Abstract
Wnt and Shh signaling pathways are critical for the development and maturation of many epithelial tissues. Both pathways have roles in stem cell maintenance, tissue development, and tumorigenesis. However, linkage between these pathways in mammalian systems had not been well established. Here, we report that Shh expression in fungiform papillae and formation of normal mature fungiform papillae depend on signaling through Wnt and β-catenin. We observed that during fungiform papilla formation in mice, Shh and components of the Wnt/β-catenin signaling pathway are expressed together in the developing placode. The elimination of Wnt/β-catenin signaling in either Lef1 or Wnt10b knockout mice resulted in down-regulation of Shh expression. In addition, the size and number of fungiform papillae were greatly reduced in Lef1 knockout mice. By examining embryonic mouse tongues in culture we determined that activation of Wnt/β-catenin signaling up-regulates Shh expression. We observed that blocking Shh signaling in cultured tongue explants enhanced papillae formation and was accompanied by an up-regulation of Wnt/β-catenin signaling, indicating that Shh inhibits the Wnt/β-catenin pathway. Exogenously added Shh suppressed expression of endogenous Shh and inhibited Wnt/β-catenin signaling (assessed in TOPGAL mice), further implicating Shh as an inhibitor of the Wnt/β-catenin pathway. Our observations indicate that Wnt/β-catenin signaling and interactions between the Wnt and Shh pathways play essential roles in the development of fungiform papillae.
Keywords: β-catenin, Lef1, Wnt10b, fungiform papilla
Wnt and Hedgehog (Hh) signaling molecules are indispensable for normal development of diverse epithelial tissues such as the gastrointestinal tract, hair follicles and teeth (for review, see refs. 1–3). Ectopic activation or elevated expression of these signaling molecules leads to tumorigenesis, whereas their inhibition causes developmental abnormalities in a variety of tissues (4, 5). Although both signaling pathways have been implicated often where epithelial–mesenchymal interactions take place (6, 7), demonstrations that these signaling pathways might interact or regulate each other are limited.
Taste tissue development in mice starts around embryonic day (E) 11.5, after the emergence of the tongue swelling on the floor of developing mandible (8, 9). This is followed by formation of the taste placode (E12.5), gustatory papillae (E13.5), and taste buds (around birth) (9). As with development of other epithelial tissues, formation and patterning of taste papillae are thought to be induced by epithelial–mesenchymal interactions (10). Expression of Shh, Bmp, Noggin, Fgf, and EGF is associated with taste papilla initiation and patterning, indicating the potential role of these factors in taste papilla development (11–14). Disruption of Shh signaling during the prenatal period by neutralizing antibody or by cyclopamine caused aberrant development and patterns of fungiform papillae (15, 16). Although Wnt signaling pathways are critical in the development of many epithelial tissues, they had not been examined in the developing taste system, nor had their interaction with Shh been previously examined in the developing taste papillae.
Results
Expression of Wnt/β-Catenin Signaling Components in Developing Taste Organ.
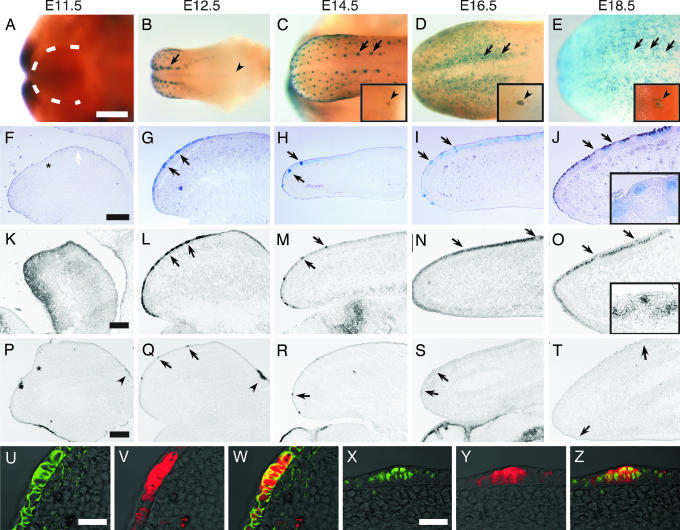
To determine whether Wnt signaling is associated with taste papilla formation in mice, we monitored Wnt/β-catenin signaling activity and expression of Wnt signaling molecules and Wnt-activated transcriptional factors in developing taste tissue. We first examined Wnt/β-catenin signaling in TOPGAL mice that carry a β-galactosidase (β-gal) reporter gene under the control of an inducible promoter that responds to the complex of β-catenin with Tcf/Lef1 transcription factors (17). At E11.5, the developing tongue presents as paired lateral swellings with no obvious Wnt/β-catenin signaling apparent, as shown by an examination of β-gal activity in TOPGAL mice (Fig. 1 A and F). β-gal activity in TOPGAL mice could first be seen in the developing dorsal tongue at E12.5, the developmental stage when placode formation is initiated (Fig. 1 B and G). This β-gal activity was present in multiple sets of rows and was strongest in the row closest to the median sulcus (Fig. 1B). β-gal activity in the fungiform papillae is most intense at E14.5 then declines in overall intensity (Fig. 1 C–E and H–J) to become highly restricted to a small region of epithelium where taste buds develop (Fig. 1J Inset). At later stages, β-gal activity appeared in developing filiform papillae (Fig. 1 D, E, I, and J) and in the circumvallate (CV) papilla (noted by arrow heads in Fig. 1 C–E Insets).
