Abstract
Werner syndrome (WS) is a rare human premature aging disease caused by mutations in the gene encoding the RecQ helicase WRN. In addition to the aging features, this disorder is marked by genomic instability, associated with an elevated incidence of cancer. Several lines of evidence suggest that telomere dysfunction is associated with the aging phenotype of the syndrome; however, the origin of the genomic instability observed in WS cells and the reason for the high incidence of cancer in WS have not been established. We previously proposed that WRN helicase activity was necessary to prevent dramatic telomere loss during DNA replication. Here we demonstrate that replication-associated telomere loss is responsible for the chromosome fusions found in WS fibroblasts. Moreover, using metaphase analysis we show that telomere elongation by telomerase can significantly reduce the appearance of new chromosomal aberrations in cells lacking WRN, similar to complementation of WS cells with WRN. Our results suggest that the genome instability in WS cells depends directly on telomere dysfunction, linking chromosome end maintenance to chromosomal aberrations in this disease.
Keywords: cancer, WRN, genome instability, aging
Werner syndrome (WS) is a genetic disease in which patients develop signs of aging prematurely in life (1). At the molecular level the syndrome results from a mutation in the gene encoding WRN, a member of the RecQ helicase family (2). The severity of this syndrome can be explained by the multiple functions of WRN, which plays a role in DNA replication, repair, and recombination (1, 3, 4). WS is also characterized by increased genomic instability, which likely is the cause of cancer in affected individuals (5–7). Cells derived from WS patients grow poorly in culture, entering a senescence phenotype after few population doublings. However, inhibiting p53- and pRB-dependent tumor-suppressive pathways allows the cells to divide more rapidly and provides a longer replicative lifespan (8), suggesting that the cells suffer from DNA damage, which in turn triggers damage checkpoints (9).
It has been well established that linear chromosomes undergo terminal sequence loss during every replicative cycle (10) and eventually telomeres become critically short, which results in cell death or permanent growth arrest (11). Signaling from critically short human telomeres depends on the p53 and pRB tumor suppressor pathways, and the signaling cascade initiated by telomere dysfunction is comparable to the cellular DNA damage response (11). Suppression of this DNA damage response allows the cells to replicate with improperly protected chromosome ends, and the cell responds with DNA repair, usually by activating the nonhomologous end-joining pathway (12). As a result of this repair mechanism chromosomes become covalently fused, and such fusions can be observed in metaphase spreads and as anaphase bridges (13). When the cell continues to cycle with fused chromosome ends, breaks appear during every cell division, eventually leading to genome instability, a hallmark of cancer (11, 14).
Replicative telomere shortening can be counteracted by the action of telomerase, the specialized reverse transcriptase capable of elongating telomeres (15–17). Consequently, stabilization of telomere length by telomerase expression allows a cell to divide indefinitely by suppressing the accumulation of critically short telomeres (18, 19).
A clear link between WRN and telomere dynamics has recently emerged. First, primary fibroblasts from WS patients displayed a shorter lifespan in culture despite exhibiting telomeres shortening rate comparable to healthy cells. However, the poor growth phenotypes of WS cells were rescued by the expression of the catalytic subunit human telomerase reverse transcriptase (hTERT) (20, 21). Second, only in the background of a deletion of the RNA subunit of telomerase mTERC was it possible to mimic the pathology of WS in mice that lack WRN. Consequently, telomere attrition has been shown to represent a key factor in the genesis of the disorder in this model system (22). Third, it has been demonstrated that WRN helicase activity is required for efficient replication of the G-rich telomeric DNA strand. Lack of WRN helicase activity led to the dramatic telomere loss from individual sister chromatids [sister telomere loss (STL)] (8), which caused a DNA damage response. Expression of the catalytic subunit of telomerase could suppress STL and the damage response. In summary, these observations suggest that telomere maintenance and telomere integrity are critical parameters in defining the pathology of WS, although a direct relation between telomere dysfunction and WS phenotypes is yet to be established.
