Abstract
When yeast cells are grown continuously at high cell density, a respiratory oscillation percolates throughout the population. Many essential cellular functions have been shown to be separated temporally during each cycle; however, the regulatory mechanisms involved in oscillatory dynamics remain to be elucidated. Through GC-MS analysis we found that the majority of metabolites show oscillatory dynamics, with 70% of the identified metabolite concentrations peaking in conjunction with NAD(P)H. Through statistical analyses of microarray data, we identified that biosynthetic events have a defined order, and this program is initiated when respiration rates are increasing. We then combined metabolic, transcriptional data and statistical analyses of transcription factor activity, identified the top oscillatory parameters, and filtered a large-scale yeast interaction network according to these parameters. The analyses and controlled experimental perturbation provided evidence that a transcriptional complex formed part of the timing circuit for biosynthetic, reductive, and cell cycle programs in the cell. This circuitry does not act in isolation because both have strong translational, proteomic, and metabolic regulatory mechanisms. Our data lead us to conclude that the regulation of the respiratory oscillation revolves around coupled subgraphs containing large numbers of proteins and metabolites, with a potential to oscillate, and no definable hierarchy, i.e., heterarchical control.
Keywords: metabolic regulation, respiratory oscillation, temporal structure, transcriptional regulation, self-organization
As we obtain a greater understanding of systems dynamics, it is becoming apparent that biological oscillators play critical roles in the organization of physiology in all time scales, ranging from milliseconds to years (1, 2). At the end of yeast growth in batch culture, there is series of cycles in respiratory activity that occur before the culture entering stationary phase (3). When continuous culture is initiated, the culture can be maintained in this oscillatory state [40 min to 5 h; see supporting information (SI) Fig. 5] for months (4–6). These dynamics result in the temporal separation of many essential cellular functions, including redox biochemistry (7, 8), transcription (9, 10), energetics (11), chromosome cycle (1), and mitochondrial function (5). Although respiration is always active, the cellular redox state cycles between an oxidative phase and reductive phase. It is known that at least two redox active compounds, acetaldehyde and H2S, mediate cell–cell communication (12), but little is known regarding how synchrony is generated within the cell and how the network is regulated.
A major problem in elucidating the oscillatory mechanism is the extent to which the cellular network is entrained, i.e., in a system where the majority of parameters oscillate how does one pull out core mechanisms. Fortunately, Saccharomyces cerevisiae has been used extensively as a proving ground for many of the new low- and high-throughput tools in modern molecular biology, leading to yeast having the best characterized cellular network among all eukaryotes. We combined this information with our data to address the regulation of oscillatory dynamics. Initially we used GC-MS data and statistical analyses of transcriptional data to identify the operation of a defined biosynthetic program during the oscillation. We then developed a simple fast Fourier transform-based algorithm to calculate a parameters correlation to a sine wave and phase of peak concentration for data from a number of experimental sources and the statistical analyses of data (13). The top oscillating components were visualized by using a reconstructed large-scale network that encompassed much of the current understanding of yeast physiology. We then tested one of the most significant network features that coordinates amino acid biosynthesis, cellular reduction, cell division and respiration, by perturbation.
Results
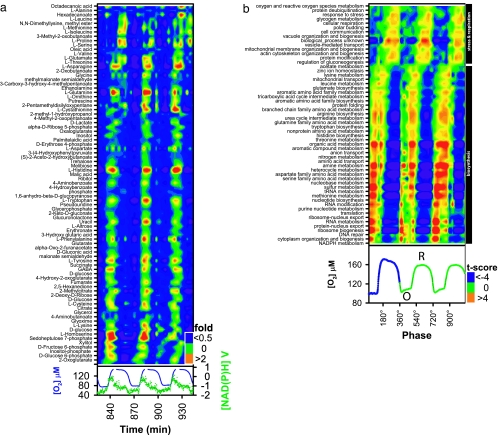
Previously we have shown that many metabolites oscillate with a phase relationship to respiration (5, 7, 12). We expanded these data by using GC-MS to show that this oscillation is metabolome-wide (Fig. 1a). Furthermore, most metabolite concentrations (70%) peak during the transition from the oxidative to reductive phase (Fig. 1a), in coincidence with the maximum in NAD(P)H concentration and the maximum DNA synthesis rate (14). The transcripts whose abundance peaked during the oxidative phase were involved in biosynthesis. Statistically grouping these transcripts according to ontology (Fig. 1b) revealed that there was a defined sequence of events or biosynthetic program between 35° and 120° (Fig. 1b). During the reductive phase of the cycle transcript, abundance was enriched with transcripts responsible for respiration, mitochondrial biogenesis, and actin arrangement. This production was compatible with our previous observation that mitochondrial cristae oscillate between orthodox (oxidative phase) and a condensed (reductive phase) state during a respiratory cycle (11). From these data we estimate that the metabolite concentrations peak 100–150° (12–18 min) after the peak in transcriptional activation of the pathway.