Fig. 1.
Wnt/β-catenin signaling elements are expressed in the developing tongue. Expression in the developing tongue of β-galactosidase (β-gal) (A–J, V, and Y), Lef1 (K–O and X), Wnt10b (P–T), and β-catenin (U) was detected by X-Gal staining in TOPGAL mice (A–E), by in situ hybridization (K–T), or by immunofluorescence (U–Z) in wild-type mice during developmental stages E11.5–E18.5. At E11.5, β-gal activity is absent from the lateral swelling (within dotted line in A and the white arrow in F). β-gal activity first appears in the placode of the dorsal tongue at E12.5 (B and G, arrows). Arrowheads in B and Insets in C–E indicate CV papillae. β-gal activity is absent from CV papillae until E14.5 (C). Lef1 expression is detected at E11.5 in both the mesenchyme and epithelium of the tongue (K), then increases in the fungiform papilla placode at E12.5 (L). At later stages, Lef1 expression continues in the fungiform papillae but at E16.5, and thereafter Lef1 also is highly expressed in the developing filiform papillae (N and O), whereas its expression still remains in fungiform papillae at later stages (O Inset) in accordance with β-gal expression (compare with J Inset). Wnt10b expression is first seen at E11.5–E12.5 in the developing CV papillae (P and Q, arrowheads), then at E12.5 in the developing placode (Q), and continues to be expressed through E14.5 (R), but its expression declines by E16.5 (S). Fluorescent micrographs of E12.5 tongue placode sections from TOPGAL mice show that β-gal expression (V and Y) associates with β-catenin (U) and Lef1 (X). W and Z are merged images of U and V and X and Y, respectively. Asterisks in F, K, and P indicate the border between the mandible and developing tongue. (Scale bars: A–E, 500 μm; F–T, 200 μm; U–Z, 100 μm.)
We next used in situ hybridization to monitor expression of Wnt-regulated transcription factors between E12.5 and E14.5, the critical time for formation of the placode and subsequently fungiform papillae. Of the four Wnt-activated transcription factors (Tcf1, Tcf3, Tcf4, and Lef1) examined during these critical stages, only Lef1 showed an expression pattern similar to that of the Wnt/β-catenin signaling detected by β-gal activity in TOPGAL mice [compare Lef1 (Fig. 1 L and M) with β-gal (Fig. 1 G and H)]. Like β-gal, Lef1 showed high expression in the developing dorsal tongue at E12.5 [compare Lef1 (Fig. 1L) with β-gal (Fig. 1G)]. Lef1 expression in the fungiform papillae remained high from E12.5 to E14.5 (Fig. 1 L and M), whereas Lef1 expression in filiform papillae started at E16.5 (Fig. 1N).
To confirm that β-gal expression in the dorsal tongue of TOPGAL mice faithfully reflected endogenous activation of Wnt/β-catenin signaling, tongues were sectioned and double immunostained with antibodies directed against β-gal and β-catenin (Fig. 1 U–W) or with antibodies against Lef1 and β-gal (Fig. 1 X–Z). Expression of Lef1 and stabilization of β-catenin in the E12.5 tongue was found to be coincident with expression of β-gal, independently identifying sites of Wnt/β-catenin signaling. Thus, the spatial–temporal pattern of Wnt/β-catenin signaling in the placode suggests that it may play a role in taste papilla development.
To find the endogenous ligand(s) that activate(s) Wnt/β-catenin signaling, we used in situ hybridization to monitor the expression of 15 Wnts (Wnt1, 2b, 3, 3a, 4, 5a, 5b, 6, 7a, 7b, 9a, 10a, 10b, 11, and 16) during the critical stages from E12.5 through E14.5 and found that only Wnt10b (Fig. 1 Q and R) showed an expression pattern similar to that of the Wnt/β-catenin signaling detected by β-gal activity in TOPGAL mice (see Fig. 1 B and C). Expression of Wnt10b in fungiform papillae generally paralleled that of β-gal, although we noted the following differences. First, after E16.5, expression of Wnt10b mRNA disappeared completely from fungiforma papillae, whereas expression of β-gal protein remained (compare Fig. 1 R–T with Fig. 1 H–J). Second, unlike β-gal, Wnt10b was not expressed in filiform papillae, indicating that Wnt10b has a temporal function in the development of fungiform but not filiform papillae.
Expression of Shh Parallels That of Wnt/β-Catenin Signaling Components.
