Abstract
We show that the Drosophila protein DSP1, an HMG-1/2-like protein, binds DNA highly cooperatively with three members of the Rel family of transcriptional regulators (NF-κB, the p50 subunit of NF-κB, and the Rel domain of Dorsal). This cooperativity is apparent with DNA molecules bearing consensus Rel-protein-binding sites and is unaffected by the presence of a negative regulatory element, a sequence previously proposed to be important for mediating repression by these Rel proteins. The cooperativity observed in these DNA-binding assays is paralleled by interactions between protein pairs in the absence of DNA. We also show that in HeLa cells, as assayed by transient transfection, expression of DSP1 increases activation by Dorsal from the twist promoter and inhibits that activation from the zen promoter, consistent with the previously proposed idea that DSP1 can affect the action of Dorsal in a promoter-specific fashion.
The Drosophila morphogen Dorsal works as both an activator and a repressor of gene transcription in the early embryo. On the ventral side of the embryo, where Dorsal concentration is highest, Dorsal activates certain genes (e.g., twist) and represses others (e.g., zen, tld, and dpp; refs. 1–7). Dorsal activation of the twist promoter is mediated by two ventral-specific enhancers called ventral activation response elements (VARs; refs. 4 and 8), each of which contains Dorsal-binding sites (8). The VAR elements of the twist promoter are also thought to bind other factors that synergize with Dorsal to drive ventral-specific activation of transcription (9, 10). Promoter elements responsible for Dorsal-mediated repression have been identified in the tld (11), zen (12, 13), and dpp (6, 14) promoters. These ventral repression elements (VREs) contain, in addition to Dorsal-binding sites, sequences proposed to bind putative cofactors required to convert Dorsal from an activator to a repressor (5, 11–14). One such sequence is the negative regulatory element (NRE), a copy of which is also found in the mammalian interferon-β promoter immediately adjacent to and partially overlapping a binding site for another member of the Rel family, NF-κB (15, 16). Genetic and biochemical experiments indicate that, in flies, Groucho (17), Cut (18), Dead Ringer (18), and NTF-1 (14) help to convert Dorsal into a repressor.
An additional candidate for such a corepressor is the Drosophila protein DSP1, which was identified by its ability to inhibit, in yeast, transcriptional activation by Dorsal working at the zen promoter (16). DSP1 interacts with the human protein SP100, and the latter interacts with hHP1, a protein required for the negative effects observed in position effect variegation in Drosophila (19). SP100 and homologues of hHP1 are found in nuclear bodies, sometimes associated with an HMG-1 derivative (20). Each of these proteins, DSP1, SP100, and hHP1, function as an efficient repressor when artificially tethered upstream at GAL4 sites of a mammalian promoter. It was further reported that, in mammalian cells, DSP1 converted Dorsal from an activator to a repressor if an NRE were present adjacent to the Dorsal-binding site (16).
DSP1 is a member of the HMG-1/2 family of DNA-binding proteins. HMG-1/2 proteins are characterized by the presence of two or more structural units called HMG domains. The carboxyl-terminal DSP1 fragment comprising residues 178–393 contains two such domains, referred to as A and B, as well as a stretch of acidic residues at its carboxyl terminus, and is closely related to a corresponding fragment of HMG-2. The amino-terminal portion of the molecule is rich in polyglutamine residues. HMG-1/2 proteins are thought to facilitate the formation of nucleoprotein complexes by bending DNA (21–24) and by interacting with sequence-specific DNA-binding proteins (25–28). In Drosophila, DSP1 is expressed uniformly until the end of germ-band retraction in embryogenesis and later becomes restricted to the ventral nerve cord and the brain. In the adult fly, DSP1 is expressed in the nurse cells and the brain (29), and a P element-induced mutation in dsp1 is lethal (29).
We show that DSP1 binds highly cooperatively to Rel sites on DNA with NF-κB, with the p50 subunit of the NF-κB heterodimer, and with the Rel domain of Dorsal and that this cooperativity is unaffected by the presence of an NRE. Cooperative binding is mediated by interactions, readily observed in the absence of DNA, between the HMG and Rel domains of the respective proteins. Two Rel proteins, the p65 subunit of NF-κB and Drosophila Dif, lack high-affinity DSP1 interaction sites and, where tested, bind cooperatively to DNA with DSP1 weakly, if at all. In transiently transfected mammalian cells, Dorsal activates transcription from both the zen and twist promoters, and additional DSP1 inhibits the former and increases the latter.
