Abstract
The ParF protein of plasmid TP228 belongs to the ubiquitous superfamily of ParA ATPases that drive DNA segregation in bacteria. ATP-bound ParF polymerizes into multistranded filaments. The partner protein ParG is dimeric, consisting of C-termini that interweave into a ribbon–helix–helix domain contacting the centromeric DNA and unstructured N-termini. ParG stimulates ATP hydrolysis by ParF ≈30-fold. Here, we establish that the mobile tails of ParG are crucial for this enhancement and that arginine R19 within the tail is absolutely required for activation of ParF nucleotide hydrolysis. R19 is part of an arginine finger-like loop in ParG that is predicted to intercalate into the ParF nucleotide-binding pocket thereby promoting ATP hydrolysis. Significantly, mutations of R19 abrogated DNA segregation in vivo, proving that intracellular stimulation of ATP hydrolysis by ParG is a key regulatory process for partitioning. Furthermore, ParG bundles ParF-ATP filaments as well as promoting nucleotide-independent polymerization. The N-terminal flexible tail is required for both activities, because N-terminal ΔParG polypeptides are defective in both functions. Strikingly, the critical arginine finger-like residue R19 is dispensable for ParG-mediated remodeling of ParF polymers, revealing that the ParG N-terminal tail possesses two separable activities in the interplay with ParF: a catalytic function during ATP hydrolysis and a mechanical role in modulation of polymerization. We speculate that activation of nucleotide hydrolysis via an arginine finger loop may be a conserved, regulatory mechanism of ParA family members and their partner proteins, including ParA-ParB and Soj-Spo0J that mediate DNA segregation and MinD-MinE that determine septum localization.
Keywords: ParA superfamily, polymerization, plasmid partition, ATPase
The precise distribution of newly replicated genomes to progeny cells is imperative for stable transmission of genetic information. In bacteria, the most well characterized segregation mechanisms are specified by low-copy-number plasmids. These systems most frequently comprise two plasmid-encoded proteins, often termed ParA and ParB, that assemble on a cis-acting centromeric site. ParB directly binds the centromere, whereas ParA is recruited by interactions with ParB. The resulting segrosome complex is a positioning apparatus that localizes the attached plasmids to specific subcellular addresses (1, 2).
The segregation locus of multidrug-resistance plasmid TP228 in Escherichia coli consists of the parF and parG genes and nearby parH centromere (3). ParG (8.6 kDa) is the prototype of a class of small proteins involved in accurate segregation that are unrelated phylogenetically to ParB, but that fulfil analogous functions as centromere-binding factors (1, 4, 5). ParG is dimeric, with symmetric C-terminal domains that interleave into a ribbon–helix–helix fold that is crucial for DNA binding, and unstructured N-terminal tails (4, 6). Additional to its role as a centromere-binding protein, ParG is a transcriptional repressor of the parFG genes: transient associations between the flexible and folded domains in complex with target DNA modulate organization of a higher-order complex critical for transcriptional repression (7).
The ParA superfamily of ATPases, widely encoded by both chromosomes and plasmids, is characterized by a variant Walker-type ATP-binding motif (8). ParF (22.0 kDa) epitomizes one clade of the superfamily (3). In common with other ParA proteins, ParF is a weak ATPase whose nucleotide hydrolysis is enhanced ≈30-fold by ParG (9). ATP binding and/or hydrolysis by ParA proteins has long been recognized as a crucial facet of the segregation process, although its mechanistic purpose was uncertain (10–12). We have recently shown that ATP binding stimulates the polymerization of ParF into extensive multistranded filaments, whereas ADP antagonizes filamentation. ParG is another key modulator of polymerization (9). Mutagenesis of the ATP-binding site in Parf perturbed DNA segregation in vivo, ATP hydrolysis, and polymerization. We envisage that segrosome formation is initiated by site-specific binding of ParG to parH, generating paired complexes of specific topology. ParF is then recruited. ParF polymerization within the complex is controlled by nucleotide binding, by ParG-mediated stimulation of ATP hydrolysis, by remodeling effects of ParG, and, more speculatively, by cell cycle signals. Polymerization, or depolymerization, invokes separation of paired plasmids and their segregation in opposite poleward directions (1, 9).