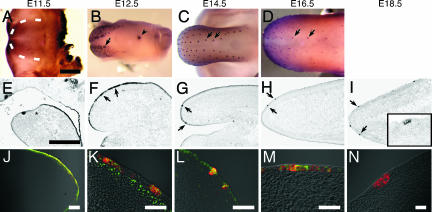
Multiple studies have shown that Shh is critical to the size, number and patterning of taste papillae during their development (11, 12, 15). Because the pattern of expression of Shh in fungiform papillae (18) appeared reminiscent of those we observed here with β-gal, Wnt10b, and Lef1 (Fig. 1), we carried out comparative studies of the expression of Shh and Wnt signaling components. The timing and restricted pattern of expression of Shh in fungiform papillae at early stages do appear to be quite similar to those of β-gal, Lef1, and Wnt10b (compare Fig. 2B and C with Fig. 1 B and C; also compare Fig. 2 F and G with Fig. 1 G, H, L, M, Q, and R). At later stages, expression of Shh, β-gal, and Lef1 in anterior tongue becomes similarly confined to circumscribed areas of epithelium, presumably where taste buds arise in the fungiform papillae (Insets in Figs. 1 J and O and 2I). By double immunostaining we determined that by E12.5, Shh and Lef1 (Fig. 2K) and Shh and β-gal (Fig. 2M) were coexpressed in the same cells of the developing fungiform papillae. The coexpression of β-gal and Wnt signaling components along with Shh in the same cells suggests the possibility of functional interaction of these signals during taste papilla formation (see below).
Fig. 2.
Shh and Wnt/β-catenin signaling elements are expressed similarly in the developing tongue. Expression in the developing tongue of Shh (A–I), Shh plus Lef1 (J–L), and Shh plus β-gal (M and N) was detected by whole-mount immunostaining (A–D), in situ hybridization (E–I), or double immunofluorescence (J–N) in wild-type (A–L) or TOPGAL (M and N) mice during developmental stages E11.5–E18.5. The dotted line in A indicates the border between the lateral swelling and the mandible. Arrows in B and F indicate the placode; arrows in C and D indicate fungiform papillae closest to the median sulcus. The arrowhead in B indicates the CV papilla. The Inset of I is a higher magnification view of a fungiform papilla. The pattern of expression of Shh is similar to that of β-gal during fungiform papillae formation (compare B–D with Fig. 1 B–D; also compare F–H with Fig. 1 G–I). The asterisk in E indicates the border between the mandible and developing tongue. Merged fluorescent micrographs of tongue placode sections from E12.0 (J), E12.5 (K and M), and E13.5 (L) stage mice show coexpression of Shh (red; J–N) with Lef1 (green; J–L), or β-gal (green; M). The CV papilla at E12.5 (N) expresses Shh (red) but not β-gal (green). The component single fluorescence images corresponding to merged images K–M are shown in supporting information (SI) Fig. 6. (Scale bars: A–I, 500 μm; J–N, 50 μm.)
Ablation of Wnt/β-Catenin Signaling Affects Fungiform Papillae Development.
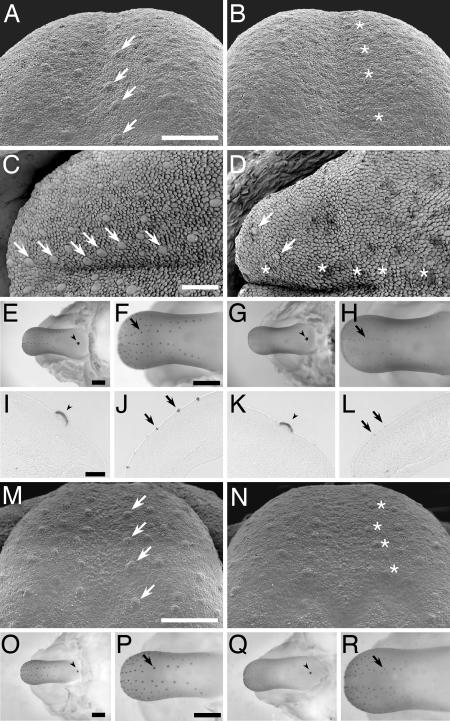
Expression of Lef1 and Wnt10b are often seen where inductive epithelial–mesenchymal interactions occur during development (e.g., hair follicles, teeth, mammary glands, and lung) (19–21). Lef1 null mice do not develop mature teeth, whiskers, or mammary glands and die shortly after birth (20). In the present study, we examined the role of Wnt/β-catenin signaling during taste papillae formation in Lef1 and Wnt10b null mice. At E14 the fungiform papillae of Lef1 null mice (Fig. 3B) were much smaller and less distinct [as assessed by scanning electron microscopy (SEM)] than were those of Lef1+/− heterozygous (Fig. 3A) or wild-type (data not shown) mice. However, fungiform papillae anlagen were present in the Lef1 null mice (asterisks in Fig. 3B). Furthermore, at postnatal day 0 the Lef1 null mice showed an even more severe phenotype in that almost all fungiform papillae were absent from the anterior half of the dorsal tongue (Fig. 3D); this contrasts markedly with the numerous papillae present in wild-type mice at this stage (Fig. 3C). Wnt10b null mice also displayed reduced size and numbers of fungiform papillae in comparison with those of wild-type mice (Fig. 3M and N), although their phenotype was somewhat milder than that of Lef1 null mice. The phenotypes of the Lef1 and Wnt10b null mice indicate that the Wnt/β-catenin pathway is necessary for the normal development of fungiform papillae. The numbers of apoptotic or proliferating cells found during fungiform papillae formation from E14.5 to E16.5 in Lef1 or Wnt10b null mice did not differ from those of either wild-type mice or of the corresponding heterozygous littermates (data not shown), suggesting that Wnt/β-catenin signaling may play a role in lineage specification.