MATERIALS AND METHODS
Plasmid Construction.
pDL (1–340) was used to express the Dorsal Rel domain (30). Expression vectors for the NF-κB subunits and glutathione S-transferase (GST)–p50 have been described (31, 32). Dorsal expression vectors were all derived from pSK-Dorsal (16, 33). Full-length Dorsal was expressed from pJBE21. pJBE21 was constructed by inserting an NdeI–NotI Dorsal fragment into the backbone of pJBE18 (16). pJBE23 was used to overexpress and purify the HMG domain of DSP1. Expression vector pJBE23 was constructed by cloning the DSP1 HMG domain-containing fragment (NdeI–NotI) from plasmid pSK-HMG-NdeI into pJBE21. GST–Dorsal was expressed from pJBE27. pJBE27 was constructed by inserting Dorsal derived from pJBE21 as an NdeI–NotI fragment with the NdeI site filled in into pGEX-5X-1 (Amersham Pharmacia), with the EcoRI site filled in, and with the NotI site left with an overhang. GST–DSP1 (1–393), GST–DSP1 (178–393), and GST–DSP1 (264–393) were expressed from pJBE29, pJBE30, and pJBE31, respectively. These vectors were constructed in the same manner as pJBE27. The p65 (1–266) (32) expression vector and p50 (1–366) expression vector (34) have been described. The twist reporter was constructed from a genomic fragment of the twist promoter (4) as a 1,200-bp fragment between the HindIII and XbaI sites of pSP73 (Promega). The entire enhancer from −1179 to +11 was inserted by directional cloning as a HindIII–XbaI fragment into pBLCAT2 (35). The chimeric PVAR–DVAR construct was made by PCR with overlapping oligonucleotides. The oligonucleotide design was based on the sequences of the twist promoter in ref. 8. The zen VRE version of pBLCAT2 contains the 180-bp minimal fragment of the zen VRE (13) inserted into pBLCAT2 (13). Other expression vectors for mammalian transfections have been described (16).
Protein–Protein Interactions and Electrophoretic Mobility-Shift Assays (EMSAs).
GST fusion interaction assays and EMSAs have been described (16, 30). The EMSA shown in Fig. 1e was performed as described (1) with modifications. DNA-binding reactions were done in 10 mM Hepes/45 mM KCl/6 mM 2-mercaptoethanol/1 mM EDTA/10% (vol/vol) glycerol/1 mg/ml BSA for 15 min at 25°C. Samples were then loaded onto a 5% polyacrylamide gel in 25 mM Tris base/190 mM glycine/1 mM EDTA. The gel and buffer were precooled to 4°C, and the gel was run for 20 min at 18 V/cm and then an additional 2 h at 8 V/cm. DNA binding assays were done over a range of Rel protein concentrations as described in ref. 16. The synthetic oligonucleotides used for the experiments of Fig. 1 a–c were GATCTGGGAAATTCCGTGGGAAATTCCT and GATCTAGGAATTTCCCACGGAATTTCCCA, two PRDII sites without an NRE, as well as GATCTGGGAAATTCCGTGGGAAATTCCTCTGAA and GATCTTCAGAGGAATTTCCCACGGAATTTCCCA, two PRDII sites with an NRE. The oligonucleotides used for Fig. 1d were GATCGGGAATTCCC annealed to itself and GATCGGGAAATTCCT and GATCAGGAATTTCCC. The oligonucleotides used for Fig. 1e are GATCTGGGAAAACCAGTGGGAAAACCAAGC and GATCGCTTGGTTTTCCCACTGGTTTTCCCA. The 110-bp zen promoter fragments that contain either wild-type or mutant AT sites were obtained from Michael Levine (described in ref. 13). Restriction fragments bearing these sequences were end labeled and used in the EMSA experiments described in Fig. 1f. The EMSA experiments shown in Fig. 2 were performed with the oligonucleotides GATCTGGGAAATTCC, GATCTGGAATTTCCCA (PRDII), GATCTGGGAAATTCCTCTGAA, and GATCTTTCAGAGGAATTTCCCA (PRDIINRE). All oligonucleotides were labeled by filling in 5′ overhangs as described (16).