Arginine fingers stimulate nucleotide hydrolysis by NTPases through the action of an arginine side chain inserted into the catalytic niche (13, 14). The arginine stabilizes the transition state through neutralization of negative charges that develop during phosphoryl transfer reactions (15, 16). This elegant charge-balancing strategy is adopted by numerous P-loop NTPases by using an arginine residue either in cis, where it belongs to the same polypeptide chain, or in trans, where it is provided by a partner protein or another monomer in oligomeric NTPases. Arginine fingers can be located in flexible loops of proteins, reflecting the mobility of the finger within the catalytic pocket. Here, we show that the N-terminal flexible tail of ParG is necessary for stimulation of nucleotide hydrolysis by ParF. In particular, a conserved arginine, R19, is absolutely required for this activation. Furthermore, the ParG N terminus promotes ParF polymerization. Interestingly, mutation of R19 abolishes ATPase stimulation but not the capacity to modulate ParF filamentation. The results reveal a crucial distinction between ParG activation of nucleotide hydrolysis by ParF, and ParG mediated remodeling of ParF polymers. Activation of nucleotide hydrolysis by ParA proteins by arginine fingers provided by cognate centromere-binding factors is likely to be a general mechanism used in bacterial chromosome and plasmid segregation.
Results
The ParG N-Terminal Tail Stimulates ATP Hydrolysis by ParF.
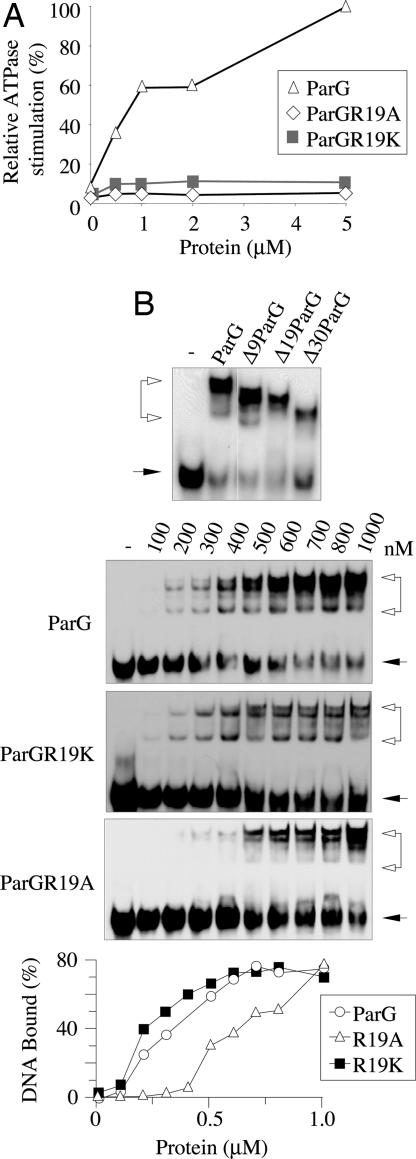
Enhancement of ATP hydrolysis is a feature of centromere-binding partners of ParA-like proteins (1). The mechanistic details underpinning this action have yet to be elucidated. The tertiary structure of ParG provides important hints for deciphering how the protein triggers enhanced nucleotide hydrolysis by ParF (6). Dimeric ParG consists of unstructured N-terminal tails (32 residues) and folded C-terminal domains (44 residues) that interweave into a ribbon–helix–helix motif. Spectral density mapping of the tails revealed that residues 17–23 show limited flexibility, probably because of transient secondary structures (6). Partial flexibility could provide entropic advantages, in comparison with completely flexible and disordered regions, when the tails are constrained during interactions with other molecules, e.g., ParF. The N-terminal tail also includes an amino acid set resembling an arginine finger loop: R19 and R23 are potential candidates for catalytic fingers delivered in trans into the ATP-binding pocket of ParF. The stretch includes closely juxtaposed phenylalanine (F15) and glutamate (E17, E22) residues often present in arginine finger motifs (15). The 17–23 region, with neighboring residues, might become transiently structured upon interaction with ParF and act as a classic arginine finger, stimulating ATPase activity. To test this possibility, activation of ParF ATP hydrolysis by ParG truncations of 9, 19, or 30 N-terminal residues (7) (Fig. 1A) was examined. Δ9ParG retained ≈25% of the activation conferred by full-length ParG (Fig. 1B). More strikingly, Δ19ParG and Δ30ParG failed to promote ParF ATPase activity (Fig. 1B). Thus, the mobile N terminus of ParG harbors the determinant for activation of ParF ATP hydrolysis.
Fig. 1.
ParG N terminus is responsible for stimulation of ParF ATPase activity. (A) Domain organization of ParG and truncated proteins. Arrow, β-sheet; cylinder, α-helix; red boxes, residues R19 and R23. (B) Relative stimulation of ParF ATP hydrolysis by ParG proteins plotted as a function of their concentration. (C) E. coli two-hybrid analysis showing interaction of ParF with full-length and truncated ParG proteins. parF and parG alleles were cloned in pT18 and pT25, respectively, and analyzed as described (4). Colonies displaying an interaction are red, whereas, in the absence of interaction, they are white.
