Abstract
Cooperative interactions are key to diverse biological phenomena ranging from multicellularity to mutualism. Such diversity makes the ability to create and control cooperation desirable for potential applications in areas as varied as agriculture, pollutant treatment, and medicine. Here we show that persistent cooperation can be engineered by introducing a small set of genetic modifications into previously noninteracting cell populations. Specifically, we report the construction of a synthetic obligatory cooperative system, termed CoSMO (cooperation that is synthetic and mutually obligatory), which consists of a pair of nonmating yeast strains, each supplying an essential metabolite to the other strain. The behavior of the two strains in isolation, however, revealed unintended constraints that restrict cooperation, such as asymmetry in starvation tolerance and delays in nutrient release until near cell death. However, the joint system is shown mathematically and experimentally to be viable over a wide range of initial conditions, with oscillating population ratio settling to a value predicted by nutrient supply and consumption. Unexpectedly, even in the absence of explicitly engineered mechanisms to stabilize cooperation, the cooperative system can consistently develop increased ability to survive reductions in population density. Extending synthetic biology from the design of genetic circuits to the engineering of ecological interactions, CoSMO provides a quantitative system for linking processes at the cellular level to the collective behavior at the system level, as well as a genetically tractable system for studying the evolution of cooperation.
Keywords: mathematical modeling, mutualism, obligate cooperation, quantitative biology, synthetic ecology
In nature, cooperation emerges under diverse conditions and over varying scales ranging from the physiological, as in the emergence of cell–cell cooperation that facilitates tumor progression (1), to the ecological, as in the evolution of mutualistic interactions between species (2, 3). In laboratory experiments, cooperation among cells of a single population has been shown to arise spontaneously under selective pressures (4), and cooperation between two populations has been attained either through mixing of organisms with natural capacities to cooperate (5, 6) or through evolution from originally parasitic associations (7, 8). All these systems relied on natural processes to establish cooperation. Here, by constructing a synthetic cooperative system, we show that it is possible to create obligatory cooperation, the most stringent form of cooperation, between two previously noninteracting yeast populations.
The existence of natural obligatory cooperative systems (9–13) is puzzling because the viability of both partners relies on cooperation. In certain cases, persistence of the system is achieved through endosymbiosis and vertical transmission of the symbiont (14). However, when both partners are free-living, as in the cases of certain flowers and their pollinators (11, 15, 16) and of metabolically coupled microbes (17–19), it is not clear how reliably viable cooperative communities can form under various initial conditions or how well they can recover from perturbations such as reductions in population size resulting from population bottlenecks. Few studies quantify how features of a cooperative system are affected by intrinsic constraints stemming from the cooperating partners, such as limited or delayed provision of supplies and imbalanced abundance of partners. This is presumably due to difficulties in measuring beneficial exchanges and population dynamics (2) and in disengaging cooperation from noncooperative interactions, such as competition and inhibition (10, 20) in natural systems.
A simplified synthetic system offers an opportunity to study an elementary ecological interaction in isolation, much like studying a single biochemical reaction outside a cell. Quantitative analysis on the synthetic system can in principle link processes at a finer scale, such as growth, death, and interactions of cooperating cells, to phenomena at a broader scale (21, 22), such as viability outcome and population dynamics of the cooperative system.
Results and Discussion
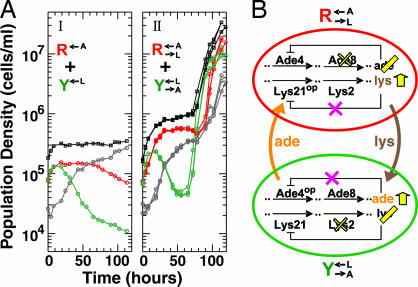
As the initial step, we genetically modified the yeast Saccharomyces cerevisiae to obtain two nonmating strains with different metabolic capabilities (Methods) so that they behave essentially as two different species. Specifically, the R←A strain, labeled with red-fluorescent protein (DsRed), synthesizes lysine at normal levels but requires adenine to grow; and the Y←L strain, labeled with yellow-fluorescent protein (YFP), synthesizes adenine at normal levels but requires lysine to grow. R←A and Y←L can be propagated in monocultures in the presence of adenine and lysine supplements, respectively. When the two strains were washed free of supplements and subsequently mixed to form a coculture, both strains initially underwent residual growth using stored metabolites (23, 24) but eventually died off (Fig. 1AI). Thus, although the two populations together have the required enzymes to synthesize both adenine and lysine, their coculture failed to achieve sustained growth.
