Abstract
Serotonin and octopamine (OA) are biogenic amines that are active throughout the nervous systems of insects, affecting sensory processing, information coding and behavior. As an initial step towards understanding the modulatory roles of these amines in olfactory processing we cloned two putative serotonin receptors (Ms5HT1A and Ms5HT1B) and one putative OA (MsOAR) receptor from the moth Manduca sexta. Ms5HT1A and Ms5HT1B were both similar to 5HT1-type receptors but differed from each other in their N-terminus and 3rd cytoplasmic loop. Ms5HT1A was nearly identical to a serotonin receptor from Heliothis virescens and Ms5HT1B was almost identical to a serotonin receptor from Bombyx mori. The sequences for homologs of Ms5HT1A from B. mori and Ms5HT1B from H. virescens were also obtained, suggesting that the Lepidoptera likely have at least two serotonin receptors. The MsOAR shares significant sequence homology with pharmacologically characterized OA receptors, but less similarity to putative OA/tyramine receptors from the moths B. mori and H. virescens. Using the MsOAR sequence, fragments encoding putative OA receptors were obtained from B. mori and H. virescens, suggesting that MsOAR is the first OA receptor cloned from a lepidopteran.
Keywords: Serotonin, Octopamine, Manduca, Receptors, Modulation
1. Introduction
There is mounting evidence to suggest that the biogenic amines serotonin and octopamine (OA) modulate neural circuits at several levels of processing in the olfactory pathway of moths. Within the antennal lobe, serotonin appears to play a role in the maintenance of a circadian rhythm in the sensitivity of the moth olfactory system. Serotonin titers in the antennal lobes are highest during periods of peak activity (Gatellier et al., 2004; Kloppenburg et al., 1999) and exogenous serotonin increases the behavioral sensitivity of male Bombyx mori to female sex pheromone (Gatellier et al., 2004) and the excitability of antennal lobe neurons of Manduca sexta by reducing two potassium currents (Mercer et al., 1995; Kloppenburg et al., 1999). While the physiological and behavioral effects of serotonin in the lepidopteran antennal lobe are well established, little is known about the functional properties of the receptors mediating these effects. Even less is known about the role of OA within the lepidopteran olfactory system. OA injection increases the sensitivity of male moths to female sex pheromone (see Roeder, 2005) and both the antennal lobes and mushroom bodies in M. sexta receive OA-immunoreactive innervation (Dacks et al., 2005), but the roles of these neurons are unknown. In this study we cloned two putative serotonin receptors (Ms5HT1A and Ms5HT1B) and one putative OA receptor (MsOAR) in order to begin to better understand the physiological and biochemical effects of serotonin and OA in the olfactory system of M. sexta. We found that Ms5HT1A and Ms5HT1B are similar to vertebrate 5HT1-type receptors and are almost identical in sequence to putative serotonin receptors from Heliothis virescens and B. mori, respectively. Using bio-informatic and molecular biological techniques, fragments of the sequence of the B. mori homolog of Ms5HT1A and of the H. virescens homolog of Ms5HT1B were obtained. Phylogenetic analysis clearly resolved the two distinct clades of lepidopteran serotonin receptors demonstrating that lepidopterans possess at least two different types of serotonin receptors. The MsOAR shared high sequence identity with pharmacologically confirmed OA receptors from other insect species, but shared less identity with OA/tyramine receptors cloned from B. mori and H. virescens (von Nickisch-Rosenegk et al., 1996), which share high sequence identity with confirmed tyramine receptors. Through bio-informatic or molecular biological techniques, respectively, we obtained fragments of sequences encoding putative OA receptors from B. mori and H. virescens, suggesting that the MsOAR is likely the first OA receptor cloned from a lepidopteran.
2. Materials and methods
2.1. Animals
M. sexta (Lepidoptera: Sphingidae) were reared at the ARLDN rearing facility at the University of Arizona. Only stage 28 pupae and 1–3-day-old adult moths were used for this study. Adult H. virescens were generously provided by Dr. Neil Vickers at the University of Utah.