Fig. 1.
Metabolic orchestration in oscillatory yeast continuous cultures. (a) The GC-MS analysis of the respiratory oscillation observed during continuous culture indicates the entire metabolome oscillates. For visualization, the data have been scaled to show the fold change. (Lower) Dissolved oxygen trace (blue) and the [NAD(P)H] trace (green) for the sample period. (b) (Lower) The respiratory oscillation consists of the cyclic switching between phases of low [oxidative (O), high oxygen uptake rate] and high [reductive (R), low oxygen uptake rate] oxygen concentrations. (Upper) The transcriptional program was visualized as a heat map constructed from the statistical analyses of gene functional ontology (13). The values were then plotted against oscillation phase. Biosynthetic processes and respiratory/stress events were clearly separated during the oscillation (black boxes), occurring (120–180°) out of phase with their respective phenotype of biosynthetic (a) and respiratory events (b Lower). Both heat maps were ordered according to the phase angle (ϕ) of the measurements' peak production.
Using fold changes to gauge how an individual parameter influences a system underestimates the importance of certain key molecules, such as transcription factors on cell physiology. Furthermore, it is difficult to integrate data from numerous experiments to produce a coherent view of complex phenomena. However, when one has densely populated time series data, one can use signal-processing techniques to derive dynamics. Here, we use a simple Fourier technique to derive two measures from a variety of raw data sets (SI Fig. 6). The peak production time of the waveform is represented by fast Fourier transform-derived phase angles (ϕ), where 0° represents the minimum first derivative of the dissolved oxygen measurements, where one cycle typically lasted ≈40 min. The correlation to sine wave or oscillation strength (O) for each measured parameter was also calculated by dividing the amplitude derived by fast Fourier transformation by the integrated raw data (SI Fig. 7). We reasoned that if the parameter had a high O, then it was likely to be coupled more closely to the oscillation regulatory loop, i.e., the further the target is away from the oscillating loop, the more the transcript would deviate from a sinusoidal waveform.
Transcriptional Core Involved in Respiratory Dynamics.
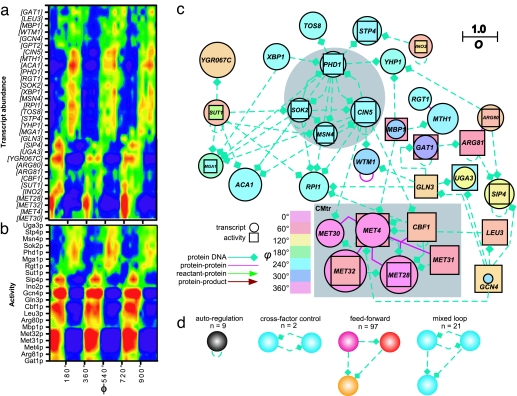
Next we asked what factors regulate the operation of the biosynthetic program. To approach this question, we reconstructed a network consisting of transcriptional regulatory and protein interactions (for details on the construction of this network, see SI Methods) that was filtered according to the oscillation strength of the transcript (O ≥ 0.75; 861 transcripts; 33 transcriptional regulators; Fig. 2a and SI Table 1) and the oscillation strength of the transcription factor activities (O ≥ 0.75; 21 transcriptional regulators; Fig. 2b) derived from statistical analyses of their targets. The filtered connected network of 312 nodes and 519 edges showed strongest oscillatory behavior. The network was further filtered to focus on the core connected transcriptional regulatory nodes and complexes (33 nodes, 74 edges; Fig. 2c). This regulatory network consisted of two out-of-phase subgraphs whose transcripts were produced in tight temporal windows.