Fig. 1.
Rational design of CoSMO. (A) Overproduction of metabolites is required for viable cooperation. At time 0, monocultures of indicated strains grown in synthetic dextrose medium (SD) with the required adenine or lysine supplement (37) were washed free of supplements and mixed. Plots show population dynamics of fluorescent live R (red), fluorescent live Y (green), nonfluorescent dead (gray), and total (black) cells of the coculture as measured by flow cytometry (Methods). (II) Data from three replicate cultures are superimposed. (B) The “wiring” diagram of CoSMO. CoSMO consists of two yeast strains: R→L←A, which lacks Ade8 enzyme and harbors Lys21op enzyme, and Y→A←L, which lacks Lys2 enzyme and harbors Ade4op enzyme. Cells lacking Ade8 (Lys2) cannot synthesize adenine (lysine) and therefore require intake (←) of the corresponding metabolite. Ade4op and Lys21op are no longer sensitive to end-product feed-back inhibition and consequently overproduce (op) the corresponding metabolite that is eventually released (→) into the medium (25, 26). Crosses represent genetic inactivation; yellow bars and arrows represent losses and gains in metabolite synthesis, respectively.
To create cooperation, we introduced an additional mutation in each strain by replacing the first enzyme in an adenine or a lysine biosynthetic pathway with an overproduction mutant that is no longer sensitive to end-product feedback inhibition (25, 26). Consequently, R←A and Y←L were respectively transformed into R→L←A, which requires adenine to grow and overproduces lysine, and Y→A←L, which requires lysine to grow and overproduces adenine (Fig. 1B).
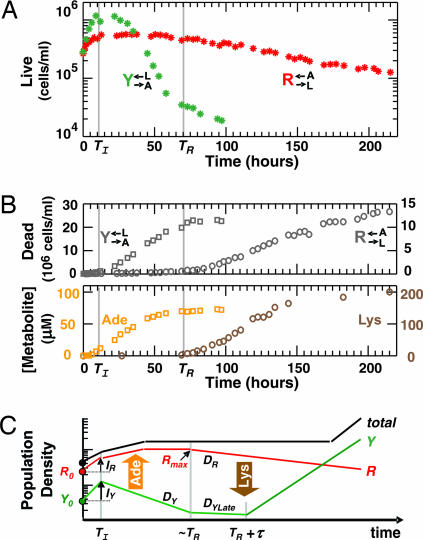
Despite our goal of creating a simple cooperative system, behavior of individual strains in monocultures reveals unintended constraints that restrict cooperation, such as asymmetric starvation tolerance between the two strains and delayed metabolite release until near cell death. After washout of the essential adenine or lysine supplement, each strain initially underwent residual growth using stored metabolites (23, 24) until time TI ≈ 10 h (Fig. 2A). Immediately afterward, Y→A←L cells entered the death phase characterized by a decrease in the number of live cells (Fig. 2A, green stars) and an increase in the number of dead cells (Fig. 2B, gray squares). R→L←A cells, in contrast, did not enter death phase until time TR, ≈70 h (Fig. 2A red stars and Fig. 2B gray circles). The release of the overproduced metabolites into the medium was associated with cell death (Fig. 2B). Consequently, the onset of lysine release by R→L←A was significantly delayed until time TR, when the majority of Y→A←L population already lost viability (Fig. 2B).
Fig. 2.