Like ParG, Δ9ParG and Δ19ParG are dimeric and interact with DNA, indicating that the deletions do not grossly affect C-terminal domain organization (7; Fig. 2B). Deletion of 30 residues perturbs the β-strand and first α-helix of the folded domain (Fig. 1A). Nevertheless, Δ30ParG is dimeric and still binds DNA, albeit with reduced efficacy (7) (Fig. 2B). Furthermore, Δ9ParG, Δ19ParG, and Δ30ParG are proficient in interacting with ParF in vivo (Fig. 1C). These observations exclude that the lack of stimulation of ParF ATPase activity by the ParG derivatives might be due to loss of interaction with ParF or perturbation of ParG tertiary structure; and they reveal that both N- and C-terminal domains of ParG contact ParF.
Fig. 2.
R19 in the ParG N terminus is a key residue for enhancement of ATP hydrolysis by ParF. (A) Relative stimulation of ParF ATPase activity promoted by ParG, ParGR19K, and ParGR19A plotted as a function of ParG proteins concentration. (B) Electrophoretic mobility shift assays in which a biotinylated 48-bp oligonucleotide corresponding to the parFG operator was incubated with ParG or mutant proteins. (Top) ParG and ΔParG mutants were added at 1 μM dimer. Filled arrows, free DNA; open arrows, nucleoprotein complexes.
R19 of ParG Behaves as an Arginine Finger-Like Residue That Enhances ParF ATPase Activity.
The ParG N-terminal tail harbors arginine residues R19 and R23 that are candidate enhancers of ParF ATPase activity. The impact on plasmid partition of mutating these amino acids to alanine or lysine was assessed (Table 1). Neither mutation at position R23 affected segregation. By contrast, the R19A and R19K mutations reduced plasmid stability ≈2- and ≈3-fold, respectively, revealing that R19 in the tail of ParG exerts a key role in DNA segregation and that substitution with another basic residue (R19K) is insufficient to maintain normal segregation. ParGR19K bound the parFG operator site displaying binding characteristics very similar to those of wild-type ParG and exhibiting a comparable half-maximal saturation point (Fig. 2B). This finding indicates that ParGR19K conformation is unaltered and that impaired DNA segregation invoked by this mutation in vivo is unlikely to be associated with a DNA-binding defect. ParGR19A displayed ≈2-fold weaker DNA binding, which may contribute to the partitioning phenotype observed in vivo.
Table 1.
Effect of mutations at ParG position R19 and R23 on plasmid segregation in vivo
| ParG mutation | Retention, % |
|---|---|
| Wild type | 60.0 ± 5.5 |
| R19K | 18.3 ± 5.7 |
| R19A | 34.7 ± 11.9 |
| R23K | 65.3 ± 9.1 |
| R23A | 54.7 ± 3.1 |
| Vector | 6.7 ± 2.9 |
The capacity of ParGR19A and ParGR19K proteins to promote ATP hydrolysis by ParF was assessed. Strikingly, both proteins failed to stimulate ATPase activity, revealing that R19 is a critical residue for enhancement of nucleotide hydrolysis (Fig. 2A). The defect induced by the R19K mutation in vivo particularly attests to the importance of this stimulation during DNA segregation. R19 in the mobile N-terminal domain of ParG exhibits hallmarks of an arginine finger residue that complements in trans the catalytic pocket of its cognate Walker-type ATPase.
A Second Function in the ParG N Terminus: Remodeling of ParF Polymers.
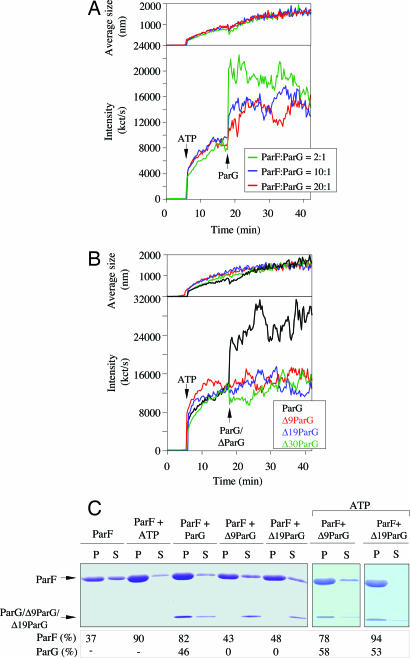
ATP-mediated polymerization of ParF can be monitored by bundling assays in which polymerized and unpolymerized ParF are separated into pellet and supernatant fractions, respectively, and analyzed by SDS/PAGE (9). ParG modulates formation of ParF filaments in these assays (9). Here, we instead used dynamic light scattering (DLS) to follow the polymerization kinetics of this interaction in real time (Fig. 3A Lower). As reported (9), addition of ATP (500 μM) to ParF (2.16 μM) induced rapid and extensive polymer formation reflected by a sharp increase in both intensity of scattered light and in particle size. Subsequent addition of ParG (1.08 μM) triggered an instantaneous enhancement of ParF polymerization, quickly reaching a stable plateau (Fig. 3A). Lower ParF/ParG ratios elicited less dramatic stimulation of polymerization. Irrespective of the ParF/ParG ratio tested, polymer size increased steadily (Fig. 3A Upper) and at a similar rate as in the absence of ParG (9). The dramatic spike in intensity of scattered light upon ParG addition was not mirrored by a corresponding surge in polymer size (Fig. 3A). This pattern supports a role for ParG in cross-bridging, bundling adjacent ParF protofilaments, rather than accelerating polymer growth. Nevertheless, we cannot exclude that addition of ParG may fuel nucleation of residual monomeric ParF still present in the reaction and thereby increase the intensity of light scattering.
