: Encyclopedia of Neuroscience. Adelman G, Smith B H, editors. Amsterdam: Elsevier; 1999. pp. 181–183. [Google Scholar]
- 2.Parkinson J S. Cell. 1993;72:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 3.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Block S M, Segall J E, Berg H C. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 5.Kehry M R, Bond M W, Hunkapiller M W, Dahlquist F W. Proc Natl Acad Sci USA. 1983;80:3599–3603. doi: 10.1073/pnas.80.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice M S, Dahlquist F W. J Biol Chem. 1991;266:9746–9753. [PubMed] [Google Scholar]
- 7.Kehry M R, Engström P, Dahlquist F W, Hazelbauer G L. J Biol Chem. 1983;258:5050–5055. [PubMed] [Google Scholar]
- 8.Nowlin D M, Bollinger J, Hazelbauer G L. J Biol Chem. 1987;262:6039–6045. [PubMed] [Google Scholar]
- 9.Nowlin D M, Bollinger J, Hazelbauer G L. Proteins. 1988;3:102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- 10.Terwilliger T C, Wang J Y, Koshland D E., Jr J Biol Chem. 1986;261:10814–10820. [PubMed] [Google Scholar]
- 11.Hazelbauer G L, Yaghmai R, Burrows G G, Baumgartner J W, Dutton D P, Morgan D G. In: Biology of the Chemotactic Response: Society for General Microbiology Symposium. Armitage J P, Lackie J M, editors. Vol. 46. Cambridge: Cambridge Univ. Press; 1990. pp. 107–134. [Google Scholar]
- 12.Park C, Dutton D P, Hazelbauer G L. J Bacteriol. 1990;172:7179–7187. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunten P, Koshland D E., Jr J Biol Chem. 1991;268:1491–1496. [PubMed] [Google Scholar]
- 14.Borkovich K A, Alex L A, Simon M I. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupas A, Stock J. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 16.Simms S A, Keane M G, Stock J. J Biol Chem. 1985;280:10161–10168. [PubMed] [Google Scholar]
- 17.Hess J F, Oosawa K, Kaplan N, Simon M I. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 18.Anand G S, Goudreau P N, Stock A M. Biochemistry. 1998;37:14038–14047. doi: 10.1021/bi980865d. [DOI] [PubMed] [Google Scholar]
- 19.Russell C B, Stewart R C, Dahlquist F W. J Bacteriol. 1989;171:3609–3618. doi: 10.1128/jb.171.7.3609-3618.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Li J, Li G, Long D G, Weis R M. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Li G, Weis R M. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 22.Le Moual H, Quang T, Koshland D E., Jr Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 23.Barnakov A N, Barnakova L A, Hazelbauer G L. J Bacteriol. 1998;180:6713–6718. doi: 10.1128/jb.180.24.6713-6718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng X, Lilly A A, Hazelbauer G L. J Bacteriol. 1999;181:3164–3171. doi: 10.1128/jb.181.10.3164-3171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazelbauer G L, Engström P. Nature (London) 1980;238:98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- 26.Feng X, Baumgartner J W, Hazelbauer G L. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weerasuriya S, Schneider B M, Manson M D. J Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkinson J S, Houts S E. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borkovich K A, Kaplan N, Hess J F, Simon M I. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gegner J A, Dahlquist F W. Proc Natl Acad Sci USA. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gegner J A, Graham D R, Roth A F, Dahlquist F W. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 32.Borczuk A, Staub A, Stock J. Biochem Biophys Res Commun. 1986;141:918–923. doi: 10.1016/s0006-291x(86)80130-9. [DOI] [PubMed] [Google Scholar]
- 33.Chelsky D, Gutterson N I, Koshland D E., Jr Anal Biochem. 1984;141:143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Swanson R W, Simon M I, Weis R M. Biochemistry. 1995;34:14626–14636. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]
- 35.Borkovich K A, Simon M I. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]
- 36.Schuster S C, Swanson R V, Alex L A, Bourret R B, Simon M I. Nature (London) 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- 37.Sherris D, Parkinson J S. Proc Natl Acad Sci USA. 1981;78:6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazelbauer G L, Engström P. J Bacteriol. 1981;145:35–42. doi: 10.1128/jb.145.1.35-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okumura H, Hishiyama S-I, Sasaki A, Homma M, Kawagishi I. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kehry M R, Doak T G, Dahlquist F W. J Biol Chem. 1984;259:11828–11835. [PubMed] [Google Scholar]
- 41.Rollins C, Dahlquist F W. Cell. 1981;25:333–340. doi: 10.1016/0092-8674(81)90051-9. [DOI] [PubMed] [Google Scholar]
- 42.Maddock J R, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]