Abstract
Automated analysis of mouse behavior will be vital for elucidating the genetic determinants of behavior, for comprehensive analysis of human disease models, and for assessing the efficacy of various therapeutic strategies and their unexpected side effects. We describe a video-based behavior-recognition technology to analyze home-cage behaviors and demonstrate its power by discovering previously unrecognized features of two already extensively characterized mouse models of neurodegenerative disease. The severe motor abnormalities in Huntington's disease mice manifested in our analysis by decreased hanging, jumping, stretching, and rearing. Surprisingly, behaviors such as resting and grooming were also affected. Unexpectedly, mice with infectious prion disease showed profound increases in activity at disease onset: rearing increased 2.5-fold, walking 10-fold and jumping 30-fold. Strikingly, distinct behaviors were altered specifically during day or night hours. We devised a systems approach for multiple-parameter phenotypic characterization and applied it to defining disease onset robustly and at early time points.
Keywords: home cage, neurodegeneration, prion protein, polyQ
The value of mouse models for human diseases has created a keen need for high-throughput behavioral analyses, as has the ambitious goal of systematic characterization of the complete mouse genome (1, 2). Variability in standard behavioral tests hinders comparative studies (3–5) and most sample “snapshots” of behavior. Anxiety caused by being handled by the researcher complicates interpretation and obscures subtle phenotypes. Testing behavior during the daytime may not reflect normal behavior, because mice are nocturnal animals. Finally, manual data collection has inherent investigator bias and high labor costs.
We tested and helped develop a conceptual framework for analysis of mouse behavior by evaluating and improving a beta version of HomeCageScan (HCS), a video-based behavior-recognition platform. The original version of the software functioned only in proof-of-principle experiments: short recordings of a single mouse. It proved unable to give any meaningful phenotypic data in the laboratory setting. By working iteratively with the software designers (Clever Systems Inc., Reston, VA) we overcame numerous video recording and hardware problems, increased the throughput of the system, and refined behavioral definitions. Further details of the modifications to the system are described in supporting information (SI) Methods. In the end, the system provided very high-resolution analysis and allowed us to characterize behavior with equally high resolution during the entire light and dark phases, with virtually no intervention by the investigator. We explored the benefits of video-based behavior recognition by conducting high-resolution automated mouse behavior analysis (AMBA) of the home cage (HC) behaviors of two mouse models of neurodegenerative disease, Huntington's disease (HD) and infectious prion disease (PrD).
HD is an autosomal dominant disorder caused by the expression of huntingtin protein with an expanded glutamine repeat (6). Degeneration of the striatum and cortex leads to severe psychological and motor abnormalities, ending in death (7). The R6/2 mouse is the most widely used HD model, showing an early, severe disease phenotype, with declines in motor performance, cognitive abilities, and weight and premature death (8–10). PrDs have different etiologies and affect distinct regions of the brain. Acquired genetically, spontaneously, or through exposure to infectious material (11), their hallmark features are the misfolding of the prion protein (PrP), dementia, severe ataxia, and death. The most common and robust PrD model involves infecting WT mice with established “strains” of prions. Prion-infected mice exhibit hunched posture, ataxia, tail rigidity, priapism, and death (12). Behavioral abnormalities such as disturbances in food intake and activity vary depending on the mouse strain as well as the prion strain (13, 14). The diversity of PrD symptoms provides excellent candidates for higher-resolution behavioral analysis.
High-resolution AMBA enabled us to characterize models of HD and PrD in unprecedented detail and to discover previously uncharacterized disease phenotypes. Using a systems approach to data analysis, we describe unique behavioral signatures for each disease. Combinatorial behavioral metrics allowed earlier assignment of disease onset. This approach will be extremely useful for phenotypic discovery and as an entry point to more specific behavioral tests.
Results
Experimental Setup.
Previously reported behavior acquisition platforms reduce the mouse to a point in space or count the number of breaks of laser beams. Both give very limited information. HCS uses video images collected at a rate of 30 frames per second in the HC and software algorithms to categorize a diverse set of mouse behaviors. The software extracts the image of the mouse as it moves and automated recovery tools adapt to changes in lighting and bedding. Information on the sequence of postures and position of body parts is used to deduce behaviors by comparisons with pretrained data sets. For example, “walking” comprises the mouse being in a particular posture and performing a concerted movement of torso and limbs that changes the position of the mouse along the horizontal axis. For “eat,” the snout must cross the plane of the food bin with a reared (forelimbs off the ground) posture. For “chew,” which is scored after “eat,” the mouse moves into a vertically huddled position with its paws in front of the snout. Detailed definitions of behaviors are in SI Methods.
We recorded control and diseased mice and assessed the accuracy of AMBA by viewing and surveying individual videos for ≈20 mice for 3–4 h each. Initially, HCS recorded one mouse at a time. We increased the throughput of the system from one cage to four cages by adding extra cameras. HCS initially also misscored behaviors so frequently that we could detect no differences between WT mice and those suffering from neurodegenerative disease. We worked reiteratively with Clever Systems Inc. to improve ease of use, data transfer capabilities, and, most importantly, the accuracy of scored behaviors through numerous modifications to the algorithms and retestings.
The final accuracy assessment was conducted by inspecting ≈100 instances of each behavior (according to HCS) in a 24-h video of a single WT C57BL/6 mouse (summary in SI Table 1). For 9 of 17 behaviors, ≥90% of the instances identified by HCS agreed with manual assessments. For example, 100 of 100 behaviors identified by HCS as “hang upside down” were confirmed, 67 of 70 for “rest,” and 95 of 100 for “walking.” For “hang vertical,” 75 of 100 instances identified by HCS were confirmed. However, most errors were for related activities. The 25 behaviors that HCS misidentified as “hang vertical” were all due to “rearing.” Both require the animal to be in a vertical posture. For the former, the forelimbs touch the wire rack and the hindlimbs lift off the ground; for the latter hindlimbs remain in contact with the ground. A small fraction of behaviors (1–2% of recording time) were unassigned because of failure of the software to recognize the mouse or a behavior (SI Figs.5 and 6 for HD and PrD, respectively). The former often occurred immediately after light/dark transitions. The misscoring of behaviors was generally unbiased between WT and diseased mice, except for the later time points for PrD, when the mice move very rapidly and erratically.
Home Cage Behavioral Abnormalities in HD Mice.
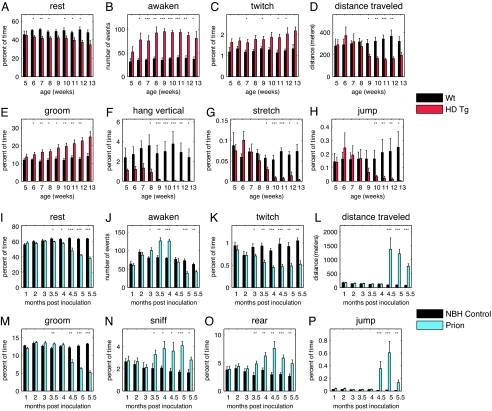
Standard methods for detecting the HD transgenic (Tg) phenotype are weight loss, clasping, and declining performance in rotarod and grip-strength tests (8, 15). In other laboratories and in our own, these metrics did not reliably detect disease onset until 9–11 weeks (10, 15). Recently earlier detection has been achieved by examining running-wheel activities (16). We recorded the behaviors of seven HD Tg mice and seven gender-matched littermate controls, from a side-view of the HC (SI Fig. 7) for two 24-h periods weekly, beginning at 5–6 weeks of age until 11–13 weeks, the terminal phase. High resolution AMBA demonstrated many abnormalities in HD Tg behaviors, many of which were previously unknown. Relative to controls, time spent “resting” progressively declined in HD Tgs (Fig. 1A). (The behavior we have defined as “resting” roughly approximates “sleep,” but because we have not validated it with electroencephalogram analysis, we use the more general term “rest.”) Commensurate with less time spent resting, awakening events were dramatically increased in HD Tgs, with Tg mice awakening from rest as much as 2.5-fold more than controls (Fig. 1B). “Twitching,” defined as a movement during rest, was elevated in HD mice at the earliest time point tested, and this difference was maintained throughout (Fig. 1C). This difference in resting and awakening between HD Tgs and controls was one of the earliest behavioral abnormalities detected and remained significantly different from 6 weeks onwards for awaken and at 6 time points for rest (Fig. 1 A and B). The early detection of rest abnormalities highlights the remarkable sensitivity of AMBA to clearly and unambiguously detect subtle phenotypic changes before more obvious disease onset at 9–10 weeks of age.
Fig. 1.
Behavioral alterations in Huntington's and PrD mice. (A–H) Mean values (±SEM) for HD and controls are shown for rest (A), awaken (B), twitch (C), distance traveled (D), groom (E), hang vertical (F), stretch (G), and jump (H). (I–P) Mean values for PrD and controls are shown for rest (I), awaken (J), twitch (K), distance traveled (L), groom (M), sniff (N), rear (O), and jump (P). P values were computed by using a two-tailed Wilcoxon rank-sum test (nonparametric) and are indicated as follows ∗, P < 0.05; ∗∗, P < 0.01; and ∗∗∗, P < 0.001 (sample sizes for HD were n = 5 HD Tg and WT control pairs for week 5, n = 7 for weeks 6–11, n = 6 for week 12, and n = 4 for week 13; for PrD, n = 8 for every time point except for 5 m.p.i., where n = 7 prion and n = 8 controls).
High resolution AMBA also revealed decreased exploratory behaviors in HD Tgs. For example, distance traveled provided a robust metric of overall activity and motor performance. By 9 weeks, there was a 30% decline in the distance traveled by HD Tgs (Fig. 1D). AMBA also detected hypergrooming in HD Tg mice. At 5 weeks, HD Tgs groom for ≈15% of their time, increasing steadily to 25% at terminal stages of disease (Fig. 1E). Control mice showed stable week-to-week grooming behavior, 12–14% of total time (Fig. 1E). HD Tgs groomed more frequently than controls rather than grooming for longer bouts (data not shown).
Consistent with previously reported motor abnormalities, high-resolution AMBA also quantified defects in behaviors requiring significant grip strength and coordination. Hanging vertically was reduced in HD Tgs even at the earliest time point tested, 5 weeks, declining ≈100-fold by 10 weeks (Fig. 1F). Despite considerable variation in hanging behaviors of diseased and control mice, statistically significant differences between HD Tgs and controls were detected at the early age of 8 weeks. Stretching was similar for HD Tgs and controls until 9 weeks, when stretching severely declined for HD Tgs (Fig. 1G). This finding is consistent with their advancing disease “hunched” posture (8), and by 13 weeks HD Tgs rarely stretched (0.003 ± 0.001% SEM of total time compared with 0.074 ± 0.02% SEM in WT controls). Jumping, another complex motor behavior, showed a dramatic decline in HD Tgs (Fig. 1H). Despite large intermouse variability of this behavior among controls, a significant difference appeared in HD Tgs by 10 weeks. Alterations in many other behaviors were detected, such as remain low, pause, walk, turn, sniff, rear, eat, chew, drink, and hang upside down (SI Fig. 5).
Home Cage Behavioral Abnormalities in PrD Mice.
Standard methods for detecting PrD rely on subjective assessments that are not readily quantified: ruffled coat, hunched posture, priapism, and ataxia (12). Consistent with a multitude of reports from other laboratories, when we injected mice with prions harvested from brains of infected animals, these symptoms were easily and reliably detected only 3–4 weeks before the endpoint of disease [5.0–5.5 months postinoculation (m.p.i.)]. For AMBA, we recorded eight similarly infected C57BL/6 males and eight mock-inoculated controls for two 24-hour periods monthly or twice monthly until the terminal phase, 5.5 m.p.i. AMBA detected alterations in PrD mice behavior much earlier than previously reported and detected many previously uncharacterized phenotypes.
By 3.5 m.p.i., prion-infected animals exhibited a significant decrease in resting, and, at the final stages of disease, PrD mice rested only half as much as mock-injected mice (Fig. 1I). Awakening from rest showed a significant increase in early stages of PrD at 3, 3.5, and 4 m.p.i. and then decreased in the late stages of disease of 5–5.5 m.p.i. (Fig. 1J). Movement during rest, or twitching, was significantly reduced from a very early time point, 3 m.p.i. (Fig. 1K). Thus, rest abnormalities were one of the most sensitive metrics of disease onset for PrD and HD.
Unexpectedly, PrD mice showed an enormous increase in activity concomitant with overt disease onset. Beginning at 4.5 m.p.i., the PrD mice traveled 1,378 (±411 SEM) meters compared with 96 (±5 SEM) meters in controls (Fig. 1L and SI Movie 1). PrD mice showed a sharp decline in grooming; by advanced disease (5.5 m.p.i.), they spent half as much time grooming as controls (Fig. 1M). In sharp contrast to HD Tg mice, the exploratory activities sniffing and rearing were highly elevated in PrD mice at 3.5 m.p.i., and this increase persisted until the final time point at 5.5 m.p.i. (Fig. 1 N and O). Despite ataxia and imbalance, PrD mice spent significantly more time jumping than controls from 4.5 to 5.5 m.p.i. (Fig. 1P). These behavioral alterations occurred at very early time points, long before classic symptoms developed. Many other behavioral alterations were detected, such as remain low, pause, walk, turn, eat, chew, drink, stretch, hang upside down, and hang vertical (SI Fig. 6).
Behaviors of HD and PrD Mice in the Light and Dark Phase.
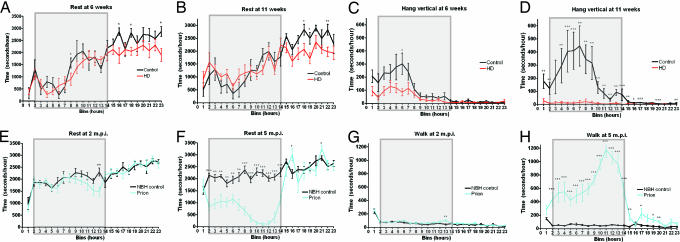
Data collection over 24 h allowed us to examine behaviors with respect to the light and dark phase, when mice are normally more active. WT mice were most active during the first half of the dark phase as demonstrated by decreased rest and increased hanging vertical behaviors (Fig. 2 A and C, respectively). In the second half of the dark phase, resting increased, and hanging vertical decreased for WT mice (Fig. 2 A and C).
Fig. 2.
Behavioral alterations with respect to light and dark phases in HD and PrD mice. (A–D) Resting at 6 weeks (A) and 11 weeks (B) and hanging vertical at 6 weeks (C) and 11 weeks (D) are displayed in 1-h bins for HD mice (red line) and controls (black line). (E–H) Resting at 2 m.p.i. (E) and at 5 m.p.i. (F) and walking at 2 m.p.i. (G) and 5 m.p.i. (H) are displayed in 1-h bins for PrD mice (blue line) and controls (black line). Shaded boxes represent the dark cycle (7 p.m. to 7 a.m.). P values were computed by using a two-tailed Mann–Whitney test (nonparametric) and are indicted as follows ∗, P < 0.05; ∗∗, P < 0.01; and ∗∗∗, P < 0.001. Sample sizes for each time point are n = 7 HD Tg and n = 7 WT controls (A–D) and n = 8 prion and n = 8 controls (E–H).
At 6 weeks, HD Tgs rested a similar amount of time as controls during the dark phase (Fig. 2A); however, even at this early stage, HD Tgs rested less during the light phase. This difference in rest during the light phase was observed again at a later stage of disease (Fig. 2B). HD Tgs spent slightly less time hanging vertical than did control mice during the dark cycle at 6 weeks, but during the light cycle, the hanging behavior was the same (Fig. 2C). By 11 weeks, the difference in hang vertical behavior with respect to light and dark cycles was much more profound (Fig. 2D). The HD Tgs showed a statistically significant decrease in hanging vertical throughout the entire dark phase. Thus, these behavioral alterations observed in HD Tgs were often observed only in light or dark phases.
PrD mice also showed remarkable changes in behaviors, distinct from HD Tgs, with respect to light and dark phases. Resting was similar between PrD and control mice at 2 m.p.i., before disease symptoms (Fig. 2E). Resting for both PrD and control mice was fairly constant throughout the light and dark phase, with a slight increase for both in the light phase (Fig. 2E). At 5 m.p.i., PrD mice barely rested during the dark phase, in striking contrast to controls, whereas little difference was observed in rest during the light phase (Fig. 2F). Upon first entering a new cage, both PrD and control mice spent time walking, indicative of exploring the new cage (Fig. 2G). In PrD and control mice at 2 m.p.i., walking was uniformly distributed during the light and dark phases (Fig. 2G). However, by 5 m.p.i., PrD mice showed a profoundly different pattern of walking; during the dark phase, PrD mice walked approximately 10-fold more than controls (Fig. 2H and SI Movie 1).
Systems Approach to Defining Disease Phenotypes.
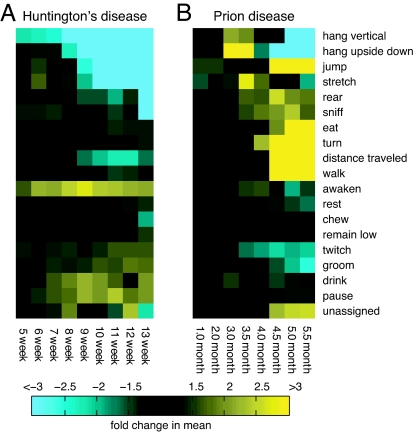
The behavioral changes in both HD and PrD mice were marked compared with control mice and were distinct between diseases. To examine the HD and PrD phenotypes globally with respect to time, we used an approach pioneered in other high-throughput high-resolution technologies such as microarray analysis. The assessment of each phenotype for each time point is represented by a box in a grid. Different colors represent behaviors that increase (yellow) or decrease (blue), with black indicating no change. Different intensities of color were used to represent different degrees of phenotypic change. To generate the phenotypic array displayed in Fig. 3, data were normalized by dividing mean diseased mouse values by mean control mouse values. In SI Fig. 8, the array is shown in terms of P values, with five behaviors significantly different between HD and control mice at the early time point of 6 weeks and eight behaviors for PrD at 3.5 m.p.i. This style of representation yields a comprehensive disease signature of HD and PrD mice and demonstrates strikingly different, easily comprehended symptomologies for these diseases at a single glance.
Fig. 3.
Phenotypic arrays of HD and PrD behaviors over time. Mean values for HD (A) or PrD (B) were divided by mean control values to give the fold increase or decrease in the behaviors listed. The fold increase or decrease is indicated in the figure as increasing intensity of blue for negative fold changes or yellow for positive fold changes.
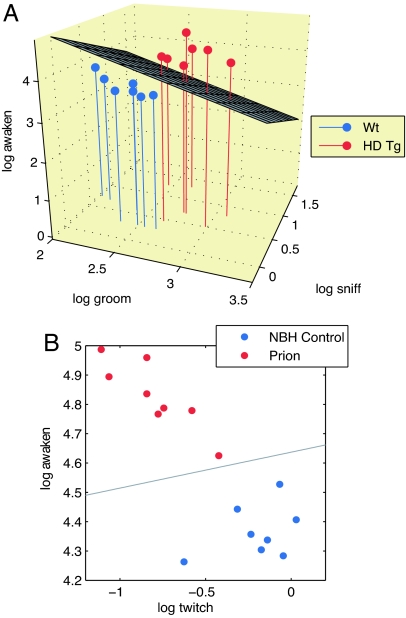
To define disease onset effectively, we took a systems-level approach, which considers the interdependencies between parts of a whole, to analyze the behavioral phenotypes. Examining combinations of behaviors rather than single behaviors in isolation allowed for early detection of disease onset. We used logistic regression to construct diagnostic rules based on linear combinations of the different behaviors. We constructed diagnostic rules independently for the nine time points in the disease progression of HD Tg mice and eight time points for PrD mice. These rules typically involved one to seven behaviors. The rules perfectly separated HD Tgs from controls at 6 weeks and subsequent time points and separated PrD mice from controls beginning at 3.5 m.p.i. The rule sufficient for diagnosing HD Tgs at week 7 encompassed the combination of awaken, groom, and sniff behaviors (Fig. 4A). The rule for predicting PrD and control mice at 4 m.p.i. used awaken and twitch behaviors (Fig. 4B). The hyperplanes in Fig. 4 A and B are the boundaries between the regions of behavior–space in which mice are classified as diseased and the regions in which they are classified as controls. Note that in Fig. 4A, none of the single features, awaken, groom, or sniff, separates HD mice from control mice, but a linear combination of the three features successfully predicts diseased mice. We used L1 regularization to control overfitting (17), the details of which can be found in SI Methods. Detailed results of cross-validation at all time points are given in SI Table 2.
Fig. 4.
Multifeature analyses to separate diseased from control mice. (A) A three-dimensional plot of the log of awaken, groom, and sniff behaviors that discriminate HD mice from control mice at 7 weeks of age. (B) A two-dimensional plot that separates PrD mice from controls at 4 m.p.i. by using the log of awaken and twitch.
Discussion
We have established a rigorous system for characterizing behavioral abnormalities in the mouse and have used it to demonstrate a contrasting array of behavioral changes in HD and PrD mouse models. Despite intense study of both models, many of these behavioral alterations were previously unreported, such as hypergrooming in HD Tgs and increased activity in PrD mice. We detected significant abnormalities in HD Tgs for most behaviors examined. Many behavioral abnormalities were observed at the reported age of disease onset for the HD Tgs, at 9–11 weeks (8–10, 18). However, the sensitivity of high-resolution AMBA detected statistically significant differences as early as 6 weeks for five behaviors despite the limited sample size (n = 7 WT and n = 7 HD Tg; Fig. 1; SI Fig. 8). In addition, we detected differences in some exceedingly rare behaviors, such as stretching (Fig. 1G), with high accuracy. With current testing paradigms for HD Tgs, a large sample size is often required for drug trials (15). High resolution behavioral analysis such as provided by HC monitoring should reduce this number, cost, and labor, particularly for mutants or models that are difficult to breed (8).
Our analysis of HC behaviors showed that the HD and PrD models reflect human symptomology better than previously appreciated. Most of the behavioral testing of HD mice is focused on their robust motor phenotypes (16), yet HD patients often present with psychological symptoms (7), and some cognitive defects have been described in HD Tgs (9, 19). Our analysis of HC activity revealed substantial psychological abnormalities in HD Tgs, such as hypergrooming, decreased resting, and altered patterns of behavior with respect to the light–dark cycle. The rest–wake abnormalities that we observed in HD Tgs have also been reported as a clinical phenotype in human HD patients, who suffer from sleep disturbances (18). Increased drinking behavior in HD Tgs may mirror the diabetes that is characteristic of some HD patients (20). In addition, we detected decreases in motor and exploratory phenotypes of HD Tgs. High-resolution AMBA was equally valuable in phenotyping PrD mice, which were distinguishable from controls at 3.5 m.p.i., with subtle phenotypic changes, followed later by more dramatic changes (Fig. 3B). The increased activity observed during prion infection is unprecedented. Mice undergoing calorie restriction, which radically increases the activity of mice, showed a ≈5-fold increase in distance traveled (21). PrD mice travel nearly 15-fold more distance at 4.5 m.p.i. For PrD mice, we established a previously undocumented rest disturbance phenotype that is likely of relevance to a genetic form of PrD in humans known as fatal familial insomnia (11) and to sleep abnormalities in Creutzfeldt–Jakob disease (22).
Because the HC provides an unperturbed laboratory environment for mice, it has the potential to reduce the variability observed in many types of behavioral testing. Our approach presents some disadvantages. Minimal bedding was used to reduce obstruction of the mouse, which eliminates the instinctive behaviors, dig and forage. Also, because we cannot discriminate between two mice in the same cage, mice must be singly housed during testing. High-resolution AMBA will never replace the cleverly designed, highly specific behavioral tests used to assess particular physical and behavioral deficits. But our approach offers advantages, including automated data collection, minimal labor, and unbiased interpretation of video data. In this study, we have measured 18 parameters from a procedure requiring minimal handling. Our approach and other automated behavioral measurement technologies will be of vital importance in detection and quantification of phenotypes in the rodent. The ability to detect a multitude of behavioral alterations and robustly diagnose both HD and PrD mice by 6 weeks and 3.5 m.p.i., respectively, will greatly aid in testing therapeutics on these mouse models of human disease.
Materials and Methods
Mice.
All experiments were approved by the Massachusetts Institute of Technology Committee on Animal Care. The HD line was obtained from The Jackson Laboratory (Bar Harbor, ME) as ovarian-transferred females. This line is maintained on a mixed C57BL/6J and CBA genetic background, and ovarian-transplanted females were crossed with WT males of the same mixed genetic background to generate the mice used in this study. The PCR genotyping protocol is described in ref. 15. C57BL/6 males were purchased from Taconic Farms (Germantown, NY) and injected intracranially with 30 μl of 0.1% uninfected mouse brain homogenate or 0.1% RML scrapie-infected mouse brain homogenate containing ≈5.5 log ID50 per 30 μl. Food and water were provided ad libitum, and mice were singly housed for the duration of the study, while being maintained on a 12:12-h light–dark cycle. During video recording, dim (25 W) red lights were used for recording in the dark phase. Because red light can entrain circadian rhythms of rodents (23), it is possible that entrainment effects were caused by our nighttime recording conditions.
Video Recording Setup.
Four JVC digital video cameras (model no. GR-D93) were mounted perpendicular to the cages (SI Fig. 8). The cameras input into a Pelco video processor connected to a Dell Dimension computer with an ATI All-In-Wonder video card. Video data were analyzed by HomeCageScan software (Clever Systems, Reston, VA) by using a Dell Dimension 450 computer. During recording, mice were housed in standard cages, with minimal bedding (80 ml) to minimize mounding, which can obscure the mouse. The cage was changed after the first 24 h of recording. Mice were recorded for 2 consecutive days per week for the duration of the HD Tg lifespan. With very few exceptions, data presented in the figures are from 46–49 h of recording; however, in a few instances, 23–25 h of recording is used because of inadequate recording and/or analysis. PrD mice and their controls were similarly recorded for two consecutive 24-h periods at 1, 2, 3, 3.5, 4, 4.5, 5, and 5.5 m.p.i. All prion-infected mice died between 5.5 and 6 m.p.i., on average 177 days (±5.7 days, SD).
Estimation of the Accuracy of HCS software.
Numerous videos were hand scored and compared with HCS scoring. For final estimation of the accuracy of HCS presented in SI Table 1, we reviewed ≈100 instances of each behavior for a single C57BL/6 mouse (as described in Results), unless the behavior occurred <100 times during the 24-h test video, giving an accuracy percentage for each behavior.
Data Analysis.
Reported P values were computed by using the nonparametric Wilcoxon rank-sum tests (two-tailed), with the MATLAB function RANKSUM. We used a nonparametric test because the sample sizes were not large enough for us to confidently conclude that the data were normally distributed. However, unpaired two-tailed Student's t tests gave very similar results. Reported P values were not adjusted for multiple-hypothesis testing. However, because we tested for differences in 19 behaviors, at each time point fewer than one is expected to have a P value <0.05 by chance. P values for classification accuracy in SI Table 2 are one-tailed, by using the exact binomial test to compute the probability of making at least the observed number of correct predictions by chance.
Supplementary Material
Acknowledgments
We thank Ken Streck and John Correa for assistance with setting up computers; Artur Topolszki, Andrew Borkowski, and Charles Yi for expert technical assistance [Whitehead Institute for Biomedical Research (WIBR)]; Gregory Raymond [National Institutes of Health (NIH)–National Institute of Allergy and Infectious Diseases, Montana] for providing RML prions; Karen Allendoerfer (WIBR) for editing and helping to write the manuscript; Caroline Yi (Harvard Medical School, Boston, MA), James Shorter (WIBR), Mu Sun (University of Minnesota, Minneapolis, MN), and Ann Graybiel (Massachusetts Institute of Technology) for critically reading the manuscript; and Vikrant Kobla and Yiqing Liang of Clever Systems, Inc. (Reston, VA) for software development, advice, technical support, and encouragement. This research was supported in part by a grant from the Ellison Medical Research Foundation (to S.L.) and an NIH National Research Service Award postdoctoral fellowship (to W.S.J.).
Abbreviations
- AMBA
automated mouse behavior analysis
- HC
home cage
- HCS
HomeCageScan
- HD
Huntington's disease
- m.p.i.
months postinoculation
- PrD
prion disease
- Tg
transgenic.
Footnotes
Conflict of interest statement: A.D.S., W.S.J., O.D.K., and S.L. initially purchased HCS and received an additional software license in exchange for assistance with further refinement of the program. None of the authors have received remuneration from Clever Systems, Inc. as a stockholder, consultant, or employee.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610779104/DC1.
References
- 1.Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, et al. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown SD, Chambon P, de Angelis MH. Nat Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- 3.Arndt SS, Surjo D. Behav Brain Res. 2001;125:39–42. doi: 10.1016/s0166-4328(01)00285-6. [DOI] [PubMed] [Google Scholar]
- 4.Kafkafi N, Benjamini Y, Sakov A, Elmer GI, Golani I. Proc Natl Acad Sci USA. 2005;102:4619–4624. doi: 10.1073/pnas.0409554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabbe JC, Wahlsten D, Dudek BC. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- 6.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 7.Vonsattel JP, DiFiglia M. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 9.Lione LA, Carter RJ, Hunt MJ, Bates GP, Morton AJ, Dunnett SB. J Neurosci. 1999;19:10428–10437. doi: 10.1523/JNEUROSCI.19-23-10428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter RJ, Lione LA, Humby T, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. J Neurosci. 1999;19:3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prusiner SB. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingsbury DT, Kasper KC, Stites DP, Watson JD, Hogan RN, Prusiner SB. J Immunol. 1983;131:491–496. [PubMed] [Google Scholar]
- 13.Dell'Omo G, Vannoni E, Vyssotski AL, Di Bari MA, Nonno R, Agrimi U, Lipp HP. Eur J Neurosci. 2002;16:735–742. doi: 10.1046/j.1460-9568.2002.02128.x. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham C, Deacon RM, Chan K, Boche D, Rawlins JN, Perry VH. Neurobiol Dis. 2005;18:258–269. doi: 10.1016/j.nbd.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Hockly E, Woodman B, Mahal A, Lewis CM, Bates G. Brain Res Bull. 2003;61:469–479. doi: 10.1016/s0361-9230(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 16.Hickey MA, Gallant K, Gross GG, Levine MS, Chesselet MF. Neurobiol Dis. 2005;20:1–11. doi: 10.1016/j.nbd.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Tibshirani R. J R Stat Soc. 1996;58:267–288. [Google Scholar]
- 18.Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. J Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy KP, Carter RJ, Lione LA, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurlbert MS, Zhou W, Wasmeier C, Kaddis FG, Hutton JC, Freed CR. Diabetes. 1999;48:649–651. doi: 10.2337/diabetes.48.3.649. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Steele AD, Lindquist S, Guarente L. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 22.Landolt HP, Glatzel M, Blattler T, Achermann P, Roth C, Mathis J, Weis J, Tobler I, Aguzzi A, Bassetti CL. Neurology. 2006;66:1418–1424. doi: 10.1212/01.wnl.0000210445.16135.56. [DOI] [PubMed] [Google Scholar]
- 23.McCormack CE, Sontag CR. Am J Physiol. 1980;239:R450–R453. doi: 10.1152/ajpregu.1980.239.5.R450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.