Here we set out to test whether WS cells accumulate chromosomal aberrations in defined culture conditions, and whether the acquisition of an unstable genome depends on telomere function. We demonstrate that telomere elongation by the expression of telomerase can limit the number of metaphases containing aberrant chromosomes to a similar extent as complementation of the WS cells with WRN. We suggest that the critically short telomeres resulting from the STL phenotype are subject to repair, leading to chromosome fusion–breakage cycles. Hence, the data presented here link the telomere loss phenotype in WS cells to the genome instability in the syndrome.
Results and Discussion
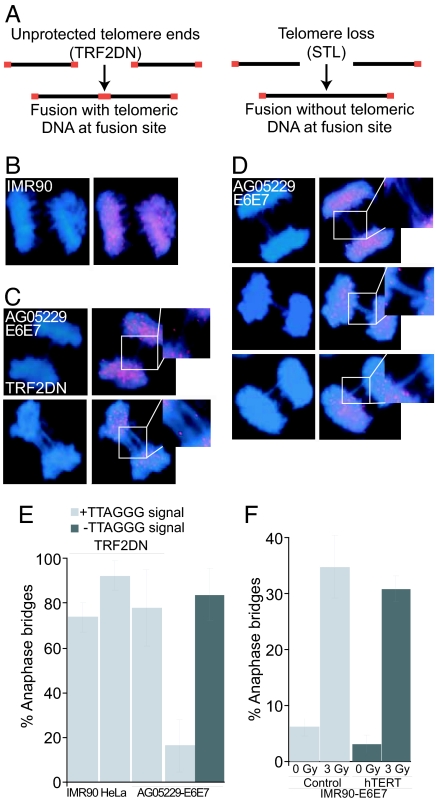
Suppression of the pRb and p53 DNA damage checkpoint pathways by expression of the human papilloma virus 16 (HPV16) E6 and E7 oncoproteins in primary fibroblasts from WS patients is sufficient to trigger anaphase bridge formation, a common indicator of telomere dysfunction (8). To test whether the chromosome fusions are caused by telomere loss, we characterized the fusions found in these cells. Indeed, when chromosome fusions are caused by telomere deprotection, e.g., by inhibition of the telomere protection factor TRF2, TTAGGG repeats are still detectable at the end of the chromosomes and can be visualized at the site of fusion by FISH. However, fusions of chromosomes due to missing telomeres at their extremities, such as STL chromosomes, are not expected to contain detectable telomeric repeats at the site of fusion (Fig. 1A). To distinguish between these two possibilities, we used FISH to identify telomeric sequences in anaphase nuclei (Fig. 1B), incorporating dominant-negative TRF2 (TRF2DN) as a positive control. TRF2DN expression in a carcinoma cell line (HeLa), in normal primary fibroblasts (IMR90), or in WS fibroblasts (AG05229) resulted in a dramatic increase of fusion frequency (13, 23). As expected, FISH revealed telomeric DNA in most of these anaphase bridges (Fig. 1E). A similar result was obtained in the AG05229 cell line, with a TTAGGG-FISH signal found at site of fusion in 78% of the anaphase bridges (Fig. 1 C and E). In contrast, anaphase bridge formation triggered quickly after expression of E6 and E7 oncoproteins in AG05229 fibroblasts could not be explained by telomere deprotection. Of >50 anaphases with bridges analyzed, 84% did not contain telomeric signals at the fusion site (Fig. 1 D and E). These results suggest that chromosomes fused because of the loss of the telomeric repeats at the extremities, consistent with the STL phenotype. These chromosomal fusions accumulated much more rapidly in WS cells expressing E6 and E7 than in normal primary fibroblasts with disrupted DNA damage checkpoints. After selection for expression of E6 and E7 only 6% of IMR90 cells exhibited anaphase bridges (Fig. 1F), as opposed to 38% of WS fibroblasts [supporting information (SI) Table 2]. Therefore, we propose that anaphase bridges in AG05229 fibroblasts were not a consequence of undetected DNA lesions due to the suppression of the p53 and pRB pathways or due to secondary effects of E6 and E7 expression such as potential break formation.
Fig. 1.