Fig. 3.
The ParG flexible tail promotes ParF polymerization. (A) ParF polymerization followed by DLS. ParF (2.16 μM) was incubated at 30°C. ATP (500 μM) and MgCl2 (5 mM) were added at 6 min and, subsequently, ParG at ratios indicated. (Lower) Increase in light-scattering intensity expressed as kct/s. (Upper) corresponding augmentation in polymer average hydrodynamic size. (B) DLS experiment in which ParF (2.16 μM) was incubated first with ATP and MgCl2 and then with ParG or ΔParG polypeptides at a 10:1 ratio. (C) Sedimentation assay in which ParF (10 μM) was incubated without nucleotides or with ATP (2 mM) and with ParG, Δ9ParG, or Δ19ParG (10 μM). In the two rightmost panels, ParF (10 μM) was incubated with ATP (2 mM) and either Δ9ParG or Δ19ParG (10 μM). After centrifugation, 100% of pellet (P) and 33% of supernatant (S) fractions were resolved on a 15% SDS gel stained with Coomassie blue. Percentages of proteins in pellets are shown. Analogous results were obtained with Δ30ParG (data not shown).
The ability of ParG truncations to promote ParF polymerization was examined (Fig. 3B). Δ9ParG, Δ19ParG, and Δ30ParG failed to enhance ParF polymerization beyond that elicited by ATP. Thus, elimination of as few as nine N-terminal residues entirely abolished the capacity of ParG to stimulate ParF polymerization. The N-terminal tip spanning the first 6–10 residues is the most flexible segment of the ParG tail (6). It is tempting to speculate that these highly mobile tips might act analogously to the flexible “tentacles” of proteins like CapZ that cap barbed ends of actin filaments (17).
In bundling assays with ATP, ParF is found predominantly in the pellet. When ParG is included, it cosediments with ParF-ATP polymers in a substoichiometric ratio. Interestingly, without added nucleotide, ParG promotes ParF polymerization, and both proteins also copellet (9), potentially reflecting an ability of ParG to nucleate and stabilize ParF polymers. To assess whether ParG deletions retain this inherent nucleotide-independent activity, cobundling assays were performed with ParF and Δ9ParG, Δ19ParG or Δ30ParG without ATP (Fig. 3C). Approximately 40% of ParF was in the pellet in the absence of nucleotide, reflecting the inherent “autopolymerization” properties of the protein. As noted (9), without added nucleotide, ParG increased the ParF polymer mass in the pellet as efficiently as did ATP and partially cosedimented with ParF. However, Δ9ParG, Δ19ParG, and Δ30ParG failed both to stimulate polymerization and to associate with ParF filaments without ATP. Therefore, the N-terminal domain of ParG is necessary for nucleotide-independent promotion of ParF polymerization.
To assess whether truncated ParG proteins associated with ParF-ATP polymers, pelleting assays were performed in the presence of nucleotide. In the presence of ATP, Δ9ParG, Δ19ParG, and Δ30ParG copelleted with ParF fibers as efficiently as full-length ParG (Fig. 3C). Thus, ΔParG polypeptides are still able to associate with ParF polymers, but only in the presence of ATP. Combining this result with those observed by DLS, it is apparent that the ParG deletions have lost the capacity to reorganize ParF-ATP filaments, even though they can still interact with them. In summary, the ParG N-terminal tail is crucial for bundling and/or nucleation of ParF-ATP filaments as well as for nucleotide-independent stimulation of ParF polymerization.
Promotion of ParF Filamentation and Stimulation of Nucleotide Hydrolysis: Distinct Functions Associated with the ParG N Terminus.