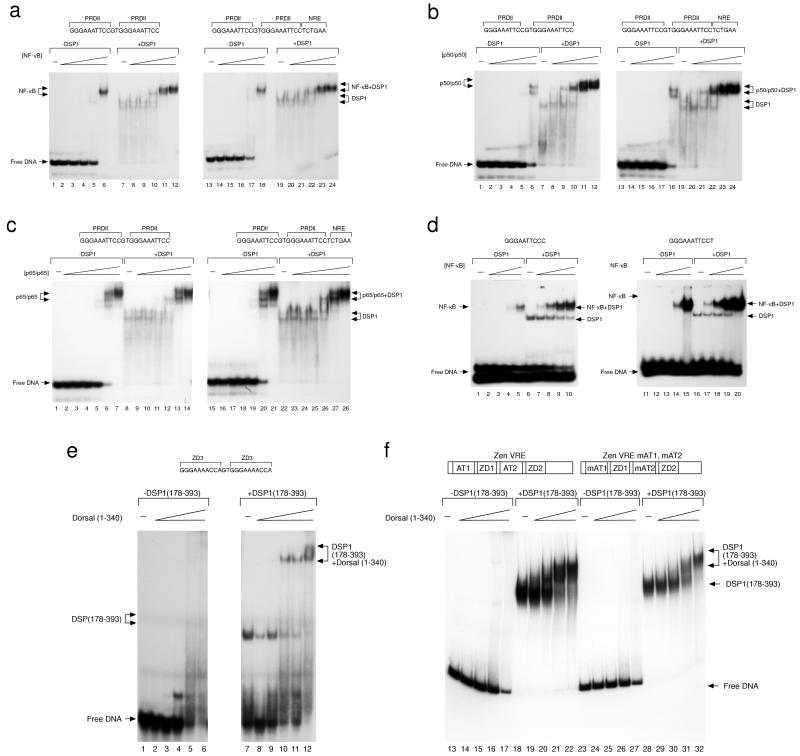
Figure 1.
EMSAs of cooperative binding of Rel proteins and DSP1 to DNA. In each case, increasing amounts of the indicated Rel protein (in 3-fold steps) were assayed with or without a constant amount of DSP1. (a–c) DSP1 helps NF-κB (a) and p50/p50 (b), but not p65/p65 (c), bind to DNA bearing PRD11 sites, and an NRE has no effect on that cooperativity. The labeled DNA molecule bore two PRD11 sites with or without an adjacent NRE as indicated. The PRDII site is the Rel site found immediately adjacent to the NRE of the mammalian interferon-β promoter. Data in a–c Right were published previously (16). (d) NF-κB binds cooperatively with DSP1 to DNA bearing solely a single consensus Rel-binding site. (e) DSP1 (178–393), which bears the HMG domains, binds cooperatively with the Rel domain of Dorsal to a synthetic oligonucleotide bearing two ZD3 sites copied from the zen promoter. (f) The effect noted in e is also observed with a DNA fragment taken from the zen promoter, and that cooperativity is unaffected by mutation of adjacent AT-rich sequences required for converting Dorsal into a repressor at this promoter (13).
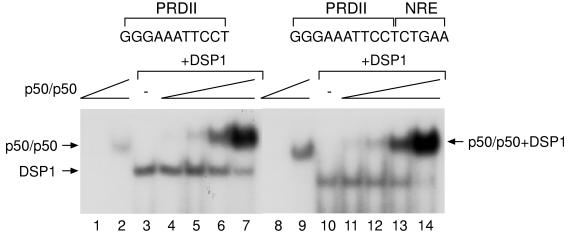
Figure 2.