Fig. 3.
Ablating the Wnt/β-catenin pathway disrupts fungiform papillae development and reduces Shh expression. Fungiform papillae in the developing tongue were examined by SEM of E14 (A, B, M, and N) or postnatal day 0 (C and D) stage mice. The fungiform papillae at E14 of either Lef1 or Wnt10b null mice are reduced in size (B and N) in comparison to those of Lef1+/− heterozygous (A) or wild-type (M) mice. Lef1 null mice at postnatal day 0 show a marked decrease in the size and number of fungiform papillae (D; asterisks indicate missing or atrophied papillae, and arrows indicate remaining papillae), in comparison to those of the wild-type mice at postnatal day 0 (C; arrows mark fungiform papillae). Shh expression in developing tongues from wild-type and null mice was monitored by immunohistochemistry in whole mounts of tongues and mandibles (E–H and O–R) and in sections (I–L). Reduced Shh expression was observed in the fungiform papillae of developing tongues from Lef1 null mice (G, H, and L) and Wnt10b null mice (Q and R) compared with wild-type mice (E, F, J, O, and P). Normal Shh expression was observed in the CV papillae of Lef1 null (G and K, arrowheads), Wnt10b null (Q, arrowhead), and wild-type (E, I, and O, arrowheads) mice. (A–L) C57B/6 mice. (M–R) FVB mice. (Scale bars: A–D, M, and N, 200 μm; E–H and O–R, 500 μm; I–L, 100 μm.)
Shh Expression in Fungiform Papillae Is Reduced by the Absence of Lef1 or Wnt10b.
Lef1 null mice showed dramatically reduced Shh protein expression in their fungiform papillae at E14.5 (Fig. 3 G, H, and L). Moreover, we observed that deletion of Wnt10b, the only Wnt ligand we found to be selectively expressed during fungiform papillae formation (see Fig. 1), caused similar effects on Shh expression and fungiform papillae formation as did the Lef1 null mutation [compare Fig. 3 Q and R (Wnt10b null) with O and P (wild type)]. Thus, the deletion of Lef1- or Wnt10b- dependent Wnt/β-catenin signaling severely diminishes the size and (eventually) the number of fungiform papillae, and decreases expression of Shh in fungiform papillae.
Although the absence of either Lef1 or Wnt10b in null mice had a profound effect on development of the fungiform papillae, these mutations had no such effect on the morphology of the CV papilla or the expression of Shh within that papilla (SI Fig. 7; also compare, respectively, Fig. 3 E, I, and O with G, K, and Q). This is consistent with previous observations that the addition of Shh inhibitors during papillae formation markedly affects the formation of fungiform but not CV papillae (15, 18). Together with our observation that expression in CV papilla of Lef1 and β-gal in TOPGAL mice starts at E14 (Fig. 1C and unpublished results), it would appear that the Wnt/β-catenin pathway does not contribute to the development of CV papilla before E14.
Wnt/β-Catenin Signaling Increases Fungiform Papillae Number.
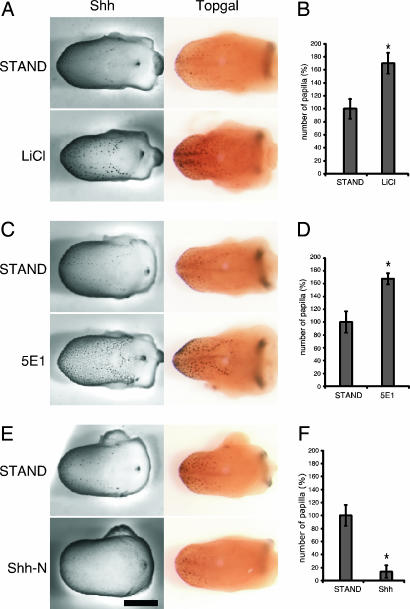
To more closely examine the interactions of Wnt and Shh signaling molecules during taste papilla development, we turned to organ culture of embryonic tongues. Rodent tongues in culture have been shown to closely mimic in vivo spatial and temporal stages of morphogenesis and molecular expression, and to support the development of taste papillae (22, 23). Because the expression of Shh and Wnt signaling components in mice is seen only after E12.5, we maintained stage E12.5 (or E11.5; data not shown) tongues in culture for up to 2 days under varying treatment conditions. When we treated cultured E12.5 tongues with LiCl, a treatment that activates Wnt/β-catenin signaling through inhibition of glycogen synthase kinase-3β (GSK3β) (24), we observed a dramatic increase in the number of fungiform papillae and number of Shh- and β-gal-expressing loci [compare tongues cultured under standard (STAND) vs. LiCl conditions] (Fig. 4 A and B). Control treatment with NaCl in place of LiCl did not alter Shh or β-gal expression (see SI Fig. 8), demonstrating the specificity of LiCl and indicating that it is activation of Wnt/β-catenin signaling by LiCl that increased both fungiform papillae number and their expression of Shh. Using in situ hybridization, we observed an increase in Shh mRNA after LiCl stimulation (data not shown), indicating that the up-regulation of Shh does not occur at the translational/posttranslational level. These results, in conjunction with the in vivo results from the Lef1 null mice, suggest that activation of Wnt/β-catenin signaling is necessary for both fungiform papillae formation and their expression of Shh.
Fig. 4.
Wnt and Shh interact in the developing tongue. Organ culture of E12.5 tongues from wild-type (Left) or TOPGAL (Right) mice treated for 2 days in culture with 5 mM LiCl (A), 50 μg/ml anti-Shh antibody (5E1) (C), or 2.5 μg/ml Shh-N (E) and examined for expression of Shh and β-gal in fungiform papillae. Control tongues (A, C, and E) were cultured for 2 days under standard culture conditions (STAND). Treatment with LiCl (A) or the anti-Shh antibody (C) up-regulated expression of both Shh and β-gal. Treatment with Shh-N (E) nearly abolished expression of Shh and down-regulated expression of β-gal. (B, D, and F) Histograms of the percent increase/decrease of the number of the Shh-immunoreactive fungiform papillae in treated vs. standard culture conditions, corresponding to images in A, C, and E, respectively (n = 6–7; ∗, P < 0.001; means ± SD). (Scale bar: 0.5 mm.)
Inhibition of Shh Signaling Increases Wnt/β-Catenin Signaling and Fungiform Papillae Formation.
Previous reports with rodent tongue cultures showed that neutralizing Shh with a monoclonal antibody (5E1) or blocking Shh's activity with cylcopamine enhanced taste papillae formation (both size and number) accompanied by increased expression of Shh (15, 16, 18). Because we have seen coexpression of Wnt signaling elements and Shh, and because we determined that Wnt/β-catenin signaling up-regulates Shh expression, we set out to more closely examine interactions of these two signaling pathways. Blocking Shh function in TOPGAL mice with either the 5E1 anti-Shh antibody or with cyclopamine both elevated levels of Shh expression and enhanced expression of β-gal in fungiform papillae (Fig. 4 C and D and SI Fig. 9).
These results suggest that endogenous Shh may inhibit the Wnt/β-catenin signaling pathway: In turn, inhibition of Wnt by Shh would provide negative feedback regulation of Shh expression. As an initial test of this hypothesis, we treated cultured embryonic tongues with the purified N-terminal domain of mouse Shh (Shh-N), which is known to possess the full biological activity of Shh (25, 26), then examined its effects on Shh expression and Wnt/β-catenin signaling. We found that culturing E12.5 tongues for 2 days with Shh-N nearly abolished Shh expression in fungiform papillae (Fig. 4 E and F). By in situ hybridization we determined that this suppression of Shh was effected by decreased mRNA levels (SI Fig. 10). In addition, E12.5 tongues cultured in the presence of Shh-N had significantly reduced β-gal expression (i.e., Wnt/β-catenin signaling) (Fig. 4E). Together, these results suggest that Shh could directly or indirectly block fungiform papilla formation by inhibiting Wnt/β-catenin signaling.
To further test our hypothesis that endogenous Shh inhibits the Wnt/β-catenin signaling pathway during fungiform papillae development, we examined effects of modulating Shh expression in cultured tongues from Lef1 null embryos. In contrast to our results with wild-type mice, wherein the addition of Shh inhibitors markedly elevated Shh expression in fungiform papillae (Fig. 4 C and D), there was no such effect on embryonic tongues from Lef1 null mice in culture from the addition of 5E1 (SI Fig. 11) or cyclopamine (data not shown).
Discussion
Wnts and Wnt signaling pathways are well known to be critical for the development of many epithelial tissues; however, they had not previously been known to play a role in the developing taste system. We initially detected Wnt/β-catenin signaling during fungiform papilla development by the use of TOPGAL reporter mice. We then confirmed expression of Wnt ligands in developing fungiform papillae by in situ hybridization and immunohistochemistry. Among the several Wnt ligands tested for expression in the developing taste organ, only Wnt10b was found to be selectively expressed in developing taste papillae where β-gal expression was detected in TOPGAL mice. Similarly, Lef1 was the only transcription factor among the Wnt-activated family of transcription factors to display such a pattern of expression. Thus, Lef1 and Wnt10b are the likely transcription factor and ligand, respectively, mediating Wnt/β-catenin signaling during fungiform papillae formation. Although Wnt10b has not been well characterized biochemically, there are several examples demonstrating Wnt10b's function as a ligand for the Wnt/β-catenin signaling pathway (27–30). In addition, we have confirmed that Wnt10b induces double axis formation when expressed in Xenopus embryos (K.I. and R.F.M., unpublished observations).
Shh is well documented to play an important role in the development of fungiform papillae (18). Yet, the underlying mechanism whereby Shh regulates taste papilla development was not known; nor was it known how disruption of Shh during fungiform papilla development led to increased expression of Shh. From our studies using knockout mice, we observed that ablation of Wnt/β-catenin signaling (by elimination of either Lef1 or Wnt10b) disrupted development of fungiform papillae and greatly reduced Shh expression therein. Also, up-regulation of the Wnt/β-catenin signaling by LiCl increased the number of fungiform papillae and their expression of Shh while addition of Shh suppressed Wnt/β-catenin signaling during fungiform papillae development in experiments using tongue cultures. These results indicate that interactions between Wnt and Shh signaling pathways play a role in fungiform papilla development.
There are precedents from developmental systems for interconnections between Wnt and Shh pathways. Experiments in Drosophila have shown that Wnt and Shh interactions occur in the embryonic epidermis and in the larval leg imaginal disk (31, 32). In the developing vertebrate spinal cord, interactions have been reported between Shh and Wnt signaling pathways in regulating patterning and cell specifications (32, 33). In mammalian systems, Lef1 null mice had been observed to have reduced expression of Shh in the developing tooth germ (34), and β-catenin mutant mice showed reduced Shh expression in hair follicles (35). In another example, Shh was shown to suppress Wnt10b mRNA expression in embryonic mandible cultures (36). Furthermore, it is intriguing that overexpression of active β-catenin causes ectopic expression of Shh during formation of hair follicles in mice and feathers in chickens (37–39).
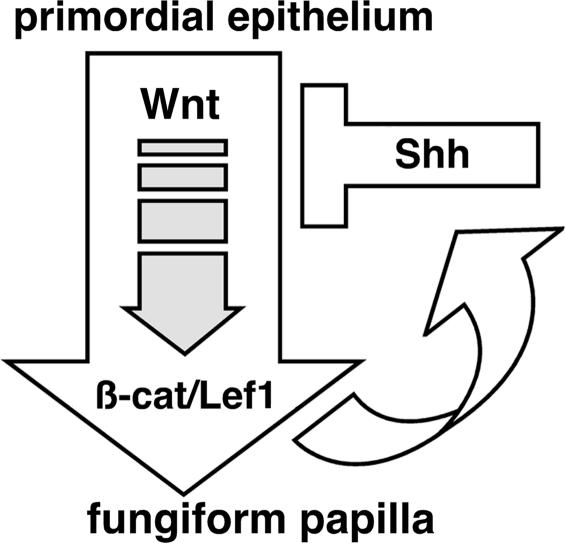
We think it likely that Wnt/β-catenin signaling regulates Shh expression during development in fungiform papillae and in other epithelial tissues, and that in turn Shh suppresses the Wnt/β-catenin pathway in these tissues. At present we do not know whether Shh directly or indirectly suppresses the Wnt/β-catenin pathway, although we note that there are multiple Lef/Tcf binding sites within the upstream region of the mouse Shh gene (data not shown), suggesting that up-regulation of Shh mRNA may be a direct effect of activation of the Wnt/β-catenin pathway. We have developed a schematic model (Fig. 5) to describe the positive and negative feedback loops that may serve to coordinate Wnt and Shh pathways during normal development of fungiform papillae. In our model, Wnt/β-catenin signaling plays a central role in regulating Shh expression and the development of fungiform papillae, while Shh itself acts as an inhibitor of Wnt/β-catenin signaling.
Fig. 5.
Schematic diagram of Wnt–Shh regulatory loop during fungiform papillae formation.
In summary, our work indicates that Wnt and Shh signaling pathways are both essential for proper development of fungiform papillae, and that these two pathways interact via feedback circuits during the development of fungiform papillae.
Materials and Methods
Animals.
TOPGAL mice, originally generated by E. Fuchs (17), were purchased from The Jackson Laboratory and maintained in the CD-1 background. Lef1 null mice are described in ref. 20. Wnt10b−/− mice were created by T. F. Lane and P. Leder (29, 40). For timed pregnancies, the day that sperm were observed in the vaginal smear was designated 0.5 days postcoitum (E0.5).
X-Gal Staining.
Tongues from TOPGAL mice at different developmental stages (E11.5, E12.5, E14.5, E16.5, and E18.5) were fixed in 0.5% glutaraldehyde in PBS for 10 min with rocking, washed in PBS, transferred into freshly prepared X-Gal solution (2.0 mM MgCl2/0.01% sodium deoxycholate/0.02% Nonidet P-40/5 mM potassium ferricyanide/5 mM potassium ferrocyanide), and incubated at 37°C for 4 h. Stained tissues were photographed with a Zeiss dissecting microscope equipped with a Zeiss AxioCam HS digital camera system.
Immunohistochemistry.
Antibodies used were rabbit anti-Lef1 antibody (20), goat anti-Shh-N (R & D Systems), anti-β-catenin (BD Biosciences), and anti-β-galactosidase (BioTrend). Tongues were fixed for 2 h at 4°C in 4% paraformaldehyde in PBS, transferred to 30% sucrose in PBS, and stored at 4°C overnight. Frozen sections (12 μm) were rinsed in PBS and blocked in 0.3% Triton X-100 and 2% donkey serum in PBS for 1 h at room temperature. The primary antibodies were added to the section in a humidified chamber and incubated overnight at 4°C. Sections were washed in PBS, incubated with the secondary antibodies (Alexa Fluor 488 or 594 IgG against primary antibodies from Molecular Probes/Invitrogen; incubation continued for 60 min), and rinsed in PBS. For immunodetection in whole embryonic tongue, tongues in organ culture were fixed in 4% paraformaldehyde for 2 h and incubated in 6% H2O2 for 2 h. After antigen retrieval with antigen unmasking solution (Vector Laboratories) for 10 min at 98°C, tongues were blocked with 0.3% Triton X-100 and 2% goat serum for 1 h at room temperature. Tongues were incubated overnight at 4°C with the antibodies against Shh-N (1 μg/ml). After washing with 0.1% Triton X-100, tongues were treated with the VECTASTAIN ABC kit for goat (Vector Laboratories), and the Biotin-Streptavidin-HRP complex was detected by DAB substrate kit for peroxidase (Vector Laboratories). The peroxidase reaction was stopped by rinsing with PBS followed by 2% paraformaldehyde fixation. Stained tissues were photographed as described above. Numbers of Shh-immunoreactive fungiform papillae were analyzed statistically across the control and experimental conditions by using the unpaired t test. A P value of <0.05 was considered significant.
In Situ Hybridization.
Shh cDNA was from H. Kimura (St. Jude Children's Research Hospital, Memphis, TN). Wnt10b cDNA was from D. Agalliu (Columbia University, New York, NY). Lef1 cDNA was cloned by RT-PCR using cDNA from CV papillae. Tongue tissues frozen in OCT compound from E11.5, E12.5, E14.5, E16.5, and E18.5 stage mice were cut into sections (12 μm) and subjected to postfixation (10 min in 4% paraformaldehyde, followed by acetylation in acetic anhydride for 10 min). After three washes in PBS, sections were prehybridized in hybridization buffer (5× SSC/50% formamide/1× Denhardt's solution/1 mg/ml salmon sperm DNA/1 mg/ml tRNA). Hybridizations were performed with digoxigenin-labeled cRNA probes in the hybridization buffer for 18 h at 72°C. Hybridization signals were detected by alkaline phosphatase-conjugated anti-digoxigenin antibodies plus NBT/BCIP substrate (Roche).
SEM Analysis.
SEM was performed as described in ref. 15.
Organ Culture of the Tongue.
Organ culture of tongues from rodent embryos was described in ref. 15. Embryonic tongues from E11.5 or E12.5 stage mice were cultured on Millipore Millicell-HA culture plate inserts in standard (STAND) culture medium [DMEM/F12 medium supplemented with 1% FBS, 2% B27 culture supplement (GIBCO/Invitrogen), and 50 μg/ml gentamicin sulfate] for 2 days. To activate Wnt/β-catenin signaling, 5 mM LiCl was supplemented to the STAND medium. To disrupt Shh signaling, either Shh-blocking antibody 5E1 (50 μg/ml; Developmental Studies Hybridoma Bank) or cyclopamine (5 μM; Toronto Research Chemicals) was added to the STAND medium. To induce Shh signaling, 2.5 μg/ml Shh N-terminal peptide (Shh-N; R & D Systems) was supplemented to the STAND medium.
Supplementary Material
Acknowledgments
We thank Dr. H. Kimura for Shh cDNA, Dr. D. Agalliu for Wnt10b cDNA, and N. Haremaki and R. Takubo for technical support. This work was supported in part by National Institutes of Health Grants DC003055 (to R.F.M.), DC003155 (to R.F.M.), and DC00456 (to C.M.M.) and a fellowship from the Japan Society for the Promotion of Science (to K.I.).
Abbreviations
- CV
circumvallate
- En
embryonic day n.