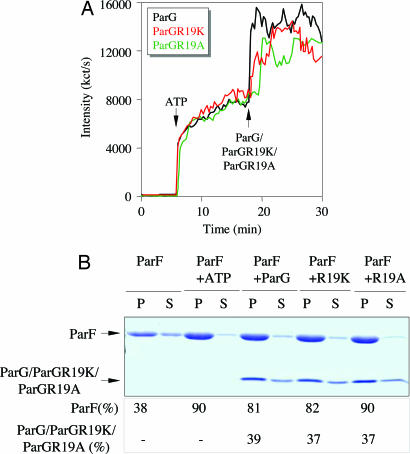
ParGR19K and ParGR19A are both entirely impaired in stimulating ParF ATP hydrolysis (Fig. 2A). The behavior of the proteins was analyzed by DLS to investigate whether they retained the ability to promote ParF polymerization, like ParG (Fig. 3A). When added to polymerizing ParF, both mutant proteins induced a steep increase in scattered light intensity, as observed with wild-type ParG (Fig. 4A). In addition, both proteins behaved similarly to ParG in bundling assays in triggering ParF polymerization and cosedimenting with ParF fibers even without ATP (Fig. 4B). Thus, ParGR19K and ParGR19A are proficient in interacting with ParF and can stabilize and reorganize ParF filaments, despite an inability to enhance nucleotide hydrolysis. Taken together with other results, this distinguo permits the discrimination of dual functions residing in the ParG tail and establishes that enhancement of ParF ATP hydrolysis and promotion of filament polymerization are distinct, independent roles played by the ParG N terminus.
Fig. 4.
ParGR19K and ParGR19A are as proficient as wild-type ParG in stimulating ParF filamentation. (A) DLS experiment in which ParG, ParGR19K, or ParGR19A was added to polymerizing ParF (2.16 μM) at a molar ratio of 10:1 (ParF/ParG). (B) Sedimentation assay. ParF (10 μM) was incubated without nucleotide, with ATP (2 mM) only, or with ParG, ParGR19K, or ParGR19A (10 μM). Reactions were centrifuged, and 100% of pellet (P) and 33% of supernatant (S) were resolved on a 15% SDS gel. Percentages of proteins in the pellet are shown.
A Conserved Arginine Residue in ParG Homologs and Orthologs.
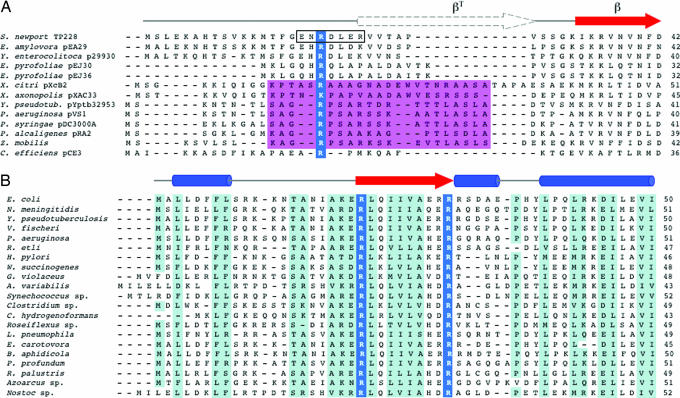
ParG proteins share similarity in the folded C-terminal domains, with fewer conserved N-terminal regions (6; Fig. 5A). However, ParG of plasmid TP228 and homologs specified by Yersinia and Erwinia plasmids also share similarity in their N termini. ParG N-terminal domains from plasmids of Pseudomonas and other species are characterized by an alanine/serine-rich patch (Fig. 5A). Significantly, both classes of ParG N termini possess an arginine residue that, potentially, is equivalent to the arginine finger-like residue in ParG of TP228.
Fig. 5.
Putative arginine finger residues in ParG and MinE proteins. (A) N-terminal regions of ParG homologs. Blue, putative arginine finger residue; magenta, alanine/serine patch. Region 17–23 in TP228 ParG, which is less mobile than the remainder of the N-terminal region, is boxed. Secondary structure features in TP228 ParG are shown above (6, 7). (B) N-terminal regions of MinE homologs. Conserved arginines that are candidate arginine finger residues are outlined in blue. Secondary structure features in E. coli and Neisseria meningitidis MinE are shown (21, 22, 36). The E. coli MinE N-terminal domain (residues 1–35) is predicted to consist of an extended or nascent helix (22).
MinD is a member of the ParA superfamily that, together with MinC, prevents placement of the cell-division septum at random locations in Escherichia coli. Inhibition is relieved specifically at midcell by the MinE protein, thereby allowing cell division at the correct site (18). MinE (10.2 kDa) stimulates ATP hydrolysis by MinD (19) and modulates polymerization of MinD into filaments (20) that are morphologically similar to ParF fibers (9). Thus, ParG and MinE are small, dimeric proteins that influence nucleotide hydrolysis and polymerization of their cognate ParA homologs. Furthermore, the C termini of both proteins are well structured, albeit with different topologies, with more disordered N termini (6, 21, 22). Intriguingly, the MinE N terminus is required for stimulation of ATP hydrolysis by MinD (19), just as for the ParG–ParF pair. Similarity in MinE N termini is extensive, and two highly conserved arginine residues (R21 and R30) in this region have been highlighted (22, 23) (Fig. 5B). Mutagenesis of R30 in a peptide spanning the first 31 aa of MinE led to impaired interaction with MinD in vivo (24). The MinER21A protein was too unstable to characterize (19). We speculate that, like ParG, the N-terminal flexible region of MinE possesses an arginine finger loop, with either R21 or R30 as catalytic residue provided in trans to the ATP-binding pocket of MinD.
Discussion
ParF is the prototype of a phylogenetically distinct subgroup belonging to the ParA family of Walker-type ATPases. This family is part of the SIMIBI superfamily of GTPases and ATPases, named after its three largest subgroups: signal recognition GTPases, the MinD class, and BioD proteins. The other large class of P-loop GTPases includes enzymes involved in translation, signal transduction (Ras-like family), and intracellular trafficking (25). Hydrolases of both classes are characterized by a Walker A motif that harbors a flexible P-loop nested between β-strand and α-helix. The loop contains a GXXXXGK[ST] signature motif that determines the molecular geometry of the catalytic niche by correctly positioning the triphosphate moiety of bound nucleotide.
ParG stimulates ParF ATPase activity through its flexible N-terminal domain (9; this work). The molecular mechanism responsible for this activation was previously unknown. Enhancement of nucleotide hydrolysis by a partner protein is a conserved regulatory feature of ParA family members, including P1 ParA (10, 26), F SopA (27), Caulobacter crescentus ParA (28), Thermus thermophilus Soj (29), E. coli cell division protein MinD (18, 19), and more broadly by Walker-type GTPases like Ras (30). Structure-prediction programs highlighted a number of potential bona fide structural homologs of ParF (1), including Soj, MinD, nitrogenase Fe protein, Ffh and FtsY signal-recognition particle GTPases, and, more distantly, human Ras. Ras has intrinsically low GTP hydrolysis, which is stimulated by a plethora of Ras GTPase-activating proteins (RasGAPs), of which p120GAP was first described (31). RasGAPs enhance hydrolysis with an invariant arginine finger residue. The residue participates in catalysis by stabilization of the transition state by neutralization of the negative charge that develops during phosphoryl transfer, presumably because of its positively charged guanidinium group (32). Ffh and FtsY are structurally homologous GTPases that associate into a heterodimeric complex (33); they share a composite catalytic chamber and mutually stimulate their GTPase activity. The orientation of two conserved arginines within the active site is consistent with a pair of reciprocal arginine fingers that stabilize the transition state in trans (34). ParF ATPase activity apparently is stimulated by the ParG protein by an equivalent arginine finger-like mechanism. Specifically, a highly conserved arginine, R19, in the ParG N-terminal flexible tail, plays a vital role in promoting ATP hydrolysis by ParF and is required for accurate DNA segregation in vivo. Replacement of R19 by lysine or alanine completely abolished enhancement of ParF ATPase activity (Fig. 2A). Thus, even a positively charged lysine residue is unable to retain the stimulatory activity of ParG.
Arginine fingers must fulfil these criteria (15): (i) they are invariant within a subfamily of GTP/ATP-activating proteins; (ii) they cannot be functionally replaced, even by a basic residue; and (iii) mutation abrogates stimulatory activity without affecting binding of the two interacting proteins. Residue R19 in ParG satisfies all of these requirements. ParGR19K and ParGR19A mutant proteins are as proficient as wild-type ParG in binding and bundling ParF polymers (Fig. 4). Thus, analogously to Ras–RasGAP interactions, mutation of the arginine finger potentially affects the transition state during catalysis but not the ground state. The location of R19 within ParG region 17–23, which is characterized by more restricted flexibility than the rest of the N-terminal tail, provides another parallel with the Ras–RasGAPp120 complex, as the postulated arginine finger R789 of p120GAP is located in a loop, designated the finger loop (32). Region 17–23 of ParG similarly might act as a finger loop, transiently folding upon interaction with ParF to properly position R19 in the catalytic niche. Nevertheless, the first nine amino acids in ParG also may play an important topological role in aligning R19 into the ParF catalytic chamber, because Δ9ParG stimulates ParF ATPase activity to one-quarter the level of wild-type ParG.
A highly conserved arginine is located in the N termini of ParG homologs from diverse bacterial hosts (Fig. 5). It is tempting to speculate that these residues might be counterparts of R19 in ParG and might similarly work by arginine finger-like mechanisms to activate their respective ParF partners. The MinCDE cell-division system presents a number of parallels with the ParFG partition machine (9), including stimulation of ATPase activities of ParF and MinD by ParG and MinE, respectively. We propose that the N-terminal domain of MinE might activate MinD ATP hydrolysis by an arginine finger-like mechanism, supplying one or both conserved arginines in trans into the ATP-binding pocket in MinD. Moreover, this charge-balancing strategy of ATPase activation might be a universally conserved feature of regulatory partners of ParA-like proteins, including Spo0J implicated in chromosome segregation that has been reported to contain an arginine residue important for stimulating ATP hydrolysis by the ParA-type Soj protein (29).
Separately from enhancement of ATP hydrolysis by ParF, the ParG flexible N terminus promotes ParF polymerization. ATP addition triggers instantaneous polymerization indicated by a surge in intensity of scattered light in DLS analysis. Subsequent addition of ParG to polymerizing ParF resulted in a further sharp augmentation in scattered light intensity (Fig. 3A). Symptomatically, the intensity increase is not paralleled by abrupt growth in polymer size, which is suggestive of a role for ParG in bundling and thus remodeling ParF protofilaments, rather than provoking an acceleration of their assembly. Thicker ParF fibers create a mesh and scatter more light, although they retain the same length as before ParG addition. However, ParG N-terminal deletions did not trigger an increase in scattered light (Fig. 3B). This finding reveals that the ability to reorganize ParF polymers resides in the N-terminal tail of ParG, and loss of as few as nine residues entirely compromises this function. The N-terminal tip, comprising the first 6–10 residues is the most flexible segment of the tail (6). This function of ParG is reminiscent of the role played by eukaryotic actin-binding proteins like CapZ. The C termini (≈30 residues) of CapZ are mobile tentacles that cap the barbed ends of actin polymers (17). For both ParG and CapZ, protein flexibility is instrumental in this role. The ability of ParG to promote ParF polymerization without ATP, a capacity lost by the N-terminal truncations (Fig. 3C), also supports ParG acting as a nucleating factor for ParF filamentation. Despite defects in ATPase stimulation, ParGR19K and ParGR19A preserve their role in promotion of ParF polymerization (Fig. 4B). This segregatio of roles establishes that enhancement of ParF ATP hydrolysis and stimulation of ParF polymerization are two separable functions performed by the N-terminal tail of ParG. The dynamic interplay between a nucleotide-dependent polymerizing protein and its cognate activating factor is likely to be a key mechanism that universally controls the DNA segregation process in bacteria.
Materials and Methods
Strains and Plasmids.
E. coli DH5α was used for plasmid construction, and BL21(DE3) was used for protein overproduction. Partition assays were performed in strain BR825 (3). Plasmids used to overproduce ParF, ParG, and ΔParG truncations were described (4, 7). Partition and overexpression constructs harboring point mutations in parG resulting in R19K, R19A, R23K, and R23A changes were generated by overlap extension mutagenesis. The overexpression vector used was pET22b(+).
Bacterial Two-Hybrid Analysis.
Assays were performed as described (4). Δ9parG, Δ19parG, and Δ30parG were cloned in pT25 (35) as described (7).
Protein Purification.
His-tagged ParF, ParG, ΔParG, ParGR19K, and ParGR19A proteins were purified as described (4, 7). His-tagged ParF and ParG are as proficient as wild-type proteins in supporting plasmid partition in vivo (4).
ATPase Assay by TLC.
ATPase assays were performed as described (9). Data shown are typical of experiments performed at least in triplicate.
DLS.
DLS experiments were performed as described (9). ParG, ΔParG truncations, ParGR19K, and ParGR19A were added at ratios indicated in figure legends.
Polymerization Assay.
Pelleting assays were carried out as described (9). ParG and ParG mutant proteins were added as indicated. In each gel, 100% of the pellet and 33% of the supernatant were loaded. Protein bands were quantitated with ImageJ (National Institutes of Health, Bethesda, MD).
Electrophoretic Mobility Shift Assay.
Band-shift assays were performed as detailed (4) by using a biotinylated 48-bp oligonucleotide corresponding to the parFG operator (FS-48) as described (7).
Partition Assay.
Plasmids replicating at low copy number in a polA strain were grown for ≈25 generations in the absence of chloramphenicol selective pressure, after which plasmid retention was assessed by replica plating to agar medium with and without antibiotic (3). Values shown are averages of at least three experiments. Plasmid pFH450 was the empty vector, and pFH547 was the construct containing the wild-type parFG cassette (3).
Acknowledgments
This work was supported by a Medical Research Council New Investigator Award (to D.B.) and by grants from The Wellcome Trust and Biotechnology and Biological Sciences Research Council (to F.H.).
Abbreviations
- DLS
dynamic light scattering
- RasGAP
Ras GTPase-activating protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Hayes F, Barillà D. Nat Rev Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 2.Ebersbach G, Gerdes K. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 3.Hayes F. Mol Microbiol. 2000;37:528–541. doi: 10.1046/j.1365-2958.2000.02030.x. [DOI] [PubMed] [Google Scholar]
- 4.Barillà D, Hayes F. Mol Microbiol. 2003;49:487–499. doi: 10.1046/j.1365-2958.2003.03564.x. [DOI] [PubMed] [Google Scholar]
- 5.Fothergill TJG, Barillà D, Hayes F. J Bacteriol. 2005;187:2651–2661. doi: 10.1128/JB.187.8.2651-2661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golovanov AP, Barillà D, Golovanova M, Hayes F, Lian L-Y. Mol Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 7.Carmelo E, Barillà D, Golovanov AP, Lian L-Y, Derome A, Hayes F. J Biol Chem. 2005;280:28683–28691. doi: 10.1074/jbc.M501173200. [DOI] [PubMed] [Google Scholar]
- 8.Motallebi-Veshareh M, Rouch DA, Thomas CM. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 9.Barillà D, Rosenberg MF, Nobbmann U, Hayes F. EMBO J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis MA, Radnedge L, Martin KA, Hayes F, Youngren B, Austin SJ. Mol Microbiol. 1996;21:1029–1036. doi: 10.1046/j.1365-2958.1996.721423.x. [DOI] [PubMed] [Google Scholar]
- 11.Bouet JY, Funnell BE. EMBO J. 1999;18:1415–1424. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Dabrazhynetskaya A, Youngren B, Austin S. Mol Microbiol. 2004;53:93–102. doi: 10.1111/j.1365-2958.2004.04111.x. [DOI] [PubMed] [Google Scholar]
- 13.Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Nat Struct Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- 14.Nadanaciva S, Weber J, Wilke-Mounts S, Senior AE. Biochemistry. 1999;38:15493–15499. doi: 10.1021/bi9917683. [DOI] [PubMed] [Google Scholar]
- 15.Scheffzek K, Ahmadian MR, Wittinghofer A. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 16.Scrima A, Wittinghofer A. EMBO J. 2006;25:2940–2951. doi: 10.1038/sj.emboj.7601171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA. Curr Biol. 2003;13:1531–1537. doi: 10.1016/s0960-9822(03)00559-1. [DOI] [PubMed] [Google Scholar]
- 18.Rothfield L, Taghbalout A, Shih YL. Nat Rev Microbiol. 2005;3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- 19.Hu Z, Lutkenhaus J. Mol Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 20.Suefuji K, Valluzzi R, RayChaudhuri D. Proc Natl Acad Sci USA. 2002;99:16776–16781. doi: 10.1073/pnas.262671699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King GF, Rowland SL, Pan B, Mackay JP, Mullen GP, Rothfield LI. Mol Microbiol. 1999;31:1161–1169. doi: 10.1046/j.1365-2958.1999.01256.x. [DOI] [PubMed] [Google Scholar]
- 22.King GF, Shih YL, Maciejewski MW, Bains NP, Pan B, Rowland SL, Mullen GP, Rothfield LI. Nat Struct Biol. 2000;7:1013–1017. doi: 10.1038/80917. [DOI] [PubMed] [Google Scholar]
- 23.Itoh R, Fujiwara M, Nagata N, Yoshida S. Plant Physiol. 2001;127:1644–1655. [PMC free article] [PubMed] [Google Scholar]
- 24.Ma LY, King G, Rothfield L. J Bacteriol. 2003;185:4948–4955. doi: 10.1128/JB.185.16.4948-4955.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leipe DD, Wolf YI, Koonin EV, Aravind L. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 26.Fung E, Bouet JY, Funnell BE. EMBO J. 2001;20:4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libante V, Thion L, Lane D. J Mol Biol. 2001;314:387–399. doi: 10.1006/jmbi.2001.5158. [DOI] [PubMed] [Google Scholar]
- 28.Easter J, Jr, Gober JW. Mol Cell. 2002;10:427–434. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- 29.Leonard TA, Butler J, Löwe J. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel US, Dixon RA, Schaber MD, Diehl RE, Marshall MS, Scolnick EM, Sigal IS, Gibbs JB. Nature. 1988;335:90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- 31.Trahey M, McCormick F. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 32.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schitz F, Wittinghofer A. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 33.Miller JD, Bernstein HD, Walter P. Nature. 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 34.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karimova G, Pidoux J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos D, Ducat T, Cheng J, Eng NF, Dillon JA, Goto NK. Biochemistry. 2006;45:4593–4601. doi: 10.1021/bi060022j. [DOI] [PubMed] [Google Scholar]