2.2. Cloning and sequence analysis of cDNAs
Total RNA was extracted from adult brains using Trizol (Gibco) and reverse transcribed for the generation of cDNA to be used for PCR as described in Nighorn et al. (1999). Degenerate oligonucleotide primers were designed based on the conserved amino acid sequences from H. virescens and B. mori for the serotonin (5HT) receptors and from B. mori, H. virescens, Anopheles gambiae and Apis mellifera for the OA receptor. For the Ms5HT1 receptors, one set of primer sequences, F5HT1 (amino acids; (I/V)(I/V)GNVFV and nucleic acids; RTCRTCGGHAAYGTNTTYGT) and R5HT1 (amino acids; DVLCCT and nucleic acids; GTGCAGCANARNACRTC), produced a band of 213 bp and a second set of primers, F5HT2 (amino acids; NVFVIAA and nucleic acids; AACGTBTTCGTBATHGCNGC) and R5HT2 (amino acids: GWKDPD and nucleic acids; GTCNGGRTCYTTCCANCC), produced a 378 bp band under PCR conditions; 35 cycles in a Hybaid PCR express thermocycler of 94 °C for 20 s, 45 °C+15 °C gradient for 20 s, and 72 °C for 30 s. The primers for the initial MsOAR fragment OAF-1 (amino acids; ASGSFYIP and nucleic acids; CCTCCTCCGGCCCTAYATHCC), and a reverse primer, OAR-1 (amino acids; CWLPFFTMYLV and nucleic acids; CCAGGTACAGGTGAAGAAAGGNARCCARCA), were designed using the CODEHOP (Rose et al., 2003) algorithm. These primers produced a 586 bp band under PCR conditions; 35 cycles 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. All of the cloned fragments were isolated and cloned into the vector pST Blue-1 (Invitrogen). The fragments were then sequenced by the sequencing facility at the University of Arizona.
Stage-18 antennal cDNA libraries were screened for the 5HT receptors and a stage-18 brain library was screened for the OA receptor. All libraries were constructed by generating oligo-dT-primed double-stranded cDNA and then packaging the cDNA into the Lambda ZapII vector (Stratagene, La Jolla, CA). Libraries were initially screened using a well-plate PCR library screening protocol as described by Dieffenbach and Dveksler (2003). Phage from positive wells were plated and the plaques were then transferred to Hybond N+ filters and hybridized overnight at 42 °C with a solution containing 50% formamide, 5X SSC, 1% SDS, 10% Dextran sulfate, 5X Denhart’s solution, 250 μg/ml sheared salmon sperm DNA and 32P-labeled probe at 106 cpm/ml. The filters were washed for 20 min in successive solutions of 2X SSC and 0.1% SDS, 0.5X SSC and 0.1% SDS, 0.1% SSC and 0.1% SDS, and 0.1X SSC and 1% SDS (50 °C). Positive clones were sequenced via primer walking in both directions using automated sequencing by the sequencing facility at the University of Arizona.
For the MsOAR, a large portion (~3900 bp) of the sequence was obtained from the cDNA library although a significant portion of the 5′ end was missing. To obtain the remainder of the OA receptor sequence, we used RNA ligase-mediated rapid amplification of cDNA ends or GeneRacer (Invitrogen). Briefly, we produced mRNA from brain tissue with specific RNA oligos ligated at each end and then used RT-PCR with one primer specific to the fragment of the OA receptor that was initially obtained from the library screen (OAGR5P 5′-AGCGTCATCTTGAACCTGTG-3′) and another primer specific for the oligo sequence ligated to the cDNA (product size: ~ 1500 bp).