Characterization of individual strains in monocultures and deduction of CoSMO growth pattern. (A and B) Asymmetry in starvation tolerance between two strains and delayed metabolite release. At time 0, monocultures of the two strains grown in the presence of the required supplement were washed free of the supplement. (A) Live population density over time for an initial population density of ≈3 × 105 cells per ml. (B) Dead population density (Upper) and the concentration of lysine or adenine released into the medium over time (Lower) as measured by a bioassay (Methods) for an initial population density of ≈6 × 106 cells per ml. The left and right scales are for experiments on Y→A←L (squares) and R→L←A (circles), respectively. Gray vertical lines mark the time TI when residual growth ends and the time TR when R→L←A enters death phase and releases lysine. (C) A schematic diagram of the initial stage of CoSMO growth deduced from A and B. R and Y denote live population densities of R→L←A and Y→A←L, respectively. Their initial values R0 and Y0 increase IR- and IY-fold, respectively, during residual growth until time TI. After TI, adenine released from dying Y→A←L enables growth of R→L←A. By time ≈TR, most of the Y→A←L population has died and R is at a local maximum Rmax. Lysine is subsequently released from dying R→L←A, and at some time τ after TR, results in an increase in Y under conditions that permit CoSMO viability. The death rate for R→L←A after TR is DR, and for Y→A←L is DY from TI to TR and DYLate from TR onward. The total cell density, which is the sum of R, Y, and dead populations, consequently takes on a pattern of “rise-plateau-rise,” with each rise resulting from net growth of at least one partner.
Despite the presence of strong constraints, cooperation can exist between R→L←A and Y→A←L, as verified through the viability of their cocultures. Coculture viability is defined here as the ability to attain saturation density (≈5 × 107 total cells per ml) in the absence of adenine and lysine supplements. We found that cocultures initiated at low density (≈105 total cells per ml) can be viable (Fig. 1AII) and that viability of cooperation requires both adenine- and lysine-overproduction mutations [supporting information (SI) Fig. 6]. Together, R→L←A and Y→A←L form a cooperative system termed CoSMO (cooperation that is synthetic and mutually obligatory), which mimics two-species obligate mutualistic systems in which cooperation is essential for the survival of both species (11, 14, 17, 19, 27, 28).
We used the individual characteristics of the two strains to compute viability conditions for CoSMO (Fig. 2C and Appendix). A fundamental requirement for system viability is that the supply of metabolites has to be sufficiently high to sustain net growth of both partners. In mathematical terms, this condition is expressed as AsLs/AcLc > 1 (Appendix, supply–consumption requirement), where As (Ls) is the total amount of adenine (lysine) supplied per Y→A←L (R→L←A) cell until its death, and Ac (Lc) is the amount of adenine (lysine) consumed to make a new R→L←A (Y→A←L) cell. For CoSMO, experimentally measured values (SI Table 1) lead to AsLs/AcLc ≈ 22, implying that CoSMO significantly exceeds this fundamental supply–consumption requirement.
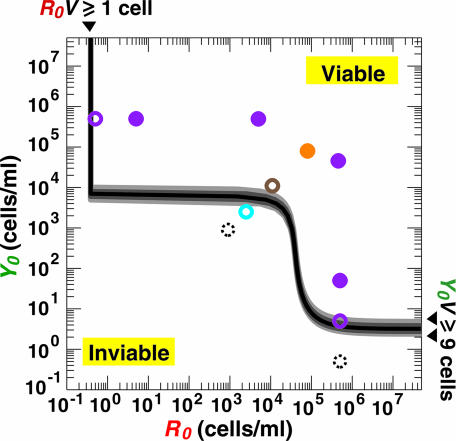
However, the system can fail to be viable if a released metabolite is too dilute and therefore its uptake rate is too slow to keep its consumer alive, or if any one strain goes extinct before its partner strain has a chance to release a substantial amount of metabolite. These two failure modes lead to constraints on the initial cell densities (Appendix, minimum initial cell density requirement) and initial cell numbers (Appendix, minimum initial cell number requirement). The two requirements can be combined to define the viability and inviability domains of CoSMO as a function of the initial densities of the two strains at a given volume. The two domains can be represented in a phase diagram (Fig. 3), which collapses multiple quantitative properties of the component strains into a concise predictive description of CoSMO system behavior.