Cooperative binding of DSP1 and Rel proteins to DNA involves formation of a ternary complex. The EMSAs show that, as above, DSP1 helps p50/p50 to bind to DNA bearing a single PRDII site and a single PRDII site with an NRE. The mobility of the DNA-bound complex formed on addition of both DSP1 and p50/p50 is slower than either the DSP1–DNA complex or the p50/p50–DNA complex. Similar results are obtained with and without an NRE. In this experiment, the free probe was run much longer than in the experiments shown in Fig. 1 and is not shown in the figure. The concentration of p50/p50 in lanes 2, 7, 9, and 14 is the same and decreases in 3-fold steps.
Transfections.
HeLa cells were transfected by the calcium phosphate method. Transfections were done according to methods described previously (16). The transfection mixture contained a total of 13 μg of DNA including 1 μg of reporter, 1 μg of a cytomegalovirus (CMV)--lacZ reporter as an internal control, and various amounts of Rel and DSP1 expression vectors or the pcDNA3 (Invitrogen) backbone vector. The cells were harvested, and chloramphenicol acetyltransferase (CAT) assays were performed 48 h after transfection (32). The numbers in Fig. 4 represent relative CAT activity. Transfections were routinely done in duplicate. HeLa cells were cultured according to standard procedures in DMEM containing 10% (vol/vol) FBS, antibiotics, and l-glutamine (2 mM) in a humidified incubator containing 4.5% (vol/vol) CO2. CAT assays and β-galactosidase assays were done as described (32).
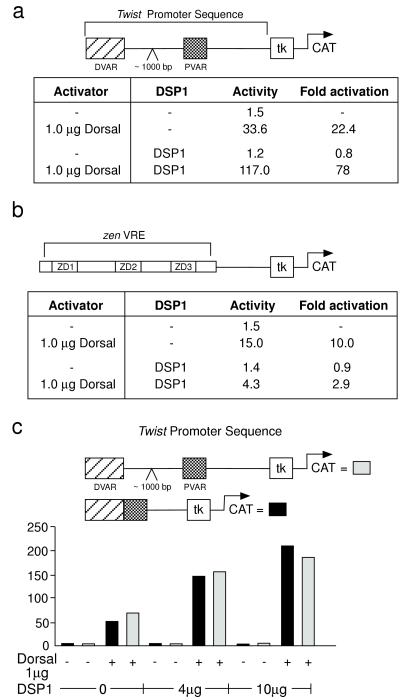
Figure 4.
DSP1 has opposite effects on Dorsal-mediated transcriptional activation on the twist and zen promoters. HeLa cells were cotransfected with plasmids expressing DSP1 and Dorsal and the depicted reporter gene. (a) DSP1 helps Dorsal-mediated activation of the twist promoter. (b) DSP1 inhibits Dorsal activity from the zen promoter. (c) DSP1 helps Dorsal to activate transcription from the minimal ventral enhancer elements of the twist promoter.
RESULTS
In the experiments of Fig. 1a, DSP1 helped NF-κB bind to DNA bearing two Rel-binding sites, and this phenomenon was unaffected by the presence of an NRE adjacent to the Rel sites. As reported previously, DSP1 also strongly increased DNA binding of the p50 homodimer but helped only slightly binding of the p65 homodimer to DNA bearing two Rel sites (Fig. 1 b and c). Fig. 1d shows that DSP1 helped NF-κB bind to DNA bearing only a single minimal Rel-protein-binding site and that the stimulatory effect of DSP1 on NF-κB was equivalent whether the Rel site was a consensus Dorsal site or the Rel site (called PRDII) found in the interferon-β promoter. Fig. 1e shows that the fragment of DSP1 comprising residues 178–393 stimulated DNA binding by the Dorsal Rel domain (residues 1–340). In this case, the DNA site contains two Rel sites and no NRE. Similar results were found when the Rel site was flanked by a mutant NRE (data not shown). Fig. 1f shows that DSP1 stimulated binding of Dorsal (1–340) to both the wild-type zen promoter and to a mutant form of the promoter that functions as a VAR in the Drosophila embryo (13).