To obtain the fragments of the putative OA and 5HT receptor from H. virescens, degenerate primers were designed based on the M. sexta receptors and mRNA was extracted from H. virescens nervous tissue as described above. The primers for the Hv5HT1B (accession number DQ839507) (Hv5HT2S 5′-ATGATHGCNTGYGTNTGG -3′ and Hv5HT2A2 5′-YTCRCARTCRCANGTNGG-3′) produced a 648 bp band under PCR conditions; 35 cycles 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The primers for the HvOAR (accession number DQ839508) (HvOASe 5′-GCNGTNGAYGTNTGGATG -3′ and HvOAAn 5′-CKCCARTARAARAANARCAT-3′) produced a 389 bp band under the same PCR conditions used for the Hv5HT1B.
2.3. Sequence analysis
All DNA sequencing was performed by the University of Arizona DNA sequencing facility (http://uofadna.arl.arizona.edu/). Sequencher (Gene Codes) was used to assemble contigs of the different sequence fragments. Basic sequence analysis was performed using the Geneworks (IntelliGenetics, Inc.) software and the Baylor College of Medicine (BCM) search launcher (http://searchlauncher.bcm.tmc.edu/seq-search/struc-predict.html). Amino acid sequences were aligned and percent identities were determined using the program ClustalW (Combet et al., 2000) (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html).
2.4. Sequence retrieval and alignment
Homologs of the OAR and 5HT1 genes from various invertebrates were retrieved from the NCBI protein database. Accession numbers for all genes used for phylogenetic analysis, with the exception of the B. mori 5HT1A sequence, are listed in the figure legends. For the B. mori 5HT1A homolog, tBLASTn searches were performed on the B. mori genome nucleotide database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=euk). Contigs retrieved with E-values greater than e-15 were assembled using Sequencher (Gene Codes). One contig of 35808 nts contained sequences with accession numbers AADK01036151, BAAB01111880, BAAB01180704, BAAB01161921, AADK01000686, BAA B01165448, BAAB01035832 AND BAAB01183084 and appeared to contain nearly the entire ORF of a B. mori 5HT1A homolog. When used as a query in a BLASTx search of the non-redundant protein database, the contig retrieved named 5HT1 homologs with a top E-value of e-71. The amino acid sequence was obtained by conceptual translation with putative introns detected as regions of nucleotide sequence that disrupt the reading frame of the sequence, contain in-frame stop codons, and fall in-between detected regions of homology in BLAST searches. These were conceptually spliced by deleting regions of sequence, which were bounded by ‘‘GT-AG’’, yielding a contiguous reading frame consistent with regions of homology identified in the BLAST analysis. The resulting protein sequence (454 amino acids), when used as a query in a BLASTp search, retrieved the H. virescens 5HT1 receptor sequence with E-values of 0.0 while retrieving the B. mori 5HT1 receptor in Genbank (B150Bom; CAA64862) with only e-values of e-95, suggesting that it is a different sequence from that already in the database.
Sequences were aligned using Clustal X (Thompson et al., 1997) with default parameters and the resulting alignments manually adjusted. Only regions of unambiguous homology were used for final analysis. This yielded two data sets. The OA/Tyramine data set contained 15 taxa and 265 positions. The arthropod specific 5HT dataset contained 13 taxa and 118 positions. All alignments are available upon request.
2.5. Phylogenetic analysis
Datasets were analyzed by three methods. The appropriate model of sequence evolution to describe each data set was estimated using the program Tree-puzzle 5.2 (Strimmer and von Haeseler, 1996). This allowed for either an 8-category gamma or gamma-plus-1 invariant model and choice of matrix of amino acid transition. For Bayesian analysis, the program Mr. Bayes 3.0 (Ronquist and Huelsenbeck, 2003) was used to obtain the optimal tree topology and posterior probability values for the nodes. Analyses ran for 500,000 generations and the burn-in value was estimated graphically by removing all trees prior to the plateau. Maximum-likelihood bootstrap values were generated as follows. The program PHYML v2.4.4 (Guindon and Gascuel, 2003) was used to analyze 100 pseudoreplicate datasets with parameters of sequence evolution estimated by Tree-puzzle 5.2. Maximum-likelihood corrected distance bootstrap values were also generated using parameters estimated by Tree-puzzle, and using Tree-puzzle and the script Puzzleboot (www.tree-puzzle.de) to calculate distance matrices from 1000 pseudoreplicate data sets. Trees were then generated using the program Neighbor from the PHYLIP package (Felsenstein, 1995) with analyses incorporating the ‘‘jumbling’’ option. While we recognize that posterior probability and bootstrap values cannot be directly compared, they each provide a measure of reliability of an observed relationship given the data and thus we consider nodes supported by 0.95 posterior probability and greater than 80% bootstrap support to be robust.