Fig. 3.
Phase diagram for CoSMO viability. The domain of viability for CoSMO at volume V of 2.6 ml is bounded by a black vertical line (single arrowhead, Appendix, inequality 6) and gray curves (Appendix, Inequality 8, with IY set to different values in the experimentally observed range from 2 to 4). The shoulder (⌉) represents the viability threshold imposed by the density requirement alone (Appendix, Eqs. 4 and 5) and is therefore not affected by the culture volume. Different volumes affect only the black vertical line (single arrowhead) and the horizontal asymptote (double arrowhead), which shift along the R0 and Y0 axis according to the initial-number requirements expressed in Appendix inequalities 6 and 8a, respectively. Circles indicate values of (R0, Y0) corresponding to different experiments (orange for Fig. 1AII; purple from top left to bottom right for Fig. 4A, from I to VI; and brown for Fig. 4B). In experiment marked with cyan, one of five replicate cultures was viable (time series not shown); in experiments marked with black, zero of four or five replicate cultures was viable (time series not shown). Overall, the viability–inviability outcome of replicate CoSMO cultures close to the calculated boundary is highly variable (open circles), whereas cultures significantly above and below the boundary show 100% viability (filled circles) and 0% viability (broken circles), respectively.
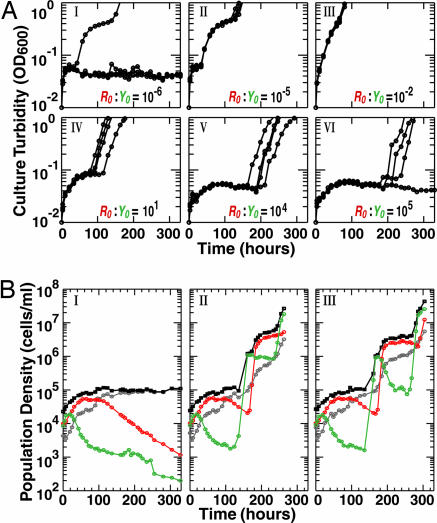
For initial conditions well within the calculated inviable domain (Fig. 3, broken circles), replicate CoSMO cultures are never viable (time series not shown). In contrast, for initial conditions well within the calculated viable domain, replicate CoSMO cultures are always viable (Fig. 3, filled circles). Specifically, when both cell-density and cell-number requirements were exceeded by at least ≈10-fold (e.g., Fig. 3, purple filled circles), CoSMO cultures initiated at R→L←A:Y→A←L ratios spanning 9 orders of magnitude from 10−5 to 104 achieved 100% viability (Fig. 4A II–V). The inherent ability to survive a wide range of partner ratios is important for natural cooperative systems because such wide ranges would be expected either as a result of initial encounters of partners at different population sizes or through intrinsic system dynamics.
Fig. 4.
Viability of CoSMO. Monocultures of the two strains were washed free of adenine and lysine, and mixed at time 0. (A) CoSMO is viable under a wide range of initial partner ratios. The two strains were mixed at the indicated R→L←A:Y→A←L ratios (R0:Y0) and at the same total initial cell density and culture volume (≈5 × 105 cells per ml × 2.6 ml = 1.3 × 106 cells per culture, four replicate cultures per condition). Plots show culture turbidity in OD600 (optical density at 600 nm) over time. OD600 of 1 corresponds to a population density from 1 × 107 to 5 × 107 cells per ml depending on the cell size. (B) Stochastic CoSMO behavior close to the initial-density requirement. Three replicate cultures (≥20 ml) were set up at 1.1 × 104 cells per ml per strain. Plots show the dynamics of live R→L←A (red), live Y→A←L (green), dead (gray), and total (black) cell densities. One of the cultures was inviable (I), whereas the other two were viable (II and III).
For initial conditions close to the boundary that separates the viable and inviable domains (black and gray curves in Fig. 3), the behavior of CoSMO is stochastic (Fig. 3, open circles), leading replicate cultures nondeterministically to either viability or inviability. For instance, when the initial-density requirement was significantly exceeded and the initial-number requirement for either strain was barely satisfied (Fig. 3, purple open circles), only a fraction of replicate CoSMO cultures were viable (Fig. 4A I and VI). Similarly, when the initial-density requirement was barely satisfied and the initial-number requirements were significantly exceeded (e.g., Fig. 3, brown open circle), CoSMO was not viable in one case (Fig. 4BI), whereas it reached saturation in the other two (Fig. 4B II and III). This stochastic behavior is often observed in systems close to a transition point between two states, where even small fluctuations can drive the system either way.
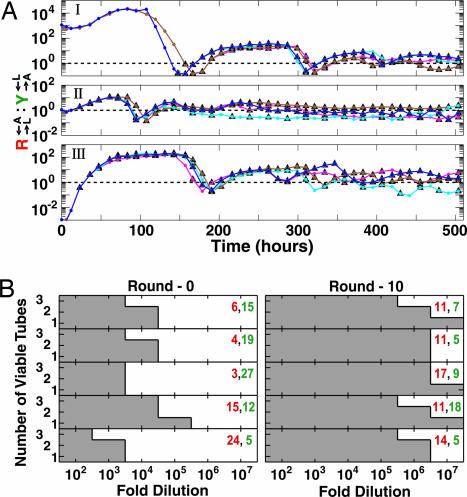
Properties of components can be used not only to determine system viability, but also to elucidate certain features of CoSMO dynamics. In particular, on long-term culturing, the wide range of initial population ratios compatible with CoSMO viability converges to a narrow range. CoSMO cultures were seeded at R→L←A:Y→A←L ratio of 103, 1, and 10−3 (Fig. 5A I, II, and III, respectively) and maintained at low density through monitoring of culture turbidities and performing dilutions at a fixed frequency (once or twice per day). Although population ratios initially spanned 6 orders of magnitude, they underwent several oscillations and eventually settled into a narrow range between 1:5 and 5:1 (Fig. 5A). In a similar experiment where dilutions were performed at high cell density, the oscillating population ratio converges to the same range (SI Fig. 7). The stabilized ratio can be computed from the supply and consumption of adenine and lysine, which can have any value in general and is on the order of 1 for CoSMO (Methods).
Fig. 5.
Long-term changes in CoSMO. (A) Stabilization of partner ratios. At time 0, duplicate CoSMO cultures (brown and blue) were initiated at OD600 of 0.01 (4.7 × 105 total cells per ml) and R→L←A:Y→A←L ratios of 103 (I), 1 (II), and 10−3 (III). When OD600 exceeded the set point of 0.06 for the first time, two 3-ml samples were taken from each culture (brown and magenta from the brown; blue and cyan from the blue), and thereafter diluted once per day (magenta and cyan) or twice per day (brown and blue) to the set point. A low set point was chosen so that nutrients other than adenine and lysine were not limiting. Plots show R→L←A:Y→A←L ratios over time, with triangles marking points of dilution. (B) Increased ability to survive reductions in population density. Five 2.6-ml CoSMO cultures, initiated at different partner ratios, were grown to near-saturation and used as Round-0 cultures for five independent series. After 10 rounds of dilution and regrowth in 2.6 ml, a near-saturation Round-10 culture was obtained for each series. Each row corresponds to a particular series and depicts the number of tubes (of three) that were viable at indicated dilutions for Round-0 (Left) and Round-10 (Right) cultures. Population densities of R→L←A (red) and Y→A←L (green) in million cells per ml for Round-0 and Round-10 cultures are shown in the Inset.
Stabilization of the population ratio irrespective of the starting ratio and the dilution regime suggests that CoSMO may be regarded as a single cooperative entity with the two partners serving as mutually dependent components. The relative quantities of components are self-adjusted to achieve a stoichiometric balance between nutrient supply and nutrient consumption. Ratio convergence has been observed in other cooperative systems (20, 29) and it is possible that similar mechanisms are important in the evolution of multicellularity where proportions of different cooperating cell types are regulated (30).