DNA bridges in WS cells expressing HPV16 E6 and E7 oncoproteins do not contain telomeric DNA. (A) Schematic of telomere dysfunction leading to fusions. Unprotected telomeres lead to fusions with telomeric DNA, whereas telomere loss leads to fusions lacking telomeric signal. (B) Normal anaphase from IMR90 cells. DNA was stained with DAPI (blue), and telomeres were visualized with a FITC-[CCCTAA]4 probe (red). (C and D) Anaphases of AG05229 cells expressing HPV16 E6 and E7 oncoproteins and TRF2DN (C) or E6 and E7 only (D). The staining was performed as in B, and a magnification of the DNA bridges is shown. (E) Quantification of DNA bridges with and without telomeric signal. An average of three independent experiments is shown. At least 50 DNA bridges were analyzed per condition per experiment, except for IMR90-E6E7 cells, where 20 bridges were analyzed per experiment. “100%” refers to all bridges detected, and error bars represent standard deviation. (F) Quantification of DNA bridges in IMR90-control-E6-E7 and IMR90-hTERT-E6-E7 cells. Cells were irradiated with 3 Gy, cultivated for 24 h, fixed, and stained with DAPI for analysis. At least 53 anaphases per condition per experiment were counted in three independent experiments. The percentage of cells with anaphase bridges is indicated, and error bars represent the standard deviation.
It was previously observed that this fusion phenotype can be rescued by the expression of the catalytic subunit of telomerase hTERT, pointing to a severe telomere defect in WS cells that can be reversed by telomere elongation (8). To strengthen this hypothesis we investigated whether anaphase bridges, resulting from lesions caused by ionizing irradiation, could also be suppressed by hTERT expression. Irradiation of young IMR90-E6E7 cells with 3 Gy did not cause cell cycle arrest, but it resulted in an increase of bridge formation from 6% to 36% of cells. Similarly, IMR90-hTERT cells expressing E6 and E7 showed anaphase bridges in 30% of cells after irradiation, indicating that telomerase overexpression is not sufficient to alleviate nontelomeric lesion phenotypes (Fig. 1F).
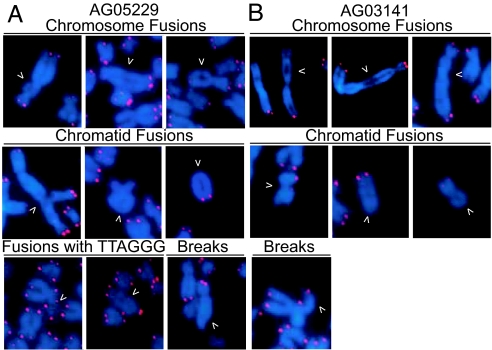
We then assessed the occurrence of fusions between metaphase chromosomes of two different WS fibroblast cell lines expressing HPV16 E6 and E7. In addition to the STL phenotype, we could detect several aberrations in AG05229 and AG03141 WS cells, including chromosome fusions, chromatid fusions, and chromosome breaks (Fig. 2). In both independent WS cell lines, we detected fused chromosomes in 21% and 20% of the metaphases analyzed (SI Table 3). Consistent with the results presented above, the majority of these fusions did not contain detectable telomeric DNA, suggesting that they result from a total or a partial telomere loss (SI Table 3).
Fig. 2.
Chromosome fusions in WS cells expressing HPV16 E6 and E7 oncoproteins. Shown is FISH of metaphase chromosomes of AG05229 (A) and AG03141 (B) WS fibroblasts. Telomeres were visualized with a FITC-[CCCTAA]4 probe (red), and DNA was stained with DAPI (blue). Arrows indicate chromosome fusions, chromatid fusions, and chromosome breaks.
Many of the chromatid fusions are sister chromatid type fusions, which is consistent with fusions resulting from STL events. A chromosome that lacks a single telomere because of the WRN-dependent replication dysfunction can be maintained in the background of p53 and pRB suppression. During the next replication cycle the telomeres of both chromatids of the daughter chromosome will be rendered dysfunctional and can consequently fuse to each other. The likelihood of this scenario is underscored by the finding that many more chromosomes lacking telomeric signal on both chromatid arms are found in AG05229 E6E7 cells (3%) than in AG05229 cells without E6E7 (0.4%) (data not shown). In summary, these data suggest that STL resulting from a telomere replication dysfunction can lead to chromosome fusions in WRN-deficient fibroblasts in subsequent division cycles.
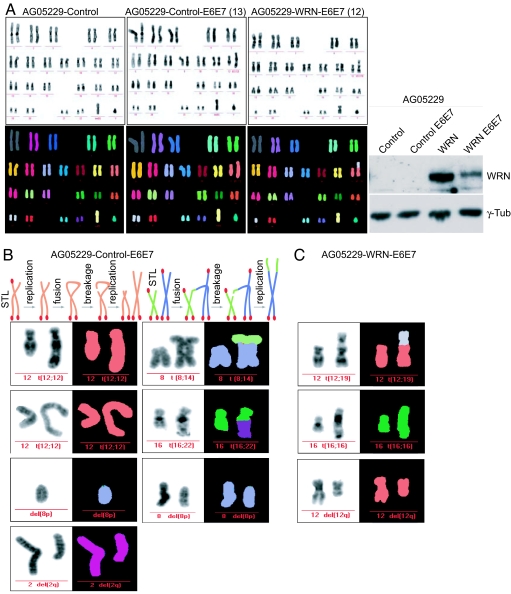
Covalent fusions between chromosomes or sister chromatids induce genomic instability, resulting from breaks that randomly occur during chromosome segregation in anaphase. To investigate whether the genomic instability observed in WS cells is a direct consequence of the loss of WRN, primary AG05229 fibroblasts were stably infected with either a control retrovirus or a retrovirus expressing the wild-type allele of WRN in two independent experiments. Genome instability was assessed by multicolor FISH (M-FISH) (24, 25) and compared between the two populations before and after stable expression of E6 and E7 oncoproteins. Suppression of DNA damage checkpoints allowed the propagation of chromosomes with missing telomeres and of chromosomal aberrations, permitting us to quantify such events after several cell divisions. M-FISH analysis was performed after every round of infection, which corresponds to approximately five population doublings between infections. In the first of two independent experiments, M-FISH analysis of AG05229 cells infected with a control virus or a virus expressing wild-type WRN (Fig. 3A) revealed no metaphases containing aberrant chromosomes (Fig. 3A and Table 1). However, when E6 and E7 oncoproteins were expressed in the cells infected with a control virus, ≈26% of metaphases displayed chromosomal aberrations, consisting of mostly chromosome translocations and chromosome deletions (Fig. 3 A and B and Table 1). This result is in accordance with previously published studies, mostly carried out in WS patient lymphocytes, where large deletions, breaks, and reciprocal translocations were reported (6, 7, 26). The appearance of these chromosomal aberrations is also consistent with the formation of chromosome fusions quickly after suppression of DNA damage checkpoints in WS cells. On the contrary, AG05229 cells expressing WRN were partially protected from the accumulation of chromosomal aberrations, because only 12% of metaphases were abnormal (Fig. 3C and Table 1). Similar results were obtained in the second experiment despite a higher basal level of genome instability in the original population of AG05229 cells, based on the observation that 10% of uninfected cells were found to carry translocations or deletions (Table 1). We attribute the difference in the baseline of aberrations compared with the WRN complementation experiment described above to a difference in cellular age at the start of the experiment. Suppression of DNA damage checkpoints further increased the frequency of metaphases containing aberrant chromosomes in the control cells to 27%. Complementation of AG05229 with WRN significantly decreased the genomic instability, because only 9% of metaphases were found to harbor aberrations after removal of DNA damage checkpoints in the complemented cell population (Table 1).
Fig. 3.
WRN expression in WS cells protects from genomic instability. (A) M-FISH pictures of one metaphase spread of AG05229-control cells, AG05229-control-E6E7 cells (image 13), and AG05229-WRN-E6E7 cells (image 12) are shown. WRN expression is shown to the right. (B) M-FISH stained aberrant chromosomes from all AG05229-control-E6E7 metaphases. Aberrant chromosomes (as listed in SI Table 4) from image 1 [del(8p)], image 6 [t(8;14)], image 13 [t(12;12)], image 14 [del(2q) and del(8p)], image 16 [t(12;12)], and image 18 [t(16;22)] are displayed. (C) M-FISH stained aberrant chromosomes from AG05229-WRN-E6E7 metaphases. Aberrant chromosomes from image 6 [del(12q)], image 10 [t(16;16)], and image 12 [t(12;19)] are shown.
Table 1.
M-FISH analysis of metaphases from WS cells
| Cell line | Vector | No. of metaphases analyzed | % of metaphases containing aberrant chromosomes |
|---|---|---|---|
| Effect of WRN expression | |||
| AG05229 | NI | NA | NA |
| Exp. 1 | Control | 29 | 0 |
| WRN | 11 | 0 | |
| Control-E6E7 | 23 | 26 | |
| WRN-E6E7 | 26 | 12 | |
| Exp. 2 | NI | 20 | 10 |
| Control | 9 | 0 | |
| WRN | NA | NA | |
| Control-E6E7 | 26 | 27 | |
| WRN-E6E7 | 23 | 9 | |
| Effect of hTERT expression | |||
| AG05229 | NI | 19 | 5 |
| Control | 19 | 5 | |
| TERT | 18 | 0 | |
| Control-E6E7 | 30 | 40 | |
| TERT-E6E7 | 27 | 7 | |
| AG03141 | NI | 19 | 11 |
| Control | 20 | 10 | |
| TERT | 30 | 10 | |
| Control-E6E7 | 30 | 40 | |
| TERT-E6E7 | 15 | 7 | |
NI, not infected; NA, not available.
All aberrations observed are listed in SI Table 4. A number of translocations were detected in the starting population of AG05229 cells and maintained stably throughout the experiment [t(5;8), t(4;7), and t(5;7/7;12)]. Consequently, these were excluded from the analysis.
In summary these data suggest that the genome instability that characterizes WS cells depends on WRN expression, because exogenous expression of the protein is sufficient to partially protect the cells from the accumulation of chromosomal aberrations.
The telomere fusion phenotype of WS cells, manifested as anaphase bridges, as well as the STL phenotype, can be rescued by telomerase expression (8). This led us to investigate whether telomerase-dependent elongation of critically short telomeres generated by STL could prevent genomic instability. Expression of hTERT in AG05229 or AG03141 fibroblasts was sufficient to efficiently elongate telomeres from 7–8 kb to 10–13 kb (SI Fig. 4A). Metaphase analysis by M-FISH revealed that uninfected primary AG05229 and AG03141 cells in this experiment contained 5% and 11% metaphases with chromosomal aberrations, respectively (Table 1).
M-FISH analysis of AG03141 revealed that translocation t(6;10) was maintained after each round of infection and cell division and was present in the genome of all cells; therefore, we excluded it from the quantification. As observed previously, the frequency of metaphases with aberrant chromosomes was strongly increased in checkpoint deficient control cells, reaching 40% in both populations (Table 1). Expression of telomerase and consequent telomere elongation in AG05229 and AG03141 cells efficiently suppressed the accumulation of chromosomal aberrations in metaphases (0% and 10%), respectively. When pRB and p53 were inhibited in WS fibroblasts expressing hTERT, the detected frequencies were 7% in both independent cell populations, which was significantly lower than the 40% found in cells lacking telomerase [Table 1, P = 0.0039 (AG05229) and P = 0.0012 (AG03141); P values were calculated by applying the Wilcoxon ranks test]. To exclude alternative roles for telomerase in the DNA damage response, cell proliferation, or chromatin modifications (27, 28), we expressed a dominant-negative allele of telomerase that is incapable of telomere elongation (29) in AG05229 cells. As expected, the inactive telomerase allele with two point mutations in the reverse transcriptase domain failed to elongate telomeres (SI Fig. 4B). Expression of this mutant had no effect on the accumulation of anaphase bridges (SI Fig. 4B Bottom), suggesting that the protection against the accumulation of anaphase bridges and metaphases containing chromosomal aberrations provided by telomerase is linked to telomere elongation.
These data demonstrate that expression of telomerase can protect the genome integrity of WS fibroblasts. We consequently hypothesize that telomerase, according to the enzyme's preference for elongating the shortest telomere available (30), acts on the unprotected and critically short telomeres resulting from STL, elongating them, and therefore rendering them functional.
It has been noted that 9 of all 44 translocations were intrachromosomal [t(12;12), t(16;16), t(17;17), t(1;1), and t(16;16)] (SI Table 4) and occurred in cells expressing E6 and E7. We interpret this phenomenon as consistent with the STL phenotype, resulting from chromatid fusions between arms of the same chromosome. In the absence of DNA damage checkpoints, such a fusion will break randomly in the next mitosis, consequently leading to a translocation (schematic in Fig. 3B).
In this study we found that telomere dysfunction has a major impact on the genome instability in cells lacking WRN. After removal of the DNA damage checkpoints primary fibroblasts from WS patients accumulate chromosome fusions rapidly, which can be observed in metaphase spreads and as anaphase bridges. Analysis of individual chromosomes from WS cells with dysfunctional checkpoints revealed a large number of deletions and translocations, suggesting that these cells become genomically unstable. It has been suggested previously that WRN plays an essential role in replication of the G-rich telomeric strand, and the lack of WRN leads to inefficient synthesis of this DNA strand, generating STL (8). Here we suggest that chromosomes containing STL are substrates for repair and ligation, leading to chromosome fusions and chromosomal aberrations in subsequent cell cycles. Consistent with the necessity to combine telomerase and WRN deletions in mice to recapitulate most clinical features of WS in an animal model (22), telomerase expression in WS cells can rescue the STL phenotype and the formation of chromosome fusions. Here we show that either complementation of the WS cells with WRN or expression of hTERT is sufficient to limit the accumulation of genome instability.
WS patients suffer from elevated cancer occurrence, and, because cancer and genome instability have been intrinsically linked, we hypothesize that telomere dysfunction is a critical feature of this human syndrome, relating a telomere replication dysfunction to the incidence of cancer in affected individuals.
Experimental Procedures
Cell Culture and Retroviral Infection.
IMR90 primary lung fibroblasts (ATCC) AG05229 and AG03141 primary WS fibroblasts (Coriell Cell Repositories, Camden, NJ) were grown in Glutamax-DMEM (Gibco/Invitrogen, Carlsbad, CA) supplemented with 15% FBS, 0.1 mM nonessential amino acids, and 100 units/ml penicillin/streptomycin at 7.5% CO2/3% O2. Retroviral infection was carried out as described (12). WRN and telomerase constructs were used as described (8, 19, 29). Inhibition of p53 and pRb pathways was obtained by retroviral expression of HPV16 E6 and E7 oncoproteins as described (31, 32). For irradiation experiments IMR90 cells were irradiated with a cobalt 60 source at 3 Gy 24 h before harvest.
Telomere FISH of Anaphase Nuclei.
Cells grown exponentially on coverslips were fixed with 2% paraformaldehyde in PBS for 10 min and permeabilized with 0.5% Triton X-100 in PBS for 10 min. After PBS washes, the cells were submitted to another fixation step by using a methanol:acetic acid (1:3) fixative solution for 30 min, then air-dried overnight. Then the coverslips were processed for telomere FISH by using the FITC-conjugated PNA probe FITC-OO-[CCCTAA]3 (Applied Biosystems, Foster City, CA) as described (12). DNA was stained with DAPI, and nuclei were visualized by using a Zeiss Axioplan II microscope. At least 50 nuclei were analyzed from three independent experiments for each cell line tested. Scoring was performed blindly. To score, pictures of random anaphases on a coverslip were acquired with the same exposure time, and anaphases were considered “telomere-free” when no red signal could be detected on the DNA strands connecting the sets of daughter chromosomes.
Telomere FISH of Metaphase Spreads.