Fig. 2.
Transcriptional regulation of the yeast respiratory oscillation. (a) The transcriptional regulatory core network involved in the regulation of the yeast respiratory oscillation was derived from the top oscillating transcript concentrations (O ≥ 0.75). (b) Activities of the transcriptional regulators were derived from the statistical analysis of their target transcripts (O ≥ 0.75). Both heat maps were ordered according to the phase angle (ϕ) of the measurements' peak production. A ball-and-stick network representation of the transcriptional regulators shown in a and b. (c) The figure key provides a guide to the network. Colored circles represent transcript abundance, and squares represent the transcription factor activity. If the transcription factor activity of a node was greater than its transcript concentration, the square was placed behind the circle, otherwise the square was placed in front of the circle. The nodes are colored according to the phase angle (ϕ; see Fig. 1b), and the oscillation strength (O) is indicated by the size of the node. The shaded box indicates the CMtr complex, and the shaded circle shows an area enriched with cross-factor control loops. (d) The gene network motifs (P < 0.01) were scored for quality (Z >7) where the number of nodes was 3. The color of the circle represents what phase in which the motif is enriched; where autoregulatory units are distributed throughout all phases they are colored black.
The gene network fanning out from the transcriptional regulatory network was then analyzed statistically for 2- and 3-node gene network motifs (15) (see SI Methods and SI Table 1). These small gene networks, in combination, are thought to regulate more complex responses (16). In the top oscillating subgraph of this network, we identified significant autoregulation, cross-factor control, feedforward, and mixed-loop control (Fig. 2d) (17). However the exact physiological meaning of these circuits can be vague because each motif exhibits a wide range of dynamics depending on interaction strength and regulation type, i.e., inhibitor or activator (18). We approached this problem by constructing a network containing much of our understanding of DNA, protein, ion, and metabolite interactions in yeast (4,912 nodes, 16,723 edges). We used this network and the oscillation data to probe for functionality. The resulting oscillatory network (O ≥ 0.75) comprised 20.8% of all of the nodes in the reconstructed yeast network (SI Fig. 6).
Transcriptional Regulation of Cellular Reduction.
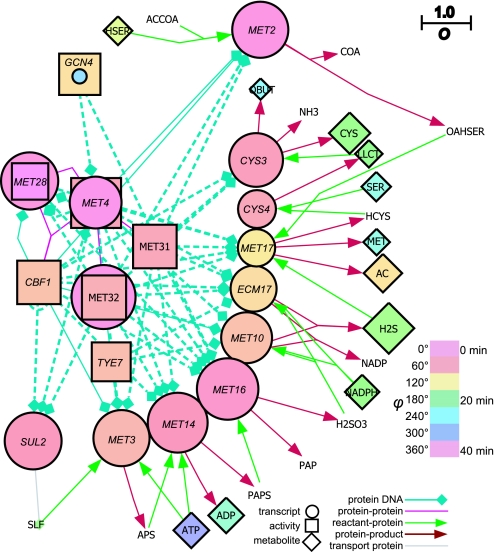
The Cbf1–Met4–Met28–Met31–Met32 transcription regulatory complex (CMtr complex for brevity) is a network feature (Fig. 2c, gray box) where the peak concentrations of MET4, MET28, MET30, and MET32 transcripts occurred in a narrow temporal window (≈4 min). Cbf1p activates this complex by transcription and physical interaction to regulate sulfur assimilation (19, 20). The transcription factor activity of Cbf1p, Met4p, Met28p, Met31p, and Met32p peaked ≈4–8 min after the maximum transcript concentration. The gene targets of the CMtr complex produced the largest amplitude oscillation of the measured transcripts; and the metabolic products, such as glutathione, hydrogen sulfide, and S-adenosylmethionine, of the target proteins of this complex are directly responsible for cellular reduction (21, 22). Therefore we examined the temporal organization of the sulfate assimilation pathway resulting from this complex (Fig. 3).
Fig. 3.