The experiments of Fig. 2 indicate that the stimulatory effect of DSP1 on DNA binding of Rel proteins is caused, in each case, by cooperative binding to the Rel sites. Thus, the putative ternary complex of DNA, p50/p50, and DSP1 migrated more slowly than did the p50/p50–DNA complex. Fig. 2 also shows that the ternary complex formed equally efficiently on oligonucleotides bearing either a single PRDII or a PRDII site with an NRE. The formation of any of these complexes requires a Rel-binding site on the DNA (data not shown).
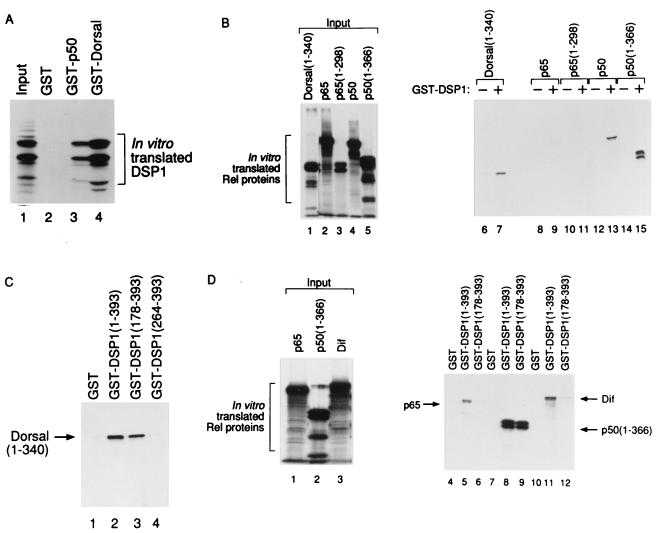
The experiments of Fig. 3 show that, in the absence of DNA, DSP1 interacted with various Rel proteins with a specificity that correlates with the observed cooperative binding to DNA. Thus, as expected, GST–Dorsal and GST–p50 both robustly bound DSP1 (Fig. 3A). Fig. 3B shows that, also as expected, GST–DSP1 bound p50 and the Rel domains of both Dorsal and p50 but bound p65 or the p65 Rel domain only very weakly. Fig. 3 C and D addresses the domain of DSP1 required to interact with different Rel proteins. Fig. 3C shows that DSP1 (178–393) is sufficient to bind p50 and Dorsal, but that a fragment of DSP1 comprising a single HMG domain, the B domain (residues 264–393), is not. Fig. 3D shows that, whereas DSP1 (178–393) bound p50 and Dorsal, the weak binding seen with p65 required intact DSP1, as did the weak interaction observed with Dif (36), another Drosophila Rel protein.
Figure 3.
DSP1 interacts with certain Rel proteins in vitro. In each case, in vitro translated 35S-labeled protein was incubated with glutathione Sepharose beads coupled to the indicated fusion protein. (a) DSP1 interacts with GST–p50 (lane 3) and GST–Dorsal (lane 4). (b) GST–DSP1 interacts with Dorsal (1–340) (lane 7), p50 (lane 13), and p50 (1–366) (lane 15) but only weakly with p65 (lane 9) or p65 (1–298) (lane 11). (c) Dorsal (1–340) interacts with GST–DSP1 (lane 2) and GST–DSP1 (178–393) (lane 3) but not with GST–DSP1 (264–393) (lane 4). (d) GST–DSP1 interacts with p50 (1–366) (lane 8) but only weakly with p65 (lane 5) or Dif (lane 11). In contrast, DSP1 (178–393) interacts only with p50 (1–366) (lane 9) and not with p65 (lane 6) or Dif (lane 12).
The experiment of Fig. 4 shows contrasting effects of DSP1 on the activity of Dorsal assayed with regulatory sequences excised from the twist and zen promoters. These experiments were performed by transiently transfecting mammalian cells in culture. Thus, reporters containing either a 180-bp fragment from zen (a fragment sufficient to mediate repression in Drosophila) or the entire regulatory region of twist (from −1,438 to +38) upstream of the tk promoter were activated by cotransfection with DNA encoding Dorsal. Cotransfection with DNA encoding DSP1 had opposite effects on this Dorsal mediated activation of the two promoters: activation from the twist promoter was stimulated 4-fold (Fig. 4a), whereas that from the zen promoter was inhibited 3-fold (Fig. 4b). DSP1’s stimulation of Dorsal-mediated activation from the twist promoter can be mapped to the defined enhancer elements or VARs. Thus, DSP1 also stimulated Dorsal-mediated activation if the template bore, instead of the intact twist promoter, a cassette that contains the two VARs that drive ventral-specific expression of the twist gene in the Drosophila embryo (Fig. 4c). The two VARs together constitute approximately 300 bp and contain multiple Rel-protein-binding sites (8). p65 also activated transcription from the twist promoter in transient transfection assays, but, consistent with the binding studies reported above, added DSP1 had no effect on that activation (data not shown).
We do not know what DNA sequences in the zen and twist promoters determine the opposite effects of DSP1 on dorsal-mediated activation in the experiments of Fig. 4. Our finding that the NRE has no effect on cooperative binding to DNA of DSP1 and various Rel proteins (see Figs. 1 and 2) prompted us to reexamine the earlier claims that, on a template bearing two synthetic Rel sites and an NRE upstream of the tk promoter, DSP1 converted Dorsal, the p50 homodimer, and the NF-κB heterodimer into repressors and that that effect required the NRE. We found that, in each case, DSP1 inhibited Rel-protein-dependent activation both in the presence and absence of an NRE, and, in no case, did we observe NRE-dependent conversion of the Rel protein to a repressor by cotransfection with DSP1. We do not understand why the current results differ from those reported previously.
DISCUSSION
We show here that DSP1 binds highly cooperatively to DNA bearing Rel-binding sites with certain Rel proteins (p50, NF-κB, and the Rel domain of Dorsal) but only weakly with another such protein (p65). DSP1 bound in parallel fashion to these Rel proteins in the absence of DNA. Two observations suggest that DSP1 can influence the action of Dorsal. First, as reported here, in transient-transfection experiments with mammalian cells, expression of DSP1 has a promoter-specific effect: it inhibits Dorsal-mediated activation at the zen promoter but amplifies that effect from the twist promoter. Consistent with the protein-interaction experiments performed in vitro, DSP1 had no effect on p65-mediated activation in vivo in such experiments. Second, as reported previously, DSP1 inhibits Dorsal-mediated activation in yeast. What sequences outside the Rel-binding sites are required for such effects remain unclear. In particular, our experiments show that an NRE sequence, a copy of which is found in the zen VRE, plays no role in the cooperative binding of DSP1 and Rel proteins. Whether the NRE plays any role in the assembly of a Dorsal repressing complex remains to be seen.
Sites of the protein–protein interactions we have described are found in the conserved Rel domains and in the fragment of DSP1 that bears both HMG domains. The Rel domains of p65 and of Dif differ from those of Dorsal and of p50 in that they lack the HMG-domain-interaction site. The HMG domain of DSP1 also interacts with the TATA-binding protein (ref. 37 and data not shown). Similar interactions have been reported for HMG-1 and HMG-2 with the steroid hormone receptors (25, 38), for HMG-1 with p53 (26), for HMG-1 with HOXD9 (28), and for HMG-2 with Oct2 (27). Thus, the HMG domain may contain a common structural motif for cooperative DNA binding and interaction with other transcription factors. The interaction between TATA-binding protein and DSP1 also seems to be influenced by the glutamine-rich amino-terminal domain in that the full-length DSP1 interacts more avidly with TATA-binding protein than does the HMG-1 domain (ref. 37 and data not shown). Our experiments (described in Fig. 3) suggest that the amino-terminal glutamine-rich domain may also potentiate the DSP1–Rel protein interaction as well, because all DSP1–Rel interactions seem stronger with full-length DSP1, particularly the weak interactions seen between DSP1 and p65 or Dif (Fig. 3D), which are observed only with GST–DSP1 and not with GST–DSP1 (178–393).
Acknowledgments
We thank Michael Levine, Dimitris Thanos, Rob Peters, Al Courey, Norbert Lehming, Shamol Saha, Tom Maniatis, Jean-Paul Vincent, Eumorphia Remboutsika, Christoph Müller, Luc Gaudreau, and Melanie Clements for plasmids and critical discussions; Renate Helmiss and the National Institute for Medical Research (London) photographics for graphics; and Mary Jo Wright for help preparing this manuscript. This work was supported by grants from the National Institutes of Health (to M.P.). J.M.B. was supported by a National Institutes of Health predoctoral fellowship.
ABBREVIATIONS
- VRE
ventral repression element
- VAR
ventral activation response element
- NRE
negative regulatory element
- GST
glutathione S-transferase
- EMSA
electrophoretic mobility-shift assay
- CAT
chloramphenicol acetyltransferase
References
- 1.Ip Y T, Kraut R, Levine M, Rushlow C A. Cell. 1991;64:439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Kosman D, Ip Y T, Levine M. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 3.Ip Y T, Park R E, Kosman D, Bier E, Levine M. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 4.Pan D J, Huang J D, Courey A J. Genes Dev. 1991;5:1892–1901. doi: 10.1101/gad.5.10.1892. [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Rushlow C, Zhou Q, Small S, Levine M. EMBO J. 1992;11:3147–3154. doi: 10.1002/j.1460-2075.1992.tb05387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J-D, Schwyter D H, Shirokawa J M, Courey A J. Genes Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- 7.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 8.Pan D, Valentine S A, Courey A J. Mech Dev. 1994;46:41–53. doi: 10.1016/0925-4773(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 9.Leptin M. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 10.Ip Y T, Park R E, Kosman D, Yazdanbakhsh K, Levine M. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 11.Kirov N, Childs S, O’Connor M, Rushlow C. Mol Cell Biol. 1994;14:713–722. doi: 10.1128/mcb.14.1.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirov N, Zhelnin L, Shah J, Rushlow C. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J, Cai H, Zhou Q, Levine M. EMBO J. 1993;12:3201–3209. doi: 10.1002/j.1460-2075.1993.tb05989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J-D, Dubnicoff T, Liaw G-J, Bai Y, Valentine S A, Courey A J. Genes Dev. 1995;9:3177–3189. doi: 10.1101/gad.9.24.3177. [DOI] [PubMed] [Google Scholar]
- 15.Nourbakhsh M, Hoffmann K, Hauser H. EMBO J. 1993;12:451–459. doi: 10.1002/j.1460-2075.1993.tb05677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehming N, Thanos D, Brickman J M, Ma J, Maniatis T, Ptashne M. Nature (London) 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 17.Dubnicoff T, Valentine S A, Chen G, Shi T, Lengyel J A, Paroush Z, Courey A J. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentine S A, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey A J. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehming N, Le Saux A, Schuller J, Ptashne M. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giese K, Cox J, Grosschedl R. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari S, Harley V R, Pontiggia A, Goodfellow P N, Lovell-Badge R, Bianchi M E. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Nature (London) 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 24.Werner M H, Huth J R, Groneborn A M, Clore G M. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 25.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M E, Taraseviciene L, Nordeen S K, Allegretto E A, Edwards D P. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayaraman L, Moorthy N C, Murthy K G, Manley J L, Bustin M, Prives C. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwilling S, Konig H, Wirth T. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zappavigna V, Falciola L, Citterich M H, Mavilio F, Bianchi M E. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 29.Mosrin-Huaman C, Canaple L, Locker D, Decoville M. Dev Genet (Amsterdam) 1998;23:324–334. doi: 10.1002/(SICI)1520-6408(1998)23:4<324::AID-DVG7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Lehming N, McGuire S, Brickman J M, Ptashne M. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du W, Thanos D, Maniatis T. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 32.Thanos D, Maniatis T. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 33.Saha S, Brickman J M, Lehming N, Ptashne M. Nature (London) 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- 34.Muller C W, Rey F A, Sodeoka M, Verdine G L, Harrison S C. Nature (London) 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 35.Luckow B, Schutz G. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ip Y T, Reach M, Engstrom L K, Gonzalez-Crespo S, Tatei K, Levine M. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 37.Kirov N C, Lieberman P M, Rushlow C. EMBO J. 1996;15:7079–7087. [PMC free article] [PubMed] [Google Scholar]
- 38.Onate S A, Prendergast P, Wagner J P, Nissen M, Reeves R, Pettijohn D E, Edwards D E. Mol Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]