WS fibroblasts were grown to ≈40% confluence and incubated with 0.1 μg/ml demecolcin for 3 h. Cells were harvested by trypsinization, resuspended in 0.075 M KCl at 37°C, and kept at 37°C for 7 min. Swollen cells were then fixed by using a methanol:acetic acid (3:1) fixative and kept overnight at 4°C. Fixed cells were spread on a water-wetted microscope slide, washed with fresh fixative, and dried on a humidified 80°C heat block. For FISH, slides were dried overnight and processed by using the FITC-conjugated PNA probe FITC-OO-[CCCTAA]3 (Applied Biosystems) as previously described (12). DNA was stained with DAPI, and nuclei were visualized by using a Zeiss Axioplan II microscope.
Southern Blotting and Detection of Telomeric Fragments.
Isolation of DNA and telomere-length measurement were carried out as described (13, 33).
M-FISH Analysis of Metaphase Chromosomes.
Metaphase spreads from WS cells were prepared as for telomeric FISH, and M-FISH analysis was performed as described (12, 24, 25). For each experiment, 9–30 metaphase spreads were acquired by using a DM RXA epifluorescence microscope (Leica Microsystems, Bensheim, Germany) equipped with a Sensys CCD camera (Photometrics, Tucson, AZ). Microscope and camera were controlled by Q-FISH software (Leica), and images were processed by using MCK software (Leica Microsystems Imaging Solutions, Cambridge, U.K.).
Supplementary Material
Acknowledgments
We thank the members of the J.K. laboratory for critical comments on the manuscript. J.K. was supported by National Institutes of Health Grants R01 069525 and R01 AG025837 and by the Mathers Foundation. A.J. was supported by the Tumorzentrum Heidelberg/Mannheim.
Abbreviations
- WS
Werner syndrome
- hTERT
human telomerase reverse transcriptase
- STL
sister telomere loss
- HPV16
human papilloma virus 16
- M-FISH
multicolor FISH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609410104/DC1.
References
- 1.Opresko PL, Cheng WH, von Kobbe C, Harrigan JA, Bohr VA. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 2.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watt PM, Hickson ID, Borts RH, Louis EJ. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto M, Miller RW, Ishikawa Y, Sugano H. Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 6.Salk D, Au K, Hoehn H, Martin GM. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 7.Melcher R, von Golitschek R, Steinlein C, Schindler D, Neitzel H, Kainer K, Schmid M, Hoehn H. Cytogenet Cell Genet. 2000;91:180–185. doi: 10.1159/000056841. [DOI] [PubMed] [Google Scholar]
- 8.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 9.Bai Y, Murnane JP. Hum Genet. 2003;113:337–347. doi: 10.1007/s00439-003-0972-y. [DOI] [PubMed] [Google Scholar]
- 10.Watson JD. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 11.de Lange T. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 12.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 13.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 14.De Lange T. Cold Spring Harbor Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 15.Lundblad V, Szostak JW. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 16.Yu GL, Bradley JD, Attardi LD, Blackburn EH. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW, Blackburn EH. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 18.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 19.Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 20.Wyllie FS, Jones CJ, Skinner JW, Haughton MF, Wallis C, Wynford-Thomas D, Faragher RG, Kipling D. Nat Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- 21.Ouellette MM, McDaniel LD, Wright WE, Shay JW, Schultz RA. Hum Mol Genet. 2000;9:403–411. doi: 10.1093/hmg/9.3.403. [DOI] [PubMed] [Google Scholar]
- 22.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 23.Celli GB, de Lange T. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 24.Speicher MR, Gwyn Ballard S, Ward DC. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 25.Eils R, Uhrig S, Saracoglu K, Satzler K, Bolzer A, Petersen I, Chassery J, Ganser M, Speicher MR. Cytogenet Cell Genet. 1998;82:160–171. doi: 10.1159/000015092. [DOI] [PubMed] [Google Scholar]
- 26.Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht KW, Jonas JB. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 27.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, et al. Proc Natl Acad Sci USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, et al. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 29.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 30.Hemann MT, Strong MA, Hao LY, Greider CW. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 31.Smogorzewska A, de Lange T. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halbert CL, Demers GW, Galloway DA. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlseder J, Smogorzewska A, de Lange T. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.