Network derived for sulfur assimilation from the top oscillating (O ≥0.750) transcripts and metabolites. The figure key provides a guide to the network. The gene descriptions are listed in SI Table 1. SLF, sulfate; LLCT, cystathione; HSER, homoserine; OASER, o-acetylhomoserine; OBUT, 2-oxobutanoate; AC, acetate.
The MET4, MET28, MET30, and MET32 peaked at ≈40°, followed by their target transcripts involved in initial sulfate activation by ATP (MET3, 72°; MET14, 60°; MET16, 53°). MET10 and ECM17 are responsible for H2S formation, and their transcript abundance peaked at 82° and 98°, respectively. MET17 catalyzes the formation of homocysteine from homoserine, and its transcript abundance peaked at 107°. Metabolically, the concentrations of homoserine, H2S, cystathione, and cysteine (products of homoserine) peaked at 135°, 178°, 186°, and 189°, respectively. Gene network analyses revealed that this temporal switching of transcripts probably occurs through multiple instances of the three nodes' feedforward control (Fig. 2d). In its simplest form, the target gene is only transcribed when two transcription factors are in place, which produces time delay circuits that were enriched in the CMtr complex; 65% of all incidences of the feedforward loop emanated from the CMtr complex.
Translational Control of Amino Acid Biosynthesis Timing.
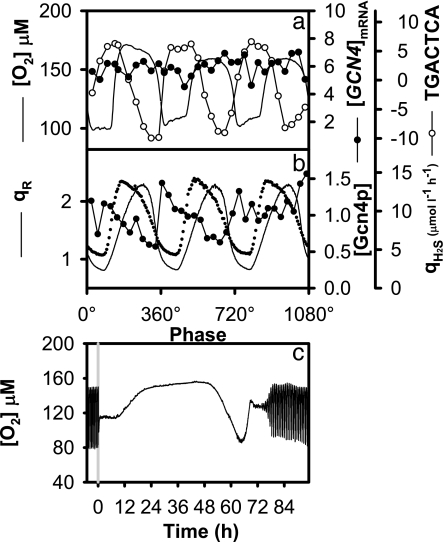
The GC-MS results presented here show distinct phase relationships between amino acid concentration and respiratory activity (Fig. 1a), where the maximum production of all detected amino acids was between 170° and 240°. The Gcn4p transcriptional activator is central to amino acid regulation and is involved in MET4 transcription (Fig. 2c) (23). The target-binding motif of Gcn4p (Fig. 4a), Gcn4p (Fig. 4b), and amino acids had strong oscillatory profiles (Fig. 1a), the [GCN4]mRNA produced a weak oscillation (O = 0.33). This apparent discrepancy may be explained by translational block of the GCN4 mRNA being blocked before translation (at μORF4) when intracellular concentrations of amino acids are high (24). The block relaxes when the intracellular concentration of amino acids decreases; therefore, the observed oscillation in intracellular amino acid concentrations creates a strong feedback on this transcriptional regulation circuitry. The activation of GCN4 translation was not the result of amino acids in the feed media becoming limited because extracellular amino acids were always below the limits of detection, and ammonia was always available for biosynthesis (25). Furthermore, addition of glutamate, cysteine, and threonine causes dose-dependent inhibition of respiration and growth (7, 26, 27), which is the opposite of what may be expected if the cell were limited by amino acids. In addition, the respiratory oscillation was sensitive to low concentrations of rapamycin, resulting in a switch to reductive phase for 60 h (Fig. 4c). Rapamycin induces the activators Gcn4p and Gat1p in a target of rapamycin (TOR) complex 1-dependent manner (24). These observations confirm that intracellular amino acid signaling plays a critical role in the regulation of the respiratory oscillation.
Fig. 4.
The dynamics of the Gen4p during the yeast respiratory oscillation. (a) The [GCN4] transcript showed a weak oscillation (O = 0.325); however, the involvement of the Gcn4p in the regulation of the respiratory oscillation in yeast was predicted by a strong oscillatory response of the TGACTCA consensus motif gene targets (O = 1.2). (b) The Gcn4p oscillated during the respiratory oscillation in phase with the TGACTCA consensus motif targets and out of phase with H2S production rate (qH2S) and respiratory quotient (qR). (c) The oscillatory dynamics were sensitive to rapamycin perturbation (200 nM).
Discussion
Once external noise has been minimized by using the precisely controlled conditions of continuous growth in a fermentor, the culture autosynchronizes physiology, to reveal a temperature-compensated oscillation (28). In this work we show the coordination of the oscillator includes core transcriptional regulation. Major outputs of the biosynthetic program were a global oscillation in amino acid biosynthesis (Fig. 1a), cellular reduction (7), and S phase initiation (9). The biosynthetic transcriptional program initiated during the oxidative phase (Fig. 1b) and our subsequent network analysis strongly suggest that the transition from oxidative to reductive phases of a cycle relies on small multiple-input gene feedforward motifs. In combination, these motifs produce time delays in transcription (Fig. 2).
The CMtr complex, whose activity peaks in the oxidative phase (Fig. 2c), produced a burst of cellular reduction tracked by sulfide and NAD(P)H concentrations. Critically, S phase initiation is regulated by the CMtr complex through production of the F box protein Met30p (29). Temporally, the Skp1/Cullin/F-box/Met30 ubiquitin ligase complex (SCFMet-30) complex acts before the G1 cyclins. A major function of this protein is to generate negative feedback by the deactivation of the Met4p through polyubiquitination, resulting in a down-regulation of S-adenosylmethionine, the major methyl donor in the cell, thus providing a strong oscillatory potential. The CMtr complex is conserved in eukaryotes and may constitute a more general redox state checkpoint in the chromosome cycle (30).
The oscillation in Gcn4p concentration and rapamycin sensitivity indicated that the CMtr complex was regulated by TOR complex 1 (24), which senses intracellular amino acid levels. The precise timing of amino acid biosynthesis involved a regulatory feedforward loop that contains the activators of amino acid biosynthesis Gln3p and Gat1p. The dynamics of this subgraph was recently explored in a computational model of amino acid synthesis regulation (31) and utilizes strong autoregulation with a strong potential to oscillate. Our data therefore indicated that Gcn4p and its translational regulation were major coordinating transcriptional regulators of the transition from oxidative to reductive phases.
Although we have focused on the regulation of the biosynthetic program, the majority (≈90%) of the mRNA species concentrations peaked during the reductive phase (9). The major oscillating cohort in this phase (≈1,500 genes) had no defined function (Fig. 1b). This poor functional classification may be partly explained by the utilization of glucose-repressed growth conditions where mitochondrial activity was greatly reduced for the majority of yeast studies (3). The core regulatory network coexpressed with this event includes an array of transcriptional regulatory hubs that were highly interconnected (Fig. 2c, gray circle). At its core are four transcription regulators (Fig. 2c) implicated in the regulation of complex phenotypic responses. The Phd1p is implicated in pseudohyphal regulation (32). Sok2p is implicated in sporulation and long-term yeast adaptation (33). Cin5p and Msn4p have a more general stress-sensing role (34). Msn4p activity by localization in the nucleus is influenced by protein kinase A (PKA) (35), and it has been suggested that Sok2p and Phd1p are involved in the regulation of cAMP metabolism (32, 36). In addition the cyclic mobilization of glycogen and oscillation in cAMP concentrations observed during oscillatory dynamics during growth on glucose media, supplies evidence that the PKA pathway is involved in the regulation of the oscillation (6). It is an attractive theory that cyclic mobilization of the reserve carbohydrate glycogen regulated by PKA in a cAMP-dependent manner, thus controlling energy metabolism, is the central oscillatory loop. However, the oscillation persists during growth on respiratory carbon sources, such as ethanol and acetaldehyde (37), whereas glycolytic activity was abolished. This persistence provides strong evidence that respiratory dynamics, not fermentative dynamics, are critical for oscillation. Also, although cAMP and glycogen oscillated when the cells were grown on glucose, no oscillation was observed in these metabolites when the cells were grown on ethanol media (38 and D.B.M., unpublished data). This finding does not rule out a role for the PKA pathway in controlling the respiratory oscillation because the TOR complex 1 and PKA signaling pathways cross-talk (39), and the PKA pathway can be phosphorylated independently of cAMP (40). The mechanistic elucidation of these transcriptional regulators is needed to elucidate their role during oscillatory dynamics.
Biological networks have been layered into genome, transcriptome, proteome, and metabolome, and these networks are then modularized. Within the cellular context, this hierarchy is at best misleading because intrinsic network fluctuations are quickly communicated throughout the highly interconnected network by molecular interactions, thus the network forms a complex heterarchy (41). The regulatory loops described here have the potential to oscillate, but in isolation they may never show stable oscillation. The regulation of the oscillatory dynamics by metabolic control of enzymes and the modification of proteins were not examined in detail in this work, but this omission does not preclude their contribution. Indeed, a purely metabolic computational model has been shown to reproduce limited aspects of the respiratory oscillation (42). Rather, it is only when metabolic, translational, transcriptional, and protein control are placed within a coupled network context that the depth of the cellular dynamics we observe can be explored. From this work, we conclude that regulation of the oscillator is not the result of a central oscillator, but rather it emerges from numerous subgraphs with the potential to oscillate, i.e., stable periodicity arises from arrays of small genetic feedforward loops, coupled together by metabolic and protein feedback loops to provide a temporal program (such as the events we described in biosynthesis). It has been postulated that this type of control is involved in circadian regulation (43), and it may be general strategy for the robust maintenance of cellular processes in eukaryotes against environmental perturbation. Indeed, there is already evidence from other eukaryotic systems such as the zebrafish development clock (44) and Protista (14) that this is the case.
Materials and Methods
Culture Techniques and Online Measurements.
S. cerevisiae strain IFO 0233 was maintained, precultured, and grown continuously on semidefined glucose medium as described previously (5) (for details, see SI Methods and SI Fig. 5). Continuous partial pressure of oxygen (PO2) and partial pressure of carbon dioxide (PCO2) off-gas measurements were carried out by using an Enoki-III analyzer (Figaro Engineering, Osaka, Japan). The partial pressure of hydrogen sulfide (PH2S) in the off-gas was measured continuously by using an electrode-based gas monitor (HSC-1050HL; Gastec, Ayase-City, Kanagawa, Japan). O2 uptake rates (qO2), CO2 production rates (qCO2), and H2S production rates (qH2S) were derived from these measurements (SI Methods). Instruments were calibrated according to the manufacturer's instructions. NAD(P)H was measured as described previously (7), except data were acquired by using a fast-acquisition USB-pod (PMD-1608; Measurement Computing, Norton, MA). Pulse additions (1 ml) of rapamycin (Wako Chemicals, Osaka, Japan) were carried out by syringe/sterile filtering (Teflon 0.2-μm pore; Millipore, Tokyo, Japan) a diluted stock solution (1 mg/ml ethanolic solution). Injections were carried out three times, and the data used show a representative perturbation. Respiratory quotient (qR) was defined as qCO2/qO2. Measurements of CO2, O2, and H2S were phase-delayed by 10.2 min to account for the measurement lag caused by the fermentor headspace (≈2 liters) and were accomplished by pulse-injecting a known concentration of the gas in question into the air supply line and subtracting the peak maxima from the time of addition.
Metabolite and Protein Measurements.
The metabolite data set used previous work (5, 7, 12, 21, 22, 25, 27, 28, 37, 38), and GC-MS analyses (SI Methods) of intracellular metabolites were used to construct the metabolite data set. All Western blots were carried out according to the manufacturer's instructions (Atto, Bunkyo-ku, Tokyo, Japan). Antibodies for Gcn4p were supplied by Santa Cruz Biotechnology (Santa Cruz, CA), and the reactions for antibody detection of proteins were carried out according to the manufacturer's instructions (Atto).
Statistical Analyses.
The analyses were carried out on each microarray time point (13). Generation of T scores used t tests to analyze for differences between groups of genes and global expression on the YS98 chip (Affymetrix, Santa Clara, CA). Before analysis, the data were scaled by dividing the transcript abundance values by the mean abundance for each experiment. All of the data were imported into MS Excel worksheets (SI Table 1). Small gene networks containing 3- and 4-node motifs were analyzed for significance and quality (15). The P value and the Z score were calculated by comparing the frequency of all occurrences of the target motif in a target network with the frequency values of the motif 1,000 random permutations of the target network. The resulting network motifs were imported into MS Excel worksheets (SI Table 1).
Signal Processing.
All calculations were carried out in MATLAB (version 6.5.1). The Affymetrix data set used was described previously by Klevecz et al. (ref. 8; Gene expression Omnibus, GSE2583). The use of the discrete Fourier transform to calculate oscillation strength from raw unscaled periodic data captured the correlation to a sine wave, therefore avoiding skewing data patterns inherent in traditional of normalization and preserving intensity information lost by scaling of high-throughput data and allowing the integration of data sets. These data included measurements of metabolites, low noise Affymetrix transcript profiles (9), and derived statistical analysis of transcription factor activity (13), online measurements, and in this study GC-MS. Therefore, we summarized waveforms by two measures: the time of maximum production (ϕ) and its correlation to a sine wave (O). The detailed methods used are shown in the SI Methods and SI Fig. 6. On the network graphs (Figs. 2 and 3), ϕ is represented by colored fills where light magenta represents 0° and light green indicates 180° (≈20 min after the minimum first derivative).
Visualization.
All graphs were constructed by using Sigmaplot 8.0 (Hulinks, Tokyo, Japan). To visualize the transcript and metabolite data, the data were first scaled to each experiment. Two independent experiments were used for the transcription data, one containing 10 microarrays and one containing 22 microarrays (the blue and green scatter plot in Fig. 1b Lower, respectively). Publicly available databases were downloaded and converted to Cytoscape (45) format before visualization (SI Methods).
Supplementary Material
Acknowledgments
D.B.M. and H.K. were supported by the Solution-Oriented Research for Science and Technology Agency to the Systems Biology Institute and the 21st Century Center of Excellence Program and Special Coordination Program of the Ministry of Education, Sports, Culture, Science, and Technology to Keio University.
Abbreviations
- CMtr complex
Cbf1–Met4–Met 28–Met31–Met32 transcription regulatory complex
- PKA
protein kinase A
- TOR
target of rapamycin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606677104/DC1.
References
- 1.Lloyd D, Murray DB. Trends Biochem Sci. 2005;30:373–377. doi: 10.1016/j.tibs.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Schibler U, Naef F. Curr Opin Cell Biol. 2005;17:223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Murray DB. Curr Genomics. 2004;5:665–671. [Google Scholar]
- 4.von Meyenburg K. Arch Microbiol. 1969;66:289–303. [Google Scholar]
- 5.Satroutdinov AD, Kuriyama H, Kobayashi H. FEMS Microbiol Lett. 1992;77:261–267. doi: 10.1016/0378-1097(92)90167-m. [DOI] [PubMed] [Google Scholar]
- 6.Xu Z, Tsurugi K. FEBS J. 2006;273:1696–1709. doi: 10.1111/j.1742-4658.2006.05201.x. [DOI] [PubMed] [Google Scholar]
- 7.Murray DB, Engelen F, Lloyd D, Kuriyama H. Microbiology. 1999;145:2739–2745. doi: 10.1099/00221287-145-10-2739. [DOI] [PubMed] [Google Scholar]
- 8.Murray DB, Engelen FA, Keulers M, Kuriyama H, Lloyd D. FEBS Lett. 1998;431:297–299. doi: 10.1016/s0014-5793(98)00777-7. [DOI] [PubMed] [Google Scholar]
- 9.Klevecz RR, Bolen J, Forrest G, Murray DB. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd D, Salgado LE, Turner MP, Suller MT, Murray D. Microbiology. 2002;148:3715–3724. doi: 10.1099/00221287-148-11-3715. [DOI] [PubMed] [Google Scholar]
- 12.Murray DB, Klevecz RR, Lloyd D. Exp Cell Res. 2003;287:10–15. doi: 10.1016/s0014-4827(03)00068-5. [DOI] [PubMed] [Google Scholar]
- 13.Boorsma A, Foat BC, Vis D, Klis F, Bussemaker HJ. Nucleic Acids Res. 2005;33:592–595. doi: 10.1093/nar/gki484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd D, Murray DB. FEBS Lett. 2006;580:2830–2835. doi: 10.1016/j.febslet.2006.02.066. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber F, Schwobbermeyer H. Bioinformatics. 2005;21:3572–3574. doi: 10.1093/bioinformatics/bti556. [DOI] [PubMed] [Google Scholar]
- 16.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 17.Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram PJ, Stumpf MP, Stark J. BMC Genomics. 2006;7:108. doi: 10.1186/1471-2164-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas D, Surdin-Kerjan Y. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent NA, Eibert SM, Mellor J. J Biol Chem. 2004;279:27116–27123. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- 21.Sohn H, Kuriyama H. Yeast. 2001;18:125–135. doi: 10.1002/1097-0061(20010130)18:2<125::AID-YEA655>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Sohn HY, Murray DB, Kuriyama H. Yeast. 2000;16:1185–1190. doi: 10.1002/1097-0061(20000930)16:13<1185::AID-YEA619>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 23.Mountain HA, Bystrom AS, Korch C. Mol Microbiol. 1993;7:215–228. doi: 10.1111/j.1365-2958.1993.tb01113.x. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch AG. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 25.Sohn H, Kuriyama H. Arch Microbiol. 2001;176:69–78. doi: 10.1007/s002030100295. [DOI] [PubMed] [Google Scholar]
- 26.Salgado LE, Murray DB, Lloyd D. Biol Rhythm Res. 2002;33:351–361. [Google Scholar]
- 27.Sohn HY, Kum EJ, Kwon GS, Jin I, Kuriyama H. J Microbiol. 2005;43:375–380. [PubMed] [Google Scholar]
- 28.Murray DB, Roller S, Kuriyama H, Lloyd D. J Bacteriol. 2001;183:7253–7259. doi: 10.1128/JB.183.24.7253-7259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser P, Flick K, Wittenberg C, Reed SI. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 30.Su NY, Flick K, Kaiser P. Mol Cell Biol. 2005;25:3875–3885. doi: 10.1128/MCB.25.10.3875-3885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boczko EM, Cooper TG, Gedeon T, Mischaikow K, Murdock DG, Pratap S, Wells KS. Proc Natl Acad Sci USA. 2005;102:5647–5652. doi: 10.1073/pnas.0501339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gimeno CJ, Fink GR. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vachova L, Devaux F, Kucerova H, Ricicova M, Jacq C, Palkova Z. J Biol Chem. 2004;279:37973–37981. doi: 10.1074/jbc.M404594200. [DOI] [PubMed] [Google Scholar]
- 34.Nevitt T, Pereira J, Rodrigues-Pousada C. Yeast. 2004;21:1365–1374. doi: 10.1002/yea.1188. [DOI] [PubMed] [Google Scholar]
- 35.Smith A, Ward MP, Garrett S. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan X, Heitman J. Mol Cell Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keulers M, Satroutdinov AD, Suzuki T, Kuriyama H. Yeast. 1996;12:673–682. doi: 10.1002/(sici)1097-0061(19960615)12:7<673::aid-yea958>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 38.Keulers M, Suzuki T, Satroutdinov AD, Kuriyama H. FEMS Microbiol Lett. 1996;142:253–258. doi: 10.1111/j.1574-6968.1996.tb08439.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen JC, Powers T. Curr Genet. 2006;49:281–293. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]
- 40.Lu A, Hirsch JP. Eukaryot Cell. 2005;4:1794–1800. doi: 10.1128/EC.4.11.1794-1800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yates FE. In: Self-Organizing Systems: The Emergence of Order (Life Science Monographs) Yates FE, editor. New York: Springer; 1987. pp. 445–457. [Google Scholar]
- 42.Wolf J, Sohn H, Heinrich R, Kuriyama H. FEBS Lett. 2001;499:230–234. doi: 10.1016/s0014-5793(01)02562-5. [DOI] [PubMed] [Google Scholar]
- 43.Roenneberg T, Merrow M. J Biol Rhythms. 2002;17:495–505. doi: 10.1177/0748730402238231. [DOI] [PubMed] [Google Scholar]
- 44.Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.