3. Results and discussion
Three putative amine receptors were cloned from M. sexta brains using degenerate oligonuleotide primers and fragments of 213 bp (clone Ms5HT1A; Genbank accession number DQ840515), 378 bp (clone Ms5HT1B; Genbank accession number DQ840516) and 586 bp (clone MsOAR; Genbank accession number DQ840514) in length were obtained from brain cDNA. The full-length clones were then obtained from stage 18 pupal cDNA libraries and the entire sequence was obtained using GENERACER (Invitrogen) to obtain flanking regions (for the MsOAR only) and primer walking.
3.1. Cloning and sequence analysis of Ms5HT1A and Ms5HT1B
The physiological and behavioral effects of 5HT on the olfactory system of moths have been extensively studied and it has been suggested that 5HT acts as a circadian modulator of olfactory sensitivity (Kloppenburg et al., 1999; Gatellier et al., 2004). In the AL of M. sexta, 5HT increases the excitability of intrinsic neurons by reducing two potassium conductances (Kloppenburg et al., 1999) and so to begin to study the cellular mechanisms by which 5HT has this effect, we cloned the 5HT receptors from M. sexta. We found that while both of the Ms5HT1 receptors contain amino acid residues characteristic of 5HT receptors, they differ significantly in their amino-terminal and 3rd cytoplasmic loop region, which indicates that they are two distinct receptors. Ms5HT1A was very similar to 5HT receptors cloned from H. virescens (CAA64863) and P. xuthus (BAD72868), but less so to the 5HT receptor cloned from B. mori (CAA64862) (Figs 1A, 3A). Conversely, Ms5HT1B shared high sequence identity with the 5HT receptor from B. mori (Fig. 1B), but much less with 5HT receptor sequences from H. virescens and P. xuthus. Overall this suggested differential paralogue sampling in the lepidopterans examined thus far and the possible presence of additional 5HT1 genes in B. mori and H. virescens as we observed for M. sexta. From the B. mori genome database, we identified several sequence fragments homologous to Ms5HT1A that were assembled in silico and we obtained the sequence for a 454 amino acid fragment of a protein that shared high sequence identity with Ms5HT1A (Figs. 1A, 3A), indicating that B. mori has a second 5HT1-type receptor. We also obtained, via RT-PCR, a 648 bp cDNA fragment putatively encoding a protein from H. virescens (Hv5HT1B; Fig. 1B) that shared high sequence identity with Ms5HT1B and the putative 5HT receptor from B. mori (Figs. 1B, 3A). Phylogenetic analysis strongly resolved two separate clades of lepidopteran 5HT1 sequences with 100% support for both nodes by all methods (Fig 3A) suggesting that, at least within the Lepidoptera, these two classes of 5HT receptors are expressed. These two 5HT receptor classes may have different patterns of expression within the nervous system of M. sexta or distinct physiological and biochemical effects, and future studies will focus on characterizing the expression patterns and physiological effects of both receptors within the context of olfactory information processing in M. sexta.
Fig. 1.

(A) The amino acid sequence of Ms5HT1A. The seven trans-membrane domains are marked by a line overtop of the amino acid sequence and numbered accordingly. Comparisons of Ms5HT1A against 5HT receptors sequences from H. virescens (K15Hel: CAA64863), P. xuthus (Px5HT: BAD72868) and B. mori (Bm5HT1A). (B) The amino acid sequence of Ms5HT1B. The seven trans-membrane domains are marked by a line overtop of the amino acid sequence and numbered accordingly. Comparison of Ms5HT1B against the 5HT receptor sequences from B. mori (B150Bom: CAA64862) and H. virescens (Hv5HT1B). Amino acid residues that were identical across species are highlighted in gray.
Fig. 3.
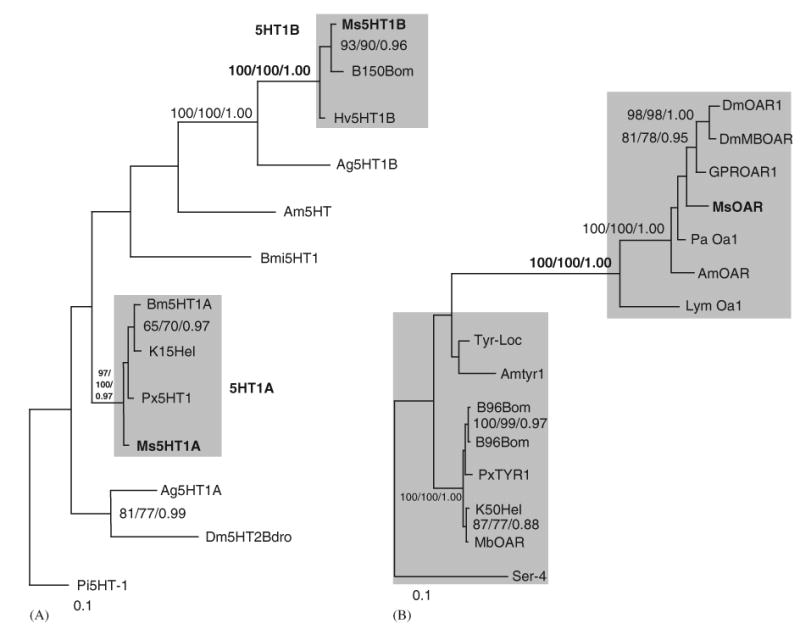
(A) Phylogenetic tree comparing the sequences for Ms5HT1A and Ms5HT1B with receptor sequences for invertebrate 5HT1 receptors. The best Bayesian topology is shown. The shaded boxes denote the two 5HT1 lepidopteran clades. The Ms5HT1A and B sequences are bolded as are relevant values for nodes supporting the two clades. This tree is displayed rooted by P. interuptus. Protein sequences used; Ag5HT1A (EAA04158), Ag5HT1B (EAA13071), genbank:Am5HT (XP_393915), Bmi5HT (AY323235.1), genbank:B150Bom (CAA64862), Dm5HTdro2B (Z11490.1), K15Hel (CAA64863), Pi5-HT1 (AY528822.1), Px5HT1 (BAD72868). Scale bar indicates 10 changes per 100 alignment positions. (B) Phylogenetic tree comparing the amino acid sequence of MsOAR with OA and tyramine receptors from other insects. Shaded areas indicate a cluster of tyramine receptor-like proteins (lower) and a cluster of OA receptor-like proteins (upper). The best Bayesian tree topology is shown. Support values for nodes are shown in the order of maximum likelihood (ML) bootstrap values, ML-corrected neighbor joining bootstrap values and Bayesian posterior probabilities. Only nodes supported by better than 0.85 posterior probability or 65% bootstrap support in 2/3 methods are shown. The M. sexta OAR sequence is bolded, as are the support values for the node separating the OA and the tyramine receptor clades. This tree should be treated as unrooted and is rooted arbitrarily on the C. elegans Tyr sequence for display purposes only. Protein sequences used; GPROAR1 (EAA06361), GPRTYR (EAA07468), AmOAR (NP_001011565), Amtyr1 (CAB76374), B96Bom (CAA64865), Ser-4 (NP_497452), DmOAR1 (CAB38026), DmMBOAR (AAC17442), K50Hel (CAA64864), Lym OA1 (AAC61296), Tyr-Loc (Q25321), MbOAR/Tyr (AAK14402), Pa OA1 (AAP93817), PxTYR1 (BAD72869).
3.2. Cloning and sequence analysis of the MsOAR
The deduced amino acid sequence for MsOAR was highly similar to functionally confirmed OA receptors from Periplaneta americana (AAP93817) and A. mellifera (XP_393915) and had an open reading frame of 502 amino acids (Fig. 2). The most highly conserved areas between these sequences fell within the predicted trans-membrane domains for these proteins (Fig. 2). Phylogenetic analysis revealed a clade of functionally confirmed OA receptors that includes MsOAR, and a separate clade that groups the tyramine receptors from various invertebrates (Fig. 3B). This strongly suggests that we have cloned a legitimate OA receptor sequence. In Fig. 3B, the node separating the tyramine and OA receptor sequences is 100% supported by all three analytical methods used, and clearly places the lepidopteran OA/tyramine receptors including (H. virescens, (CAA64864), B. mori (CAA64865) and Mamestra brassicae (AAK14402) in the tyramine receptor clade (Fig. 3B). Several recent studies and reviews have suggested that the two putative lepidopteran OA/tyramine receptors (von Nickisch-Rosenegk et al., 1996) are most likely tyramine receptors (Roeder, 2005; Ohta et al., 2003). Indeed, the B96Bom protein was recently isolated and functionally expressed demonstrating its ligand specificity to tyramine (Ohta et al., 2003).
Fig. 2.
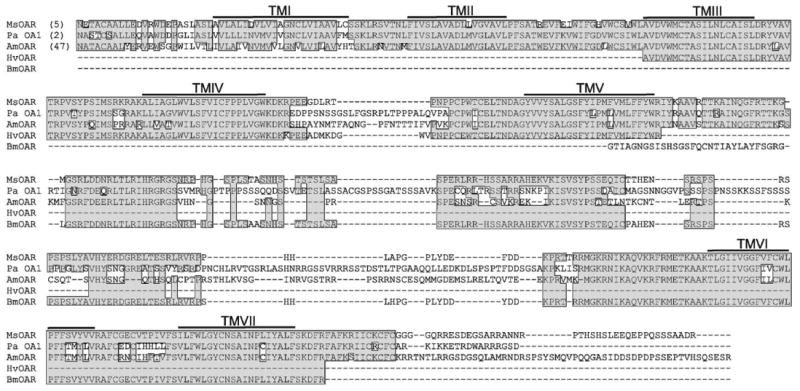
(A) The amino acid sequences and genomic structure of MsOAR. The seven trans-membrane domains are marked by a line overtop of the amino acid sequence and numbered accordingly. Comparisons of MsOAR are against OA receptor sequences from P. americana (Pa OA1: AAP93817), A. mellifera (AmOAR: NP001011565), B. mori (BmOAR) and H. virescens (HvOAR). Numbers in parenthesis indicate the amino acid position at the beginning of the alignment. Amino acid residues that were identical across species are highlighted in gray.
To establish that B. mori and H. virescens have OA receptors distinct from the OA/tyramine receptors previously cloned by von Nickisch-Rosenegk et al. (1996), we used tBLASTn searches with MsOAR to identify a genomic contig fragment from the B. mori genome (AADK01027192) which encodes a protein fragment of 244 amino acids (Fig. 2). The sequence for the new putative B. mori OA receptor (BmOAR) shared high identity with MsOAR, and the identified OA receptors from P. americana and A. mellifera and relatively little identity with the OA/tyramine receptors from B. mori and H. virescens. We also obtained a 386 bp cDNA fragment of a putative H. virescens OA receptor (HvOAR) via RT-PCR which again shared high identity with MsOAR, but relatively little with the OA/tyramine receptors from B. mori and H. virescens.
There has been mounting evidence that while tyramine is the precursor for the biosynthesis of OA, tyramine has physiological and behavioral effects distinct from those of OA (Roeder et al., 2003; Roeder 2005). While all octopaminergic neurons are by definition also tyraminergic, there have been tyraminergic neurons described in D. melanogaster, L. migratoria and C. elegans that do not contain OA (Nagaya et al., 2002; Donini and Lange, 2004; Alkema et al., 2005, respectively). Furthermore, several studies have demonstrated that tyramine has physiological effects that are independent, and often antagonistic to, OA (see Roeder et al., 2003).
3.3. The detection of amine receptor mRNA in various nervous tissues by RT-PCR
As a broader measure of the expression of these receptors, RT-PCR was performed using cDNA extracted from antennae, abdominal ganglia and thoracic ganglia. The mRNA for Ms5HT1AR, Ms5HT1BR and MsOAR were expressed in the antennae and in both the abdominal and thoracic ganglia (Fig. 4). PCR was performed on mRNA lacking reverse transcriptase to control for genomic DNA contamination. Both 5HT and OA have been reported throughout the nervous system of M. sexta (Maxwell et al., 1978) and thus it was not surprising that we were able to detect mRNA for all three receptors in the antennae, thoracic and abdominal ganglia.
Fig. 4.
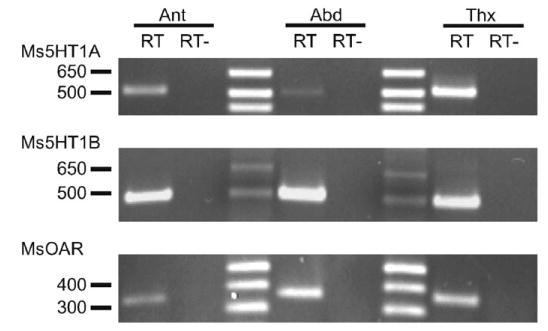
Expression of Ms5HT1A, Ms5HT1B MsOAR mRNA in the antennae (Ant), abdominal ganglia (Abd) and thoracic ganglia (Thx) detected by RT-PCR. The predicted size of the PCR products were 514 bp for Ms5HT1A, 495 bp for Ms5HT1B and 361 bp for MsOAR. The RT+ (reverse transcriptase included in the cDNA preparation) PCR products are shown in the left column for each tissue type examined and the RT-(no reverse transcriptase included in the cDNA preparation) PCR products are in the right column. The position of the molecular weight columns are indicated on the left-hand side of each gel shown.
Biogenic amines have been shown in many invertebrate species to modulate the functionality of neural circuits involved in a variety of behaviors and physiological processes including olfaction. However, little is known about the biochemical effects of 5HT and OA on AL circuits. The cloning of 5HT and OA receptors is an essential first step towards furthering our understanding of how the cellular effects of these two biogenic amines modulate the physiology of the AL and how this in turn affects the olfactory-guided behavior of M. sexta.
Acknowledgments
The authors would like to thank C. Collmann, S. Fernando, Dr. N. Gibson, L. Hua, and M. Higgins for invaluable technical assistance and S. Mackzum for rearing the M. sexta. We would like to thank the B. mori, and A. melifera genome projects for making their data publicly available. The authors would also like to thank Dr. N. Vickers for graciously supplying the H. virescens adults and Dr. J.G. Hildebrand for valuable discussions and support. Funding was provided by the National Institute on Deafness and Other Communication Disorders Grants DC-05652 (to TAC) and DC-02751 (to AJN), the National Science and Engineering Research Council of Canada PGS B-244345-2003 (to AMD) and a joint CIHR/ Welcome Trust Traveling Research Fellowship (to JBD).
References
- Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25 (3):147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Dacks AM, Christensen TA, Agricola HJ, Wollweber L, Hildebrand J. Octopamine-immunoreactive neurons in the brain and subesophageal ganglion of the hawkmoth Manduca sexta. J Comp Neurol. 2005;488 (3):255–268. doi: 10.1002/cne.20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieffenbach CW, Dveksler GS. PCR Primer: A Laboratory Manual. second ed. Cold Spring Harbor Laboratory Press; Coldspring Harbor, NY: 2003. [Google Scholar]
- Donini A, Lange A. Evidence for a possible neurotransmitter/ neuromodulator role of tyramine on the locust oviducts. J Insect Physiol. 2004;50:351–361. doi: 10.1016/j.jinsphys.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Department of Genetics, University of Washington; Seattle: 1995. [Google Scholar]
- Gatellier L, Nagao T, Kanzaki M. Serotonin modifies the sensitivity of the male silkmoth to pheromone. J Exp Biol. 2004;207:2487–2496. doi: 10.1242/jeb.01035. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 2003;52 (5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Kloppenburg P, Ferns D, Mercer A. Serotonin enhances central olfactory neuron responses to female sex pheromone in the male sphinx moth Manduca sexta. J Neurosci. 1999;19 (19):8172–8181. doi: 10.1523/JNEUROSCI.19-19-08172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell GD, Tait JF, Hildebrand JG. Regional synthesis of neurotransmitter candidates in the CNS of the moth Manduca sexta. Comp Biochem Physiol. 1978;61C:109–119. doi: 10.1016/0306-4492(78)90120-x. [DOI] [PubMed] [Google Scholar]
- Mercer A, Hayashi J, Hildebrand JG. Modulatory effects of 5-hydroxytryptamine on voltage-activated currents in cultured antennal lobe neurons of the sphinx moth Manduca sexta. J Exp Biol. 1995;198:613–627. doi: 10.1242/jeb.198.3.613. [DOI] [PubMed] [Google Scholar]
- Nagaya Y, Kutsukake M, Chigusa SI, Komatsu A. A trace amine, tyramine, functions as a neuromodulator in Drosophila melanogaster. Neurosci Lett. 2002;329 (3):324–328. doi: 10.1016/s0304-3940(02)00596-7. [DOI] [PubMed] [Google Scholar]
- Nighorn A, Byrnes K, Morton D. Identification and characterization of a novel beta subunit of soluble guanylyl cyclase that is active in the absence of a second subunit and is relatively insensitive to nitric oxide. J Biol Chem. 1999;274 (4):2525–2531. doi: 10.1074/jbc.274.4.2525. [DOI] [PubMed] [Google Scholar]
- Ohta H, Utsumi T, Ozoe Y. B96Bom encodes a Bombyx mori tyramine receptor negatively coupled to adenylate cyclase. Insect Biochem Mol Biol. 2003;12 (3):217–223. doi: 10.1046/j.1365-2583.2003.00404.x. [DOI] [PubMed] [Google Scholar]
- Roeder T. Tyramine and octopamine: ruling behavior and metabolism. Annu Rev Entomol. 2005;50:447–477. doi: 10.1146/annurev.ento.50.071803.130404. [DOI] [PubMed] [Google Scholar]
- Roeder T, Seifert M, Kahler C, Gewecke M. Tyramine and octopamine: antagonistic modulators of behavior and metabolism. Arch Insect Biochem Physiol. 2003;54 (1):1–13. doi: 10.1002/arch.10102. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucl Acids Res. 2003;31 (13):3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Nickisch-Rosenegk E, Krieger J, Kubick S, Laage R, Strobel J, Strotmann J, Breer K. Cloning of biogenic amine receptors from moths (Bombyx mori and Heliothis virescens) Insect Biochem Mol Biol. 1996;26 (8–9):817–827. doi: 10.1016/s0965-1748(96)00031-8. [DOI] [PubMed] [Google Scholar]