An intriguing aspect of CoSMO is that, upon long-term culturing, the cell-density requirement for viability can undergo drastic reduction. The cell-density requirement was estimated by performing a series of dilutions of a viable CoSMO culture and evaluating the minimum density at which viability was retained. Such an experiment is a laboratory analog of a natural system recovering from population bottlenecks, a commonly occurring perturbation. Five CoSMO cultures were initiated, grown to near-saturation (Round 0), and subjected to 10 rounds of dilution-and-regrowth, ending in Round-10 cultures (protocol illustration in SI Fig. 8). The population density of both Round-0 and Round-10 cultures was on the order of 107 total cells per ml. The Round-0 cultures typically tolerated 103- to 104-fold dilution (Fig. 5B Left), and therefore a total population density on the order of 103 to 104 cells per ml was required for the viability of a diluted CoSMO culture. In contrast, the Round-10 cultures typically tolerated 105- to 106-fold dilution (Fig. 5B Right), and therefore a total population density on the order of 101 to 102 cells per ml was sufficient for viability. Thus, although the initial requirements for viable cooperation can be accurately predicted from properties of components (Fig. 3), the density requirement underwent 100-fold relaxation over a relatively short period (≈70 generations). This phenomenon may result from changes in one or both strains that aid the survival of the strain itself (e.g., through increased starvation tolerance) or the survival of its partner (e.g., through increased overproduction or expedited release of metabolites). Unlike natural systems in which partner coevolution has rendered the evolutionary history of cooperation difficult to retrace (11), multiple CoSMO cultures can be initiated and their evolutionary trajectories compared. Uncovering the nature of these changes will elucidate the adaptation pathways of cooperation and the diversity in adaptive strategies (6).
Our results show that persistent cooperation between two populations can be created de novo through a small set of targeted genetic modifications. In fact, each population is essentially only one mutation step away from being a cooperator. Despite its artificial origin, CoSMO closely mimics aspects of naturally occurring cooperative systems such as exchange of essential nutrients between partners (14, 19, 28), death of a fraction of one partner population to support the reproduction of the other partner (11, 27), and delayed reward for a particular investment (11, 27). Even with obstacles such as severe delays in nutrient release, significant asymmetry in partners' starvation tolerance, and skewed population ratios resulting from intrinsic dynamics, the range of conditions permissible for cooperation is wide, consistent with the diversity observed in naturally occurring cooperative systems (2, 3). Although the interplay between cooperative organisms in natural systems must be far richer and deeper, we show that even in a simplified synthetic cooperative system, novel properties such as increased ability to stay alive could emerge. Future work is required to analyze the viability requirements, the population dynamics, and the evolution of CoSMO when challenged by “cheater” strains such as Y←L and R←A, which consume metabolites and release none. This would allow a quantitative assessment of a key question in the evolution of cooperation: the persistence of cooperation in the face of individuals that reap benefits without paying the cost of cooperation.
Our work highlights the importance of quantitatively linking processes on finer scales to system behavior at larger scales and underscores the challenges in predicting the behavior of an adapting biological system.
Methods
Construction of CoSMO Components.
Yeast strains of desired genotypes were obtained through genetic crosses. The complete genotype for WY811 (R→L←A) is MATa ste3Δ::kanMX4 ade8Δ0 LYS21op trp1–289::pRS404(TRP)-ADHp-DsRed.T4 and that for WY833 (Y→A←L) is MATa ste3Δ::kanMX4 ADE4op lys2Δ0 trp1–289::pRS404(TRP)-ADHp-venus-YFP.
lys2Δ and ade8Δ mutations were derived from BY4743 (Euroscarf Y20000) (31) and SY9913 (32), respectively. Yeast cells of the same mating type do not mate. ste3Δ::KanMX4 (Euroscarf Y05028) cells lack Ste3, the receptor for a-mating factor (33). Thus, in the rare occasion where a cell of MATa ste3Δ genotype switches mating type to MATα ste3Δ, it still cannot mate.
ADE4op mutant is the PUR6 allele of ADE4 (26). LYS21op was isolated in an MNNG (1-methyl-3-nitro-1-nitrosoguanidine, Sigma–Aldrich, St. Louis, MO) mutagenesis screen as a mutation that was resistant to the lysine analog thialysine (l-4-thialysine hydrochloride, Sigma–Aldrich) and that also cross-fed lysΔ cells (34). One lysine-releasing mutation was dominant, showed tight linkage to LYS21 (20/20 tetrads), and was therefore assigned LYS21op. Both ADE4op and LYS21op mutations were backcrossed into the S288C background five times.
To introduce fluorescent protein markers, WSB37 and WSB41 were constructed after ligating three DNA fragments: the TRP1-integrating plasmid pRS404 (American Type Culture Collection, Manassas, VA) digested with SacI and XhoI, a SacI–HindIII fragment harboring the ADH promoter from pKW431 (35), and a HindIII–XhoI PCR fragment containing either Venus-YFP amplified from pDH6 (http://depts.washington.edu/∼yeastrc/) or RGS-His6-DsRed.T4 amplified from pQE81-L-DsRed.T4 (36). The resulting plasmids were linearized with XbaI and transformed into a yeast strain harboring trp1–289. Among TRP+ transformants, a stable integrant was selected such that all its progeny cells expressed the expected fluorescent protein even when grown in nonselective media containing tryptophane.
Measurement of Metabolite Concentration Using a Bioassay.
A series of SD media (37) supplemented with various amounts of metabolite adenine (lysine) and inoculated with a test strain auxotrophic for adenine (lysine) were grown to saturation (≈20 h). A linear regression of saturation OD600 values against concentrations of the metabolite was performed (correlation coefficient >0.99). To measure the metabolite concentration in a culture, the culture was filtered through a 0.2-μm filter, and the supernatant was supplemented with 1/10 volume of 10× SD and inoculated with the appropriate test strain. The metabolite concentration was obtained from the saturation OD600 value through interpolation.
Measurement of Population Dynamics Using Flowing Cytometry.
For every round of measurement on FACS Calibur (with 488-nm and 633-nm lasers; BD Biosciences, Franklin Lakes, NJ), the flow rate k of the instrument (μl/sec) was determined using a dilution series of a bead stock. Specifically, the concentration of a 6-μm bead stock (≈2 × 106 per ml; Duke Scientific, Fremont, CA, catalog no. 35-2) was measured using a hemacytometer. The bead stock was diluted 25-, 10-, 5-, and 2.5-fold to a standard 0.5-ml series of bead samples and processed by Calibur for 65 sec. The cumulative event counts at 5.2, 10.0, 15.2, 20.0, 25.2, and 30.0 sec were plotted against time, and the event rate (events per sec) for each bead sample was deduced from the slope. Event rates (events per sec) were plotted against bead densities (beads per μl) for the standard series, and the linear regression line was forced through the origin. The slope k was the flow rate of Calibur (μl/sec). The correlation coefficients of all linear regressions were >0.999.
To measure the population composition of a culture, a sample was diluted into H2O to OD600 ≈0.01 and briefly sonicated. S, the event rate of the sample (events per sec), was determined as described above for bead samples. The total cell density is S/k (events per μl). Clusters of DsRed-positive, YFP-positive, and dark cells were clearly segregated (SI Fig. 9), and the percentages of each cluster were calculated using FlowJo software (TreeStar, Ashland, OR). Dark cells accumulated during starvation and were considered dead because >99% (sample size >5,000) had lost colony-forming ability in a FACS analysis. Fluorescent cells are considered alive because all of them retain the ability to exclude the nucleic acid dye propidium iodide (sample size >150).
Calculation of the Steady-State Population Ratio.
When a finite nonzero steady state ratio is achieved, R→L←A and Y→A←L grow with the same rate Ḡ. Furthermore, let D̄R and D̄Y represent the death rates of R→L←A and Y→A←L at this stage, respectively.
Because ΔR̃/ΔR = D̄RRΔt/ḠRΔt and ΔỸ/ΔY = D̄YYΔt/ḠYΔt, Eq. 1 in Appendix becomes
Solving for ΔR/ΔY after replacing Ḡ, we obtain
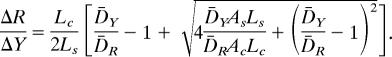 |
If we assume D̄Y/D̄R = DYlate/DR, then ΔR/ΔY ≈ 1 for CoSMO (SI Table 1). R/Y = (R0 + ΔR)/(Y0 + ΔY) tends to ΔR/ΔY when t is large because R0 and Y0 are small compared with ΔR and ΔY. Therefore, R/Y is on the order of 1 for CoSMO. In CoSMO, D̄Y/D̄R − 1 is small compared with (4D̄YAsLs/D̄RAcLc)1/2, in which case R/Y can be simplified to (D̄YAsLc/D̄RAcLs)1/2, a function of adenine and lysine supply rates (D̄YAs and D̄RLs) and consumption (Ac and Lc).
Supplementary Material
Acknowledgments
W.S. is particularly indebted to the following individuals at The Rockefeller University: S. Leibler, whose laboratory hosted most of the experiments; F. R. Cross for making his laboratory facilities available; and S. Mazel for superb assistance with flow cytometry. We thank S. G. Oliver, R. A. Woods, B. Glick, E. Dubois, D. Botstein, and the Yeast Resource Center at University of Washington for reagents and C. I. Bargmann, J. L. Bronstein, R. Chait, J. E. Cohen, F. R. Cross, D. A. Harrington, M. Heirtzler, J. N. Holland, M. Kampmann, M. J. B. Krieger, S. Leibler, R. Losick, Y. Lu, T. G. Marr, G. P. Moore, A. W. Murray, J. Robbins, L. Saiz, M. Schroeder, S. Shahriari, and H. Zu Dohna for discussions. W.S. was supported by the Damon Runyon Cancer Research Foundation and the Leibler laboratory.
Appendix: Three Requirements for CoSMO Viability
Supply–Consumption Requirement.
Supply of metabolites must be sufficiently high to sustain net growth of both components. Let As (Ls) be the total amount of adenine (lysine) supplied per Y→A←L (R→L←A) cell until its death, and let Ac (Lc) be the amount of adenine (lysine) consumed to make a new R→L←A (Y→A←L) cell. Assuming that all released metabolites are completely consumed, changes (Δ) in population densities of live R→L←A and Y→A←L, denoted R and Y, and of the corresponding dead cells, denoted R̃ and Ỹ, are related through
Positive growth of both components requires ΔỸ(As/Ac) > ΔR̃ and ΔR̃(Ls/Lc) > ΔỸ, which leads to AsLs/AcLc > 1. This condition is analogous to those derived in mathematical models of obligate mutually cross-feeding systems in chemostats at steady state (38).
Minimum Initial Cell-Density Requirement.
The growth rate of Y→A←L, GY, must exceed the death rate DYLate at a finite time τ after the initiation of lysine release from dying R→L←A at time ≈TR (Fig. 2C). If each Y→A←L cell uptakes lysine at concentration L in the medium following Michaelis–Menten kinetics with half-saturation constant KmL and maximum rate VmaxL, and produces a new cell after acquiring a quantity Lc of lysine, we obtain
Note that measured L is small compared with KmL. L is given by
where Rmax is the population density of R→L←A at time TR, and (1 − e−DR·τ) is the fraction of R→L←A cells that have died from time TR to TR + τ. Rmax is related to R0 and Y0, the initial population densities of the two partners, through
accounting for increase in R resulting first from IR-fold residual growth from R0 and then from adenine released on the death of almost the entire Y→A←L population, which has undergone IY-fold residual growth from Y0. From inequality 2, Eq. 3, and the measured parameters (SI Table 1), we obtain the minimal Rmax required for CoSMO viability as
Minimum Initial Cell-Number Requirement.
For a coculture of volume V, the initial number of R→L←A cells must be at least 1
In addition, there must be at least one Y→A←L cell alive at time TR + τ:
From inequalities 2 and 7 and Eqs 3–5, