Abstract
Continuous treatment of malignancies with tyrosine kinase inhibitors (TKIs) may select for resistant clones (ie, imatinib mesylate). To study resistance to TKIs targeting FLT3, a receptor tyrosine kinase that is frequently mutated in acute myelogenous leukemia (AML), we developed resistant human cell lines through prolonged coculture with FLT3 TKIs. FLT3 TKI-resistant cell lines and primary samples still exhibit inhibition of FLT3 phosphorylation on FLT3 TKI treatment. However, FLT3 TKI-resistant cell lines and primary samples often show continued activation of downstream PI3K/Akt and/or Ras/MEK/MAPK signaling pathways as well as continued expression of genes involved in FLT3-mediated cellular transformation. Inhibition of these signaling pathways restores partial sensitivity to FLT3 TKIs. Mutational screening of FLT3 TKI-resistant cell lines revealed activating N-Ras mutations in 2 cell lines that were not present in the parental FLT3 TKI-sensitive cell line. Taken together, these data indicate that FLT3 TKI-resistant cells most frequently become FLT3 independent because of activation of parallel signaling pathways that provide compensatory survival/proliferation signals when FLT3 is inhibited. Anti-FLT3 mAb treatment was still cytotoxic to FLT3 TKI-resistant clones. An approach combining FLT3 TKIs with anti-FLT3 antibodies and/or inhibitors of important pathways downstream of FLT3 may reduce the chances of developing resistance.
Introduction
Constitutive activation of the class III receptor tyrosine kinase, FLT3, plays important roles in leukemogenesis.1–4 Internal tandem duplications in the juxtamembrane region (FLT3-ITD) or point mutations in the kinase domain (FLT3-PM) lead to constitutively activated FLT3.5–7 FLT3 is also activated by coexpression of FLT3 ligand (FL) through intracrine, paracrine, and/or autocrine pathways.8–10 The presence of FLT3-ITD mutations is associated with a poor prognosis in acute myelogenous leukemia (AML).11–14 Activated FLT3 mediates signaling through at least 3 major downstream signaling pathways: signal transducers and activators of transcription (STAT5), PI3K/Akt, and Ras/mitogen-activated protein (MAP) kinase.15–25 These signaling pathways have overlapping roles in cell differentiation, proliferation, and survival. FLT3 is expressed in most acute leukemias, including 94% of B-lineage acute lymphoblastic leukemia (ALL), 34% of T-lineage ALL, and 89% of AML cases.26–28 These observations strongly suggest FLT3 as a candidate for molecularly targeted therapy.
In fact, a number of FLT3 tyrosine kinase inhibitors (TKIs) have been developed. Some of the best studied to date include CEP-701 (lestaurtinib), PKC412, MLN518, SU11248 (sunitinib malate), and AG1295.23,29–33 Although these inhibitors vary in their potency and selectivity for FLT3, all are able to induce cytotoxicity in FLT3-expressing cells in vitro and/or in vivo. Furthermore, clinical trials with some of these inhibitors have demonstrated their ability to decrease peripheral blood and bone marrow blast counts in some patients.34–36 CEP-701 is currently being tested on relapsed patients with FLT3 mutant AML in a randomized phase 2 clinical trial in combination with chemotherapy.
Although FLT3 inhibitors demonstrate preclinical and clinical activity, they possess a number of limitations. Clinical trials have revealed that FLT3 TKIs used as single agents are able to significantly reduce peripheral blood and bone marrow blasts only in a minority of patients, and the effect is transitory.34–36 This may be due to achieving insufficient levels of FLT3 inhibition in these patients, a lack of dependence of these cells on FLT3 signaling for proliferation and survival, and/or selection of resistant cell populations. Furthermore, most cases of AML and ALL do not express mutant FLT3, and it is unclear to what degree these cells depend on FLT3 signaling for sustaining the leukemic phenotype. At drug concentrations necessary to inhibit FLT3 phosphorylation past a critical threshold required to induce cytotoxicity, a varying spectrum of other kinases are frequently also inhibited, which can lead to toxicities.
Even when cells are dependent on FLT3 signaling for survival and proliferation, prolonged exposure to TKIs are likely to select for resistant clones, as has been seen with imatinib mesylate (Gleevec), a TKI targeting BCR-ABL in chronic myelogenous leukemia.37 Cells have the potential to develop several mechanisms of resistance: (1) Cells may acquire FLT3 mutations that prevent drug binding.38,39 (2) Expression of cell-surface transport proteins may reduce the intracellular drug concentration and thereby interfere with FLT3 inhibition. (3) FLT3 overexpression may reduce the efficacy of FLT3 TKIs to inhibit target. (4) Cells can become FLT3 independent by activating compensatory signaling pathways.
Anti-FLT3 immunotherapy is an alternative to small molecule inhibitors. Antibodies are very specific and therefore are usually less toxic than TKIs and have the potential added advantage of recruiting the host's immune system in clearing leukemic cells. We recently demonstrated that IMC-EB10, an unconjugated monoclonal antibody (mAb) that binds FLT3, blocks (FL) binding and in turn affects downstream STAT5, Akt, and MAPK signaling pathways.40–42 Although IMC-EB10 has no in vitro cytotoxicity, IMC-EB10 is able to prolong survival and reduce the level of engraftment of AML and ALL-derived cell lines and of primary samples in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice without affecting the engraftment of normal cord blood CD34+ human stem cells.41,42 Unlike TKIs, the in vivo efficacy of IMC-EB10 depends on antibody-dependent cell-mediated cytotoxicity (ADCC) and is therefore not dependent on inhibition of FLT3 signaling.41,42
To investigate some of the mechanisms used by human leukemic cells to overcome FLT3 TKI-mediated cytotoxicity, we selected for resistant cells by exposing 3 AML- or ALL-derived FLT3-dependent human cell lines to increasing concentrations of FLT3 inhibitors. We then examined signal transduction in the resistant cells. We also investigated whether the TKI-resistant cell lines would also be resistant to anti-FLT3 mAb treatment.
Methods and materials
Reagents
Mouse monoclonal antiphosphotyrosine antibody (4G10) and recombinant protein A-agarose were purchased from Upstate Biotechnology (Lake Placid, NY). Rabbit anti–P-FLT3, anti–P-STAT5, anti–-P-Akt, anti-FLT3, anti-Akt, anti–P-STAT1, anti-STAT1 anti–P-MAPK p44/42, and anti-MAPK p44/42 antibodies were obtained from Cell Signaling Technologies (Beverly, MA). Rabbit anti-FLT3 and anti-STAT5 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Humanized anti-EGFR antibody, IMC-C225 (cetuximab; ImClone Systems, New York, NY) and IMC-EB10 were generously provided by ImClone Systems. Horseradish peroxidase–conjugated secondary antibody and enhanced chemiluminescence (ECL) detection system were purchased from Amersham (Arlington Heights, IL).
FLT3 inhibitors, CEP-5214 and CEP-701, were generously provided by Cephalon (Frazer, PA). PKC412 was generously provided by Novartis (East Hanover, NJ). AG1295 was purchased from Sigma-Aldrich (St Louis, MO). The following human ligands were obtained from PeproTech (Rocky Hill, NJ): IL-2, IL-3, IL-6, IL-11, GM-CSF, SCF, and EGF. Insulin, IGF-1, and GDNF human ligands were obtained from Santa Cruz Biotechnology. MEK inhibitor (U0126) was obtained from Promega (Madison, WI) and PI3K inhibitor (LY294002) from Cell Signaling Technologies.
Cell lines
Human cell lines were cultured in RPMI-1640 (GIBCO, Rockville, MD), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gimini Bio-Products, Woodland, CA) and penicillin/streptomycin (GIBCO) at 37°C with 5% CO2. The Molm14 cell line was derived from an AML primary sample and expresses FLT3-ITD and MLL-AF9 mutant proteins. Both Hb1119 and SEM-K2 cells are derived from ALL primary samples. Hb1119 cells express FLT3-D835H and MLL-ENL mutant protein, whereas SEM-K2 cells express amplified wild-type FLT3 along with the MLL-AF4 fusion protein.
Development of FLT3 TKI-resistant cell lines
Log phase growing MOLM14, Hb1119, and SEM-K2 cell lines were cocultured with increasing concentrations of CEP-5214 or CEP-701 for 2 to 4 months. Cell cultures were deemed resistant when cells proliferated in the presence of 50 to 120 nM CEP-5214 or CEP-701, depending on the sensitivity of the parent cell line to the FLT3 TKI. The selection for resistant cells was repeated a total of 4 separate times for each cell line to ensure reproducibility. FLT3 TKI-resistant cell lines were grown without CEP-5214 or CEP-701 for at least 2 days prior to experimentation.
Human primary samples
Human samples from patients with AML and patients with ALL were obtained under an institutional review board–approved protocol at the Sidney Kimmel Cancer Center at Johns Hopkins Hospital. Informed consent was provided according to the Declaration of Helsinki. Leukemic blasts were separated by Ficoll-Hypaque (Amersham, Piscataway, NJ) density gradient centrifugation as previously described.43
Western blot analysis
Cell viability assays
MTT (Roche, Indianapolis, IN) and annexin V (BD-Bioscience, Palo Alto, CA) binding assays were performed according to the manufacturer's recommendations. Cell lines were plated at 300 000 cells/mL and primary samples at 5 × 106 cells/mL for the MTT and annexin V binding assay. Cells were treated as indicated for 48 hours. MTT results are presented as the percentage of OD570 of the control sample not treated with a FLT3 TKI.
PCR: identification of FLT3 and MLL (mixed-lineage leukemia) mutations
Total RNA was isolated using the RNeasy Mini kit (Qiagen Sciences, Valencia, CA) and converted into cDNA using Superscript III (Invitrogen, Carlsbad, CA) and the manufacturer's protocol. Polymerase chain reaction (PCR) used primers spanning the FLT3 mutation or MLL translocation sites and 40 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 60 seconds. The primer sequences will be provided on request. Half of the PCR product for FLT3-D835H analysis was digested with EcoRV (New England BioLabs, Beverly, MA). PCR products were run on a 2% agarose gel.
Heteroduplex analysis: kinase and phosphatase mutational screen
Total RNA and genomic DNA were separated by Trizol (Invitrogen) extraction, and RNA was further isolated with the RNeasy Kit (Qiagen Sciences). cDNA was prepared from RNA using Superscript III. PCR primers were synthesized to amplify the catalytic and juxtamembrane domain (where applicable) of each of the genes to be screened (see Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). Each primer pair was previously optimized to ensure a PCR product. Using a denaturation step of 95°C followed by a slowly decreasing temperature gradient (80°C-25°C), PCR products were annealed and formed duplexes. Capillary electrophoresis was performed on the SpectruMedix Reveal machine (SpectruMedix, State College, PA) using temperature gradients of 45°C to 55°C and 50°C to 60°C. The CCD camera of the SpectruMedix Reveal machine captures images of the product in the capillary, and the software (SpectruMedix Revelation) converts these images into peaks, based on the intensity of the ethidium bromide signal. The peaks were analyzed to determine which samples contained heteroduplexes that might indicate changes in the DNA sequence between the 2 strands. These samples were further analyzed by sequencing to determine the nature of the change. If mutations were found, they were confirmed using the original RNA and genomic DNA.
Quantitative real-time PCR (Q-PCR)
Q-PCR was performed as previously described.42 GAPDH was used to normalize gene expression.
DNA construct and lentiviral transduction
HA-tagged full-length cDNA coding for wt N-Ras or N-Ras-G12V was obtained from the UMR cDNA Resource Center (www.cdna.org) and cloned into a lentiviral vector with an EF1 promoter and a CMV promoter driving GFP expression. This lentiviral vector was kindly provided by Linzhao Cheng. Lentiviral constructs along with VSVG and PMV helper plasmids were transfected into 293T cells using Lipofectamine 2000 (Invitrogen). The supernatant was collected for 48 hours, starting 16 hours after tranfection of 293T cells. Lentivirus containing supernatant was concentrated using Centricon Plus-70 filters (Milipore, Billerica, MA). Cells were transduced with a given lentivirus in media containing 8 μg/mL Polybrene (Sigma-Aldrich) for 48 hours. At 96 hours after transduction, cells with equal levels of GFP expression were sorted on a BD FACSvantage SF cell sorter (BD-Bioscience).
Flow cytometry
In vivo models of human leukemia: engraftment and survival
NOD/SCID mice were bred at the Johns Hopkins University School of Medicine. Mice were sublethally irradiated (280 cGy) and injected with 0.5 × 106 cells in 200 μL PBS via tail-vein injection. Mice were given intraperitoneal injections of 400 μg IMC-C225 or IMC-EB10 at 24, 72, and 120 hours after cell injection. All animal procedures were conducted in conformity with institutional guidelines.
Results
Culture of leukemia-derived cell lines with CEP-5214 or CEP-701 leads to FLT3 TKI cross-resistance
To investigate mechanisms of drug resistance, the human leukemia-derived cell lines, SEM-K2, Molm14, and Hb1119, expressing amplified wt FLT3, FLT3-ITD, and FLT3-D835H, respectively, were cocultured with increasing concentrations of CEP-5214 or CEP-701. After 2 to 4 months of coculture with FLT3 TKIs, these cell lines were able to proliferate in the presence of 50 to 120 nM CEP-701 or CEP-5214, although their growth rates were somewhat slower in the presence of FLT3 TKIs than in their absence (data not shown). Resulting resistant cell lines are named with an “(R)” followed by the FLT3 inhibitor used during coculture (eg SEM(R)5214). PCR analysis of FLT3 mutations, MLL translocations, and cytogenetic analysis confirmed that the FLT3 TKI-resistant cell lines were derived from the parental line and still expressed those defining characteristics after prolonged drug exposure (data not shown).
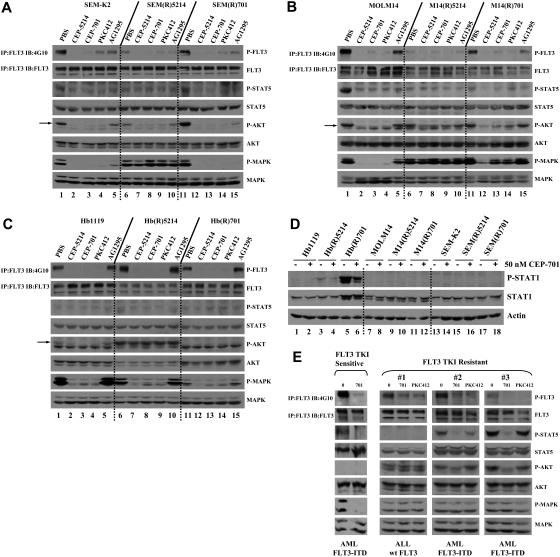
Both the MTT and annexin V binding assays were used to determine the cytotoxic effects of CEP-5214, CEP-701, PKC412, and AG1295 on the sensitive parental versus resistant cell lines. MTT assays revealed that prolonged coculture of SEM-K2, Molm14, and Hb1119 cell lines with CEP-5214 or CEP-701 led to cross-resistance to CEP-5214, CEP-701, PKC412, and, a structurally dissimilar FLT3 inhibitor, AG1295 (Table 1)The annexin V binding assays demonstrated that the M14(R)5214 and M14(R)701 cell lines are resistant to FLT3 TKI-induced apoptosis compared with the Molm14 parental cell line (Figure 1A-D). Similar annexin V binding results were also observed with the FLT3 TKI-sensitive and -resistant Hb1119 and SEM-K2 cell lines (data not shown). The lack of cytotoxicity observed with AG1295 treatment of the parental Hb1119 cells was expected because of the presence of the D835H-activating mutation in FLT3, which is resistant to AG1295 inhibition.
Table 1.
IC50 Values for FLT3 TKI sensitive and resistant cell lines
| CEP-5214, nM | CEP-701, nM | PKC412, nM | AG1295, μM | |
|---|---|---|---|---|
| SEM-K2 | 29 | 23 | 96 | 10 |
| SEM(R)5214 | > 250 | 238 | > 250 | > 20 |
| SEM(R)701 | > 250 | 65 | > 250 | > 20 |
| MOLM14 | 3 | 12 | 27 | 1.6 |
| M14(R)5214 | > 250 | 170 | 165 | > 20 |
| M14(R)701 | > 250 | 51 | 158 | > 20 |
| Hb1119 | 104 | 14 | 125 | > 20 |
| Hb(R)5214 | > 250 | 85 | 226 | > 20 |
| Hb(R)701 | 147 | 88 | 199 | > 20 |
Prolonged exposure to CEP-5214 or CEP-701 selects for cell lines that are cross-resistant to CEP-5214, CEP-701, PKC412 and AG1295. Cellular activity/proliferation, as indicated by the IC50, was determined using the MTT assay in triplicates. Cells (300 000 cells/mL) were incubated with 0 to 250 nM CEP-5214, CEP-701, or PKC412, or 0 to 20 μM AG1295 for 48 hours at 37°C. IC50 values were calculated using CalcuSyn software (Cambridge, United Kingdom) and had correlation coefficients > 0.95. The results are representative of 3 independent studies.
Figure 1.
M14(R)5214 and M14(R)701 are resistant to CEP-5214-, CEP-701-, PKC412-, and AG1295-induced apoptosis compared with the parent Molm14 cell line. (A-D) Cells (300 000 cells/mL) were incubated with 0 to 50 nM CEP-5214 or CEP-701, 0 to 100 nM PKC412, or 0 to 20 μM AG1295 for 48 hours at 37°C. Induction of apoptosis was assessed by measuring annexin V binding by flow cytometry. Values indicate percentage of annexin V positive above the untreated samples, which were 5% to 15% annexin V positive. The data are representative of 2 independent experiments.
FLT3 TKI-resistant cell lines maintain activation of downstream signaling pathways despite inhibition of FLT3 phosphorylation by FLT3 TKIs
Western blot analysis was performed to compare the ability of CEP-5214, CEP-701, PKC412, and AG1295 to inhibit phosphorylation of FLT3 and downstream signaling proteins in FLT3 TKI-resistant and parental cell lines. Western blots revealed that CEP-5214, CEP-701, PKC412, and/or AG1295 inhibit FLT3 phosphorylation in both FLT3 TKI-sensitive and -resistant cell lines (Figure 2A-C, lanes 1-15). Treatment of the parental SEM-K2, Molm14, and Hb1119 cell lines with FLT3 TKIs led to inhibition of downstream STAT5, Akt, and MAPK phosphorylation that correlated with inhibition of FLT3 phosphorylation (Figure 2A-C, lanes 1-5). However, Akt and/or MAPK signaling proteins remained phosphorylated in most FLT3 TKI-resistant cell lines on FLT3 TKI treatment, even when FLT3 was inhibited (Figure 2A, lanes 6-10; Figure 2B, lanes 6-15; Figure 2C, lanes 6-10). Furthermore, STAT1 was overexpressed and phosphorylated in the Hb(R)701 cell line but not in the parental Hb1119 cell line (Figure 2D, lane 5 versus 1). Wild-type or mutant forms of FLT3 do not appear to signal through the STAT1 pathway (Figure 2D, lanes 1, 7, and 13). STAT1 phosphorylation was not completely inhibited when the Hb(R)701 cell line was treated with 50 nM CEP-701, a dose that completely inhibits FLT3 phosphorylation (Figure 2D, lanes 6 versus 5; Figure 2C, lanes 1 and 3). A lack of STAT5 phosphorylation was observed in the M14(R)5214 and M14(R)701 cells lines, possibly because of selection of a resistant clone with deficiencies in protein(s) required for phosphorylation of STAT5. These data indicate that prolonged exposure to FLT3 TKIs selects for cells with activated Akt and/or MAPK pathways no longer dependent on FLT3 signaling, as well as for new pathways not typically regulated by FLT3. Continued activation of downstream signaling proteins that play roles in cell survival and proliferation, despite inhibition of FLT3 phosphorylation, may then explain the resistance to FLT3 TKIs observed in cell lines cocultured with CEP-5214 or CEP-701.
Figure 2.
FLT3 phosphorylation in FLT3 TKI-sensitive and -resistant cell lines and primary samples decreases with FLT3 TKI treatment, although STAT5 and/or Akt and/or MAPK pathways remain activated in most resistant cells. Cell lines expressing (A) wt FLT3 [SEM-K2], (B) FLT3-ITD [MOLM14], or (C) FLT3(D835H) [Hb1119] were treated with PBS, 50 nM CEP-5214, 50 nM CEP-701, 50 nM PKC412, or 1 μM AG1295 for 1 hour at 37°C. Arrow points to P-Akt. (D) Indicated cell lines were treated with PBS or 50 nM CEP-701 for 1 hour at 37°C. (E) Primary samples (20 × 106 cells) were treated with PBS, 50 nM CEP-701, or 50 nM PKC412 for 1 hour at 37°C. Immunoprecipitates and total protein extracts were resolved by 8% SDS–polyacrylamide gel electrophoresis (PAGE) or 10% SDS-PAGE, respectively, and subjected to immunoblot analysis with the indicated phospho-specific antibodies. The same blots were then stripped and reprobed with protein-specific antibodies.
Inhibition of FLT3 phosphorylation does not always translate into inhibition of downstream signaling pathways in FLT3 TKI-resistant primary samples
Several samples from patients with AML and patients with ALL were also examined to determine whether a similar phenomenon might explain resistance to FLT3 TKIs that we have observed previously in approximately 20% of FLT3-ITD AML samples.29,34 Treatment of a FLT3 TKI-sensitive primary sample (IC50 value < 50 nM CEP-701) with CEP-701 led to inhibition of FLT3 phosphorylation that correlated with inhibition of downstream signaling pathways (Figure 2E; data not shown). However, primary samples resistant to CEP-701–induced cytotoxicity (IC50 value > 50 nM CEP-701) exhibited continued phosphorylation of STAT5 and/or Akt and/or MAPK signaling proteins even with inhibition of FLT3 phosphorylation (Figure 2E; data not shown). A lack of P-STAT5 and P-Akt was observed in 2 tested primary samples, possibly because of deficiencies in upstream signaling proteins responsible for their activation.
Data from the resistant cell lines and primary samples thus suggest that alternative pathways not inhibited by FLT3 TKIs may sustain the activation of downstream signaling pathways that provide compensatory survival/proliferation signals during inhibition of FLT3 phosphorylation. Thus, prolonged exposure to FLT3 TKIs selects for FLT3-independent clones. Because we never observed the spontaneous selection for mutations within wt FLT3, FLT3-ITD, or FLT3-D835H that would render them resistant to inhibition of kinase activity, this may be a much more frequent mechanism to explain why patients' responses to FLT3 TKIs are often short lived.
Genes regulated by constitutively activated FLT3 and required for FLT3-mediated cellular transformation are expressed in FLT3 TKI-resistant cell lines despite FLT3 inhibition
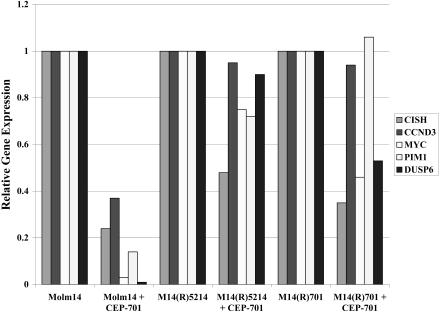
Recently, we identified a number of genes regulated by constitutively activated wt FLT3 and FLT3-ITD that play a role in FLT3-mediated cell survival. (K.-T.K., Kristin Baird, Sean Davis, O.P., M.L., Li Li, Peili Chen, Paul Meltzer, and D.S., manuscript submitted). We investigated the expression of 5 of these genes by Q-PCR in FLT3 TKI-sensitive and -resistant cell lines treated with 50 nM CEP-701. The M14(R)5214, M14(R)701, SEM(R)5214, and SEM(R)701 cell lines maintained higher expression levels of the tested genes involved in cell survival/proliferation compared with the FLT3 TKI-sensitive Molm14 or SEM-K2 cell lines (Figure 3; data not shown). These data further demonstrate that FLT3 TKI-resistant cell lines activate parallel signaling pathways unaffected by FLT3 TKIs that lead to the altered gene expression profile, normally dependent on FLT3 activation.
Figure 3.
FLT3 TKI-resistant M14(R)5214 and M14(R)701 cell lines maintain higher expression levels of 5 genes regulated by FLT3 signaling and required for FLT3-mediated cellular transformation despite FLT3 inhibition. Molm14, M14(R)5214, and M14(R)701 cell lines were treated with 50 nM CEP-701 for 4 hours prior to isolation of total RNA. The RNA was converted into cDNA and used for Q-PCR analysis. Target gene expression was normalized to GAPDH.
Mutations within analyzed kinases or phosphatases do not completely account for the observed resistance to FLT3 TKIs
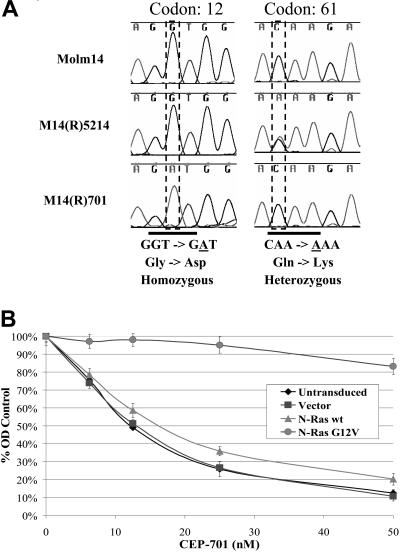
It is possible that prolonged exposure to FLT3 TKIs could select for mutations in kinases, other than FLT3, that activate downstream signaling pathways and thereby render cells FLT3 independent. Western blot analysis of immunoprecipitated tyrosine phosphorylated proteins from FLT3 TKI-sensitive versus -resistant cell lines revealed increased protein phosphorylation within the FLT3 TKI-resistant cells, even when cells were treated with 50 nM CEP-701 (data not shown). Mutation(s) within kinase(s) and/or phosphatase(s) could account for these findings and would explain the constitutive activation of Akt and/or MAPK observed in most FLT3 TKI-resistant cell lines. To examine this possibility, we performed a mutational screening of 100 tyrosine or serine/threonine kinases (including FLT3 and all major human RTKs) and 8 phosphatases using heteroduplex analysis of the PCR-amplified genes from the resistant cell lines. M14(R)5214 and M14(R)701 cell lines were each found to contain activating mutations within N-Ras, at positions Q61K and G12D, respectively, both of which are known to constitutively activate N-Ras (Figure 4A). Constitutive activation of Ras within M14(R)5214 and M14(R)701 cells helps explain why the Akt and MAPK signaling proteins remain activated despite inhibition of FLT3 phosphorylation. To determine whether constitutive activation of Ras, by itself, can lead to FLT3 TKI resistance, Molm14 cells were transduced with constitutively activated N-Ras-G12V. MTT assays demonstrate that Molm14 cells transduced with N-Ras-G12V are resistant to CEP-701 treatment compared with untransduced Molm14 cells or Molm14 cells transduced with vector only or wt N-Ras (Figure 4B). No mutations within the tested set of kinases and phosphatases were observed in 5 FLT3 TKI-resistant AML or ALL samples nor were Ras mutations identified in 10 FLT3 TKI-resistant primary AML or ALL samples tested (data not shown).
Figure 4.
Activating N-Ras mutations were found in the M14(R)5214 and M14(R)701 cell lines and play a role in resistance to FLT3 TKIs. (A) Sequence analysis of cDNA from the Molm14, M14(R)5214, and M14(R)701 cell lines. (B) Molm14 and Molm14 cells (300 000 cells/mL) transduced with the lentiviral vector, wt N-Ras or N-Ras G12V, were treated with CEP-701 for 48 hours at 37°C. Cellular activity/proliferation was determined using the MTT assay in triplicates. Error bars represent standard error of the mean (SEM).
Overexpression of wild-type RTKs may play a role in resistance to FLT3 TKIs
Overexpression of RTKs may contribute to the activation of compensatory downstream signaling pathways. To explore this possibility, the expression level of RTKs was determined using Q-PCR analysis using cDNA from FLT3 TKI-sensitive and -resistant cell lines with expression normalized to GAPDH expression. These studies revealed that most FLT3 TKI-resistant cell lines overexpress a number of RTKs compared with FLT3 TKI-sensitive cell lines (Figure 5).
Figure 5.
FLT3 TKI-resistant cell lines overexpress a number of RTKs compared with FLT3 TKI-sensitive cell lines. The RNA isolated from FLT3 TKI-sensitive and -resistant cell lines was converted into cDNA and used for Q-PCR analysis in triplicates. Target gene expression was normalized to GAPDH. Error bars indicate SEM.
Ligand stimulation of receptor tyrosine kinases (RTKs) do not account for the observed resistance to FLT3 TKIs
Ligand-mediated RTK phosphorylation could also maintain the activation of various signaling pathways, including Akt and/or MAPK, during inhibition of FLT3 phosphorylation leading to FLT3-independent growth. To test this hypothesis, the parental SEM-K2, Molm14, and Hb1119 cell lines were treated with 10 human ligands (IL-2, IL-3, IL-6, IL-11, GM-CSF, SCF, EGF, insulin, IGF-1, and GDNF) and were then exposed to various concentrations of CEP-701. Receptors that bind a number of these ligands were confirmed to be expressed on SEM-K2, Molm14, and/or Hb1119 cell lines by Q-PCR and Western blotting (Figure 5; data not shown). MTT assays demonstrated that ligand stimulation of these 10 RTKs did not result in significant resistance to CEP-701 treatment, even when cells were simultaneously stimulated with a combination of 2 to 4 ligands (data not shown). To determine whether other ligands might play a role in resistance to FLT3 TKIs, the parental SEM-K2, Molm14, and Hb1119 cell lines were grown in human bone marrow stromal cell–conditioned media and treated with CEP-701. Conditioned media resulted in only minimal changes to CEP-701–mediated cytotoxicity (data not shown). Overall, these data indicate that resistance to FLT3 TKIs is not usually the result of ligand activation of wild-type RTKs, at least under these conditions.
Inhibition of constitutively activated Akt or MAPK signaling pathways in FLT3 TKI-resistant cell lines restores some degree of sensitivity to FLT3 inhibition
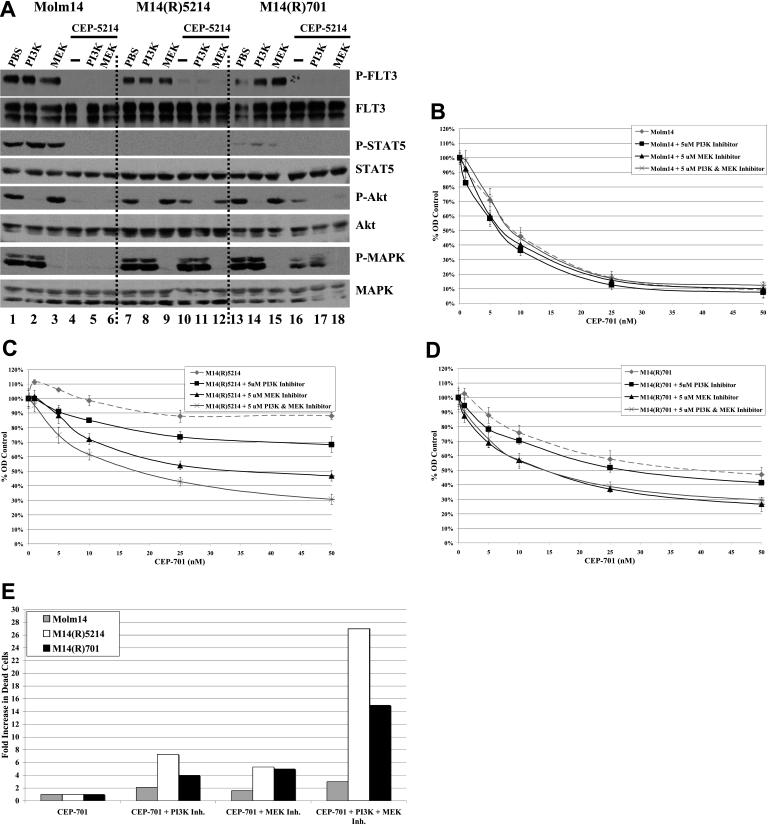
The selection for activating Ras mutations and the continued activation of Akt and/or MAPK signaling pathways in the TKI-resistant cell lines despite inhibition of FLT3 phosphorylation suggest that these pathways may play critical roles in FLT3 TKI resistance. To test this hypothesis, FLT3 TKI-resistant cell lines were treated with PI3K and/or MEK inhibitors while simultaneously exposing them to increasing concentrations of CEP-5214 or CEP-701. Western blots confirmed that the PI3K and MEK inhibitors inhibited Akt and MAPK phosphorylation, respectively, without affecting the phosphorylation of FLT3 or STAT5 (Figure 6A). MTT assays demonstrated that inhibition of Akt and/or MAPK phosphorylation with PI3K or MEK inhibitors restored some sensitivity to CEP-701 and CEP-5214 in several FLT3 TKI-resistant cell lines (Figure 6B-D; data not shown). Only those resistant cell lines showing continued activation of Akt and/or MAPK pathways in the face of FLT3 inhibition showed this restoration of sensitivity (Figure 2B, lanes 6-15; Figure 6B-D; data not shown). Furthermore, inhibition of both the Akt and MAPK signaling pathways led to a greater decrease in MTT signal than inhibition of either pathway alone when both pathways were activated in the absence of FLT3 phosphorylation, as seen with the M14(R)5214 cell line (Figure 2B, lanes 6-10; Figure 6C). In addition, annexin-V/7-AAD binding assays demonstrated that inhibition of FLT3 along with the PI3K and/or MEK signaling pathways resulted in a greater induction of apoptosis in FLT3 TKI-resistant M14(R)5214 and M14(R)701 cell lines that constitutively activate these pathways than FLT3 TKI treatment alone (Figure 6E). Taken together, these data demonstrate that activation of the Akt and/or MAPK signaling pathways through FLT3-independent pathways play a role in resistance to FLT3 TKIs.
Figure 6.
Resistance to FLT3 TKIs is partially mediated through activation of Akt and/or MAPK signaling pathways. (A) Molm14, M14(R)5214, and M14(R)701 cells (10 × 106 cells) were treated with 5 μM PI3K or MEK inhibitors and/or 50 nM CEP-5214 for 1 hour at 37°C. Immunoprecipitates and total protein extracts were resolved by 8% SDS-PAGE or 10% SDS-PAGE, respectively, and subjected to immunoblot analysis with the indicated phospho-specific antibodies. The same blots were then stripped and reprobed with protein-specific antibodies. (B) Molm14, (C) M14(R)5214, and (D) M14(R)701 cells (300 000 cells/mL) were incubated with 0 to 50 nM CEP-701 and/or 5 μM PI3K or MEK inhibitors for 48 hours at 37°C. Cellular activity/proliferation was determined using the MTT assay in triplicates and normalized to control cells that were not treated with CEP-701. (E) Cells (300 000 cells/mL) were incubated with 25 nM CEP-701 and/or 5 μM PI3K inhibitor and/or 5 μM MEK inhibitor for 48 hours at 37°C. Induction of apoptosis was assessed by measuring annexin V/7-AAD binding by flow cytometry. Values indicate relative fold increase in the percentage of cells that were annexin V/7-AAD positive. Results are representative of 2 independent experiments. Error bars indicate SEM.
Treatment with PI3K and/or MEK inhibitors did not result in a significant increase in cytotoxicity if Akt and MAPK proteins were already dephosphorylated because of FLT3 TKI treatment. This was seen with the SEM-K2, SEM(R)701, Molm14, Hb1119, and Hb(R)701 cell lines (Figure 2A, lanes 1-5, 11-15; Figure 2B, lanes 1-5; Figure 2C, lanes 1-5, 11-15; Figure 6B; data not shown). Phosphorylation of Akt and MAPK was fully inhibited in cells treated with 5 μM PI3K and MEK inhibitors, and no increase in cytotoxicity was observed when a higher concentration of PI3K or MEK inhibitor was used (Figure 6A; data not shown). This indicates that the increase in cytotoxicity observed when PI3K and/or MEK inhibitors are combined with FLT3 TKIs is likely due to inhibition of Akt and/or MAPK signaling pathways and not likely due to off target inhibition.
Inhibition of constitutively activated Akt or MAPK signaling pathways in FLT3 TKI-resistant primary samples does not restore sensitivity to FLT3 inhibition
To determine whether inhibition of constitutively activated Akt and/or MAPK sensitizes FLT3 TKI-resistant primary samples to FLT3 inhibition, 3 FLT3 TKI-resistant AML or ALL primary samples were used. MTT assays revealed that combining PI3K and/or MEK inhibitors with FLT3 TKIs does not result in increased cytotoxicity in the tested primary samples (data not shown). Western blot analysis confirmed that treatment of FLT3 TKI-resistant primary samples with PI3K and MEK inhibitors inhibited the phosphorylation of Akt and MAPK, respectively (data not shown).
FLT3 TKI-resistant cell lines still respond to anti-FLT3 mAb treatment in NOD/SCID mice
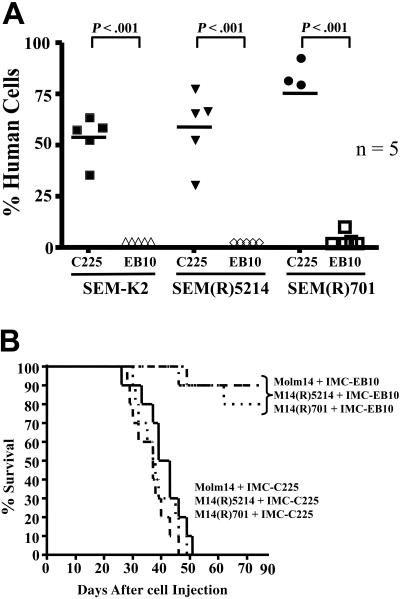
The data shown in these studies indicate that prolonged exposure to FLT3 TKIs often selects for cells that are FLT3 independent; therefore, inhibition of FLT3 fails to induce cytotoxicity. We have previously shown that the efficacy of anti-FLT3 mAb treatment does not depend on FLT3 inhibition and thus could still prove cytotoxic to FLT3 TKI-resistant cell lines. This hypothesis was tested by injecting NOD/SCID mice via tail vein with SEM-K2, Molm14, or FLT3 TKI-resistant cells derived from SEM-K2 or Molm14 cell lines. Mice were given intraperitoneal injections of 400 μg IMC-C225 or IMC-EB10 at 24, 72, and 120 hours after cell injection. Fluorescence-activated cell sorting (FACS) analysis of the bone marrows 30 days after cell injection revealed that IMC-EB10 treatment reduced the engraftment of SEM-K2 as well as the FLT3 TKI-resistant SEM(R)5214 and SEM(R)701 cell lines to below 1% (Figure 7A). In contrast, mice treated with the control mAb, IMC-C225, had 50% to 70% human cells in their bone marrow. Furthermore, IMC-EB10 treatment significantly prolonged the survival of NOD/SCID mice injected with Molm14, M14(R)5214, and M14(R)701 cell lines compared with control treatment (Figure 7B). IMC-EB10 is not cytotoxic to Hb1119 cells in vivo; therefore, it was not included in these studies.42
Figure 7.
IMC-EB10 treatment still reduces leukemic engraftment and prolongs the survival of NOD/SCID mice injected with FLT3 TKI-sensitive or -resistant cell lines. (A) NOD/SCID mice, in groups of 5, were injected with 0.5 × 106 SEM-K2, SEM(R)5214, or SEM(R)701 cells via tail-vein injection. Starting 24 hours after cell injection, mice were injected intraperitoneally a total of 3 times with 400 μg IMC-C225 or IMC-EB10, given every other day. Mice were killed 30 days after cell injection, and bone marrow cells were obtained from femurs. Cells harvested from femurs were stained with hCD19-FITC, mCD45-PE, and hCD45-APC, and human engraftment was determined by flow cytometry. (B) NOD/SCID mice, in groups of 10, were injected with 0.5 × 106 Molm14, M14(R)5214, or M14(R)701 cells via tail-vein injection. Starting 24 hours after cell injection, mice were injected intraperitoneally a total of 3 times with 400 μg IMC-C225 or IMC-EB10, given every other day. Mice were monitored daily for survival.
Discussion
Clinical trials using FLT3 TKIs have shown clinical responses in some patients, and the responses typically persist for only a few months.34–36 The inability of FLT3 TKIs to sustain suppression of leukemic blasts in these patients suggests that prolonged exposure to FLT3 TKIs may select for resistant cell populations. Several FLT3 mutations occurring with FLT3-ITD mutants were found to render transfected Ba/F3 cells resistant to SU5614 and/or PKC412.39,44,45 Furthermore, activation loop mutations of FLT3 have variable sensitivities to MLN518, AG1296, PKC412, and SU5614.33,46 However, to date, only a single patient with an acquired FLT3 mutation leading to FLT3 TKI resistance has been identified in clinical trials of these agents.38 Therefore, the inability of FLT3 TKIs to suppress the levels of leukemic blasts in patients for prolonged periods of time is likely due to alternative resistance mechanisms.
To investigate mechanisms of FLT3 TKI resistance in human leukemic cell lines, we cocultured SEM-K2, Molm14, and Hb1119 cells, each sensitive to FLT3 TKIs, with increasing concentrations of CEP-5214 or CEP-701 for 2 to 4 months. The resulting cell lines exhibited cross-resistance to all tested FLT3 TKIs. Western blot analysis revealed that FLT3 TKI treatment of FLT3 TKI-sensitive and -resistant cell lines resulted in inhibition of FLT3 phosphorylation. Thus, in this setting, resistance to FLT3 TKIs is not due to FLT3 mutations interfering with drug binding or p-glycoprotein–mediated export of FLT3 TKIs across the plasma membrane. Furthermore, sequencing of FLT3 confirmed no mutations within the resistant cell lines, and there was no increase in expression of p-glycoprotein observed by flow cytometry. Western blots and flow cytometry also confirmed that FLT3 TKI-resistant cell lines were not overexpressing FLT3 as a mechanism of drug resistance.
Western blot analysis of FLT3 TKI-resistant cell lines and primary samples did show continued activation of a number of signaling pathways normally dependent on FLT3 signaling, despite inhibition of FLT3. This pattern was previously observed in a number of FLT3 TKI-resistant primary samples.29,47 Furthermore, recent studies reveal that inhibition of FLT3 phosphorylation does not always correlate with induction of cytotoxicity.48,49 Therefore, continued activation of these pathways despite inhibition of FLT3 phosphorylation in FLT3 TKI-resistant cells suggests that FLT3-independent pathways phosphorylate/activate STAT1, STAT5, Akt, and/or MAPK along with possibly other unidentified signaling proteins and maintain the expression of genes that play a role in cell survival/proliferation. Activation of these and/or other signaling proteins through pathways not inhibited by FLT3 TKIs may explain the observed resistance to FLT3 TKIs.
In fact, inhibition of the PI3K/Akt and/or MEK/MAPK pathways by their respective inhibitors increased the sensitivity of FLT3 TKI-resistant cell lines to CEP-5214 and CEP-701. The data indicate that both of these pathways independently contribute to the observed resistance to FLT3 TKIs. However, FLT3 TKI-resistant cell lines vary in their degree of dependence on a given signaling pathway and may depend on more than just the Akt and MAPK signaling pathways for survival. This might explain why inhibition of the Akt and/or MAPK signaling pathways does not completely restore FLT3 TKI sensitivity. Furthermore, the lack of constitutive activation of Akt and/or MAPK in SEM(R)701 and Hb(R)701 cell lines, activation of a new signaling pathway (STAT1) in Hb(R)701 cells, and continued activation of STAT5 in some primary resistant samples supports the conclusion that pathways other than Akt and MAPK also play a role in FLT3 TKI resistance. Moreover, the observation that inhibition of Akt and/or MAPK phosphorylation in FLT3 TKI-resistant primary samples does not sensitize these cells to FLT3 TKIs suggests that other signaling pathways also play a role in FLT3 TKI resistance.
Activation of compensatory survival pathways may be due to mutations in a number of upstream signaling proteins, including kinases and phosphatases. Western blots showing a general increase in tyrosine-phosphorylated proteins from FLT3 TKI-resistant cells compared with FLT3 TKI-sensitive cells in the presence of FLT3 TKIs support this hypothesis. In fact, activating Ras mutations were found in two of the resistant cell lines (M14(R)5214 and M14(R)701), but they were not present in the FLT3 TKI-sensitive parent cell line. Mutant Ras is likely contributing to the activation of the Akt and MAPK signaling pathways in these cells and thus plays a role in FLT3 TKI resistance. Besides N-Ras, no mutant kinase or phosphatase was found within the FLT3 TKI-resistant cell lines or primary samples when 100 tyrosine and serine/threonine kinases and 8 phosphatases were screened. This does not exclude the possibility of activating mutations in one of more than 300 kinases and phosphatases not examined. Alternatively, signaling proteins can be phosphorylated through ligand stimulation of wild-type RTKs and thereby potentially lead to FLT3 TKI resistance. However, growth in human stromal cell–conditioned media or ligand stimulation of 10 selected RTKs did not result in resistance to CEP-701. It is still possible that under proper growth conditions cell-cell interactions and a combination of paracrine activation could contribute to resistance.
It is not clear how the signaling pathways are constitutively activated in the absence of FLT3 phosphorylation in the non–Ras-mutated FLT3 TKI-resistant cells. Overexpression of RTKs was observed in the FLT3 TKI-resistant cell lines by Q-PCR analysis. These RTKs could play a role in the activation of signaling pathways that render cells FLT3 independent. The role of additional mutations, Ras and overexpression of RTKs in resistance to FLT3 TKIs and TKIs targeting other RTKs warrants further investigation.
The data demonstrate that the combination of a FLT3 TKI with PI3K and/or MEK inhibitors can partially overcome some examples of FLT3 TKI resistance. In contrast to RTK inhibitors, PI3K and MEK inhibitors have the added advantage of inhibiting their pathway regardless of the upstream protein(s) responsible for their activation. However, inhibiting downstream pathways is also more likely to prove cytotoxic to normal cells that use these pathways for growth and survival. In contrast to TKIs, IMC-EB10, as a single therapeutic agent, still proved cytotoxic to FLT3 TKI-resistant cell lines in vivo. Anti-FLT3 mAb treatment of FLT3-expressing leukemias could potentially overcome a variety of drug resistance mechanisms because IMC-EB10–mediated cytotoxicity is not dependent on the phosphorylation status of FLT3 or downstream signaling proteins.
In summary, we demonstrate that prolonged exposure of human leukemic cell lines to FLT3 TKIs most frequently results in FLT3 independence and cross-resistance to multiple FLT3 TKIs without mutations in FLT3. Resistance to FLT3 TKIs is mediated, at least in some FLT3 TKI-resistant cell lines, via phosphorylation of downstream signaling proteins by FLT3-independent pathways. In 2 cases, this was due to selection of activating Ras mutations. Furthermore, treatment of NOD/SCID mice with anti-FLT3 mAbs is still effective in reducing the engraftment and prolonging survival when injected with FLT3 TKI-resistant cell lines. We hypothesize that combining FLT3 TKIs with PI3K and/or MEK inhibitors and/or anti-FLT3 mAbs may reduce the incidence of resistance and thereby increase the efficacy of FLT3 TKIs in patients.
Supplementary Material
Acknowledgments
We thank Susan Jones-Bolin and Bruce Ruggeri of Cephalon, Inc, for providing CEP-701 and CEP-5214, Pam Cohen from Novartis, Inc, for providing PKC412, and Yiwen Li of ImClone Systems, Inc, for providing IMC-EB10. We also thank Allen Williams for productive discussions.
This work was supported by grants from the Burroughs Wellcome Trust and NCI (grants CA90668, CA70970, and CA100632) (D.S.), (grant CA95600-03) (M.L.), and (grant CA111728) (P.B.). D.S. is also supported by the Kyle Haydock Professorship in Oncology. ImClone Systems, Inc, provided some of the funding for the studies reported here.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: O.P. performed the research, analyzed the data, and wrote the paper; M.W., P.B., K.-T.K., and M.L. performed the research; D.S. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Donald Small, Johns Hopkins University, School of Medicine, 1650 Orleans St, CRB I 251, Baltimore, MD 21202; e-mail: donsmall@jhmi.edu.
References
- 1.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17:1738–1752. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 2.Brown P, Small D. FLT3 inhibitors: a paradigm for the development of targeted therapeutics for paediatric cancer. Eur J Cancer. 2004;40:707–724. doi: 10.1016/j.ejca.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 3.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 4.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 5.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 6.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br J Haematol. 2001;113:983–988. doi: 10.1046/j.1365-2141.2001.02850.x. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 8.Zheng R, Levis M, Piloto O, et al. FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood. 2004;103:267–274. doi: 10.1182/blood-2003-06-1969. [DOI] [PubMed] [Google Scholar]
- 9.Hawley TS, Fong AZ, Griesser H, Lyman SD, Hawley RG. Leukemic predisposition of mice transplanted with gene-modified hematopoietic precursors expressing flt3 ligand. Blood. 1998;92:2003–2011. [PubMed] [Google Scholar]
- 10.Drexler HG. Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells. Leukemia. 1996;10:588–599. [PubMed] [Google Scholar]
- 11.Iwai T, Yokota S, Nakao M, et al. Internal tandem duplication of the FLT3 gene and clinical evaluation in childhood acute myeloid leukemia. The Children's Cancer and Leukemia Study Group, Japan. Leukemia. 1999;13:38–43. doi: 10.1038/sj.leu.2401241. [DOI] [PubMed] [Google Scholar]
- 12.Kondo M, Horibe K, Takahashi Y, et al. Prognostic value of internal tandem duplication of the FLT3 gene in childhood acute myelogenous leukemia. Med Pediatr Oncol. 1999;33:525–529. doi: 10.1002/(sici)1096-911x(199912)33:6<525::aid-mpo1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Kottaridis PD, Gale RE, Linch DC. Prognostic implications of the presence of FLT3 mutations in patients with acute myeloid leukemia. Leuk Lymphoma. 2003;44:905–913. doi: 10.1080/1042819031000067503. [DOI] [PubMed] [Google Scholar]
- 14.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 15.Lavagna-Sevenier C, Marchetto S, Birnbaum D, Rosnet O. FLT3 signaling in hematopoietic cells involves CBL, SHC and an unknown P115 as prominent tyrosine-phosphorylated substrates. Leukemia. 1998;12:301–310. doi: 10.1038/sj.leu.2400921. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Broxmeyer HE. Flt3 ligand induces tyrosine phosphorylation of gab1 and gab2 and their association with shp-2, grb2, and PI3 kinase. Biochem Biophys Res Commun. 2000;277:195–199. doi: 10.1006/bbrc.2000.3662. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Mantel C, Broxmeyer HE. Flt3 signaling involves tyrosly-phosphorylation of SHP-2 and SHIP and their association with Grb2 and Shc in Baf3/Flt3 cells. J Leukoc Biol. 1999;65:372–380. doi: 10.1002/jlb.65.3.372. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Broxmeyer HE. p85 subunit of PI3 kinase does not bind to human Flt3 receptor, but associates with SHP2, SHIP, and a tyrosine-phosphorylated l00-kDa protein in Flt3 ligand-stimulated hematopoietic cells. Biochem Biophys Res Commun. 1999;254:440–445. doi: 10.1006/bbrc.1998.9959. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Fukuda S, Lee Y, et al. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med. 2000;192:719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosnet O, Buhring HJ, deLapeyriere O, et al. Expression and signal transduction of the FLT3 tyrosine kinase receptor. Acta Haematol. 1996;95:218–223. doi: 10.1159/000203881. [DOI] [PubMed] [Google Scholar]
- 21.Kiyoi H, Towatari M, Yokota S, et al. Internal tandem duplication of the FLT3 gene is a novel modality of elongation mutation which causes constitutive activation of the product. Leukemia. 1998;12:1333–1337. doi: 10.1038/sj.leu.2401130. [DOI] [PubMed] [Google Scholar]
- 22.Kiyoi H, Ohno R, Ueda R, Saito H, Naoe T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene. 2002;21:2555–2563. doi: 10.1038/sj.onc.1205332. [DOI] [PubMed] [Google Scholar]
- 23.Tse KF, Novelli E, Civin CI, Bohmer FD, Small D. Inhibition of FLT3-mediated transformation by use of a tyrosine kinase inhibitor. Leukemia. 2001;15:1001–1010. doi: 10.1038/sj.leu.2402199. [DOI] [PubMed] [Google Scholar]
- 24.Tse KF, Allebach J, Levis M, Smith BD, Bohmer FD, Small D. Inhibition of the transforming activity of FLT3 internal tandem duplication mutants from AML patients by a tyrosine kinase inhibitor. Leukemia. 2002;16:2027–2036. doi: 10.1038/sj.leu.2402674. [DOI] [PubMed] [Google Scholar]
- 25.Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14:1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- 26.Carow CE, Levenstein M, Kaufmann SH, et al. Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias. Blood. 1996;87:1089–1096. [PubMed] [Google Scholar]
- 27.Rosnet O, Buhring HJ, Marchetto S, et al. Human FLT3/FLK2 receptor tyrosine kinase is expressed at the surface of normal and malignant hematopoietic cells. Leukemia. 1996;10:238–248. [PubMed] [Google Scholar]
- 28.Birg F, Courcoul M, Rosnet O, et al. Expression of the FMS/KIT-like gene FLT3 in human acute leukemias of the myeloid and lymphoid lineages. Blood. 1992;80:2584–2593. [PubMed] [Google Scholar]
- 29.Levis M, Allebach J, Tse KF, et al. A FLT3-targeted tyrosine kinase inhibitor is cytotoxic to leukemia cells in vitro and in vivo. Blood. 2002;99:3885–3891. doi: 10.1182/blood.v99.11.3885. [DOI] [PubMed] [Google Scholar]
- 30.Yee KW, O'Farrell AM, Smolich BD, et al. SU5416 and SU5614 inhibit kinase activity of wild-type and mutant FLT3 receptor tyrosine kinase. Blood. 2002;100:2941–2949. doi: 10.1182/blood-2002-02-0531. [DOI] [PubMed] [Google Scholar]
- 31.Weisberg E, Boulton C, Kelly LM, et al. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell. 2002;1:433–443. doi: 10.1016/s1535-6108(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 32.O'Farrell AM, Abrams TJ, Yuen HA, et al. SU11248 is a novel FLT3 tyrosine kinase inhibitor with potent activity in vitro and in vivo. Blood. 2003;101:3597–3605. doi: 10.1182/blood-2002-07-2307. [DOI] [PubMed] [Google Scholar]
- 33.Clark JJ, Cools J, Curley DP, et al. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518. Blood. 2004;104:2867–2872. doi: 10.1182/blood-2003-12-4446. [DOI] [PubMed] [Google Scholar]
- 34.Smith BD, Levis M, Beran M, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103:3669–3676. doi: 10.1182/blood-2003-11-3775. [DOI] [PubMed] [Google Scholar]
- 35.Fiedler W, Serve H, Dohner H, et al. A phase 1 study of SU11248 in the treatment of patients with refractory or resistant acute myeloid leukemia (AML) or not amenable to conventional therapy for the disease. Blood. 2005;105:986–993. doi: 10.1182/blood-2004-05-1846. [DOI] [PubMed] [Google Scholar]
- 36.Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105:54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 37.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 38.Heidel F, Solem FK, Breitenbuecher F, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 39.Cools J, Mentens N, Furet P, et al. Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer Res. 2004;64:6385–6389. doi: 10.1158/0008-5472.CAN-04-2148. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Li H, Wang MN, et al. Suppression of leukemia expressing wild-type or ITD-mutant FLT3 receptor by a fully human anti-FLT3 neutralizing antibody. Blood. 2004;104:1137–1144. doi: 10.1182/blood-2003-07-2585. [DOI] [PubMed] [Google Scholar]
- 41.Piloto O, Levis M, Huso D, et al. Inhibitory anti-FLT3 antibodies are capable of mediating antibody-dependent cell-mediated cytotoxicity and reducing engraftment of acute myelogenous leukemia blasts in nonobese diabetic/severe combined immunodeficient mice. Cancer Res. 2005;65:1514–1522. doi: 10.1158/0008-5472.CAN-04-3081. [DOI] [PubMed] [Google Scholar]
- 42.Piloto O, Nguyen B, Huso D, et al. IMC-EB10, an anti-FLT3 monoclonal antibody, prolongs survival and reduces NOD/SCID engraftment of some ALL cell lines and primary leukemic samples. Cancer Res. 2006;66:4843–4851. doi: 10.1158/0008-5472.CAN-06-0018. [DOI] [PubMed] [Google Scholar]
- 43.Levis M, Tse KF, Smith BD, Garrett E, Small D. A FLT3 tyrosine kinase inhibitor is selectively cytotoxic to acute myeloid leukemia blasts harboring FLT3 internal tandem duplication mutations. Blood. 2001;98:885–887. doi: 10.1182/blood.v98.3.885. [DOI] [PubMed] [Google Scholar]
- 44.Bagrintseva K, Geisenhof S, Kern R, et al. FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x(L). Blood. 2005;105:3679–3685. doi: 10.1182/blood-2004-06-2459. [DOI] [PubMed] [Google Scholar]
- 45.Bagrintseva K, Schwab R, Kohl TM, et al. Mutations in the tyrosine kinase domain of FLT3 define a new molecular mechanism of acquired drug resistance to PTK inhibitors in FLT3-ITD-transformed hematopoietic cells. Blood. 2004;103:2266–2275. doi: 10.1182/blood-2003-05-1653. [DOI] [PubMed] [Google Scholar]
- 46.Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102:646–651. doi: 10.1182/blood-2002-11-3441. [DOI] [PubMed] [Google Scholar]
- 47.Brown P, Meshinchi S, Levis M, et al. Pediatric AML primary samples with FLT3/ITD mutations are preferentially killed by FLT3 inhibition. Blood. 2004;104:1841–1849. doi: 10.1182/blood-2004-03-1034. [DOI] [PubMed] [Google Scholar]
- 48.Knapper S, Burnett AK, Littlewood T, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108:3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 49.Levis M, Brown P, Smith BD, et al. Plasma inhibitory activity (PIA): a pharmacodynamic assay reveals insights into the basis for cytotoxic response to FLT3 inhibitors. Blood. 2006;108:3477–3483. doi: 10.1182/blood-2006-04-015743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.