Footnotes
Conflict of interest statement: R.F.M. has a personal financial interest in the form of stock ownership in Linguagen Corp., receives consulting fees from Linguagen Corp., and is an inventor on patents and patent applications in the area of taste signal transduction that have been licensed to Linguagen Corp. Linguagen Corp. carries out work in the area of taste modification and taste signaling. The work described in the present manuscript would appear to be only peripherally related to the interests of Linguagen Corp.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607399104/DC1.
References
- 1.Hooper JE, Scott MP. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 2.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 3.Gregorieff A, Clevers H. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 4.Taipale J, Beachy PA. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 5.McMahon AP, Ingham PW, Tabin CJ. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 6.Bitgood MJ, McMahon AP. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 7.Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. Mech Dev. 2001;107:69–82. doi: 10.1016/s0925-4773(01)00452-x. [DOI] [PubMed] [Google Scholar]
- 8.Paulson RB, Hayes TG, Sucheston ME. J Craniofac Genet Dev Biol. 1985;5:59–73. [PubMed] [Google Scholar]
- 9.Kaufman MH. The Atlas of Mouse Development. London: Academic; 1992. [Google Scholar]
- 10.Mistretta CM. Ann NY Acad Sci. 1998;855:1–13. doi: 10.1111/j.1749-6632.1998.tb10542.x. [DOI] [PubMed] [Google Scholar]
- 11.Jung HS, Oropeza V, Thesleff I. Mech Dev. 1999;81:179–182. doi: 10.1016/s0925-4773(98)00234-2. [DOI] [PubMed] [Google Scholar]
- 12.Hall JM, Hooper JE, Finger TE. J Comp Neurol. 1999;406:143–155. doi: 10.1002/(sici)1096-9861(19990405)406:2<143::aid-cne1>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Liu HX, Mistretta CM. Dev Biol. 2006;297:198–213. doi: 10.1016/j.ydbio.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Mistretta CM, Grigaliunas A, Liu HX. Chem Senses. 2005;30(1):i52–i53. doi: 10.1093/chemse/bjh109. [DOI] [PubMed] [Google Scholar]
- 15.Mistretta CM, Liu HX, Gaffield W, MacCallum DK. Dev Biol. 2003;254:1–18. doi: 10.1016/s0012-1606(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 16.Hall JM, Bell ML, Finger TE. Dev Biol. 2003;255:263–277. doi: 10.1016/s0012-1606(02)00048-9. [DOI] [PubMed] [Google Scholar]
- 17.DasGupta R, Fuchs E. Development (Cambridge, UK) 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 18.Liu HX, Maccallum DK, Edwards C, Gaffield W, Mistretta CM. Dev Biol. 2004;276:280–300. doi: 10.1016/j.ydbio.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 19.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Development (Cambridge, UK) 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 20.van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 21.Zhou P, Byrne C, Jacobs J, Fuchs E. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 22.Mbiene JP, Maccallum DK, Mistretta CM. J Comp Neurol. 1997;377:324–340. doi: 10.1002/(sici)1096-9861(19970120)377:3<324::aid-cne2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Nosrat CA, MacCallum DK, Mistretta CM. Cell Tissue Res. 2001;303:35–45. doi: 10.1007/s004410000271. [DOI] [PubMed] [Google Scholar]
- 24.Klein PS, Melton DA. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 26.Ingham PW, McMahon AP. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 27.Lane TF, Leder P. Oncogene. 1997;15:2133–2144. doi: 10.1038/sj.onc.1201593. [DOI] [PubMed] [Google Scholar]
- 28.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 29.Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Proc Natl Acad Sci USA. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouji Y, Yoshikawa M, Shiroi A, Ishizaka S. Biochem Biophys Res Commun. 2006;342:1063–1069. doi: 10.1016/j.bbrc.2006.02.028. [DOI] [PubMed] [Google Scholar]
- 31.Heemskerk J, DiNardo S. Cell. 1994;76:449–460. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 32.Roelink H. Curr Opin Neurobiol. 1996;6:33–40. doi: 10.1016/s0959-4388(96)80006-7. [DOI] [PubMed] [Google Scholar]
- 33.Lei Q, Jeong Y, Misra K, Li S, Zelman AK, Epstein DJ, Matise MP. Dev Cell. 2006;11:325–337. doi: 10.1016/j.devcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. Genes Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- 35.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 36.Dassule HR, McMahon AP. Dev Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- 37.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 38.Silva-Vargas V, Lo Celso C, Giangreco A, Ofstad T, Prowse DM, Braun KM, Watt FM. Dev Cell. 2005;9:121–131. doi: 10.1016/j.devcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Noramly S, Freeman A, Morgan BA. Development (Cambridge, UK) 1999;126:3509–3521. doi: 10.1242/dev.126.16.3509. [DOI] [PubMed] [Google Scholar]
- 40.Vertino AM, Taylor-Jones JM, Longo KA, Bearden ED, Lane TF, McGehee RE, Jr, MacDougald OA, Peterson CA. Mol Biol Cell. 2005;16:2039–2048. doi: 10.1091/mbc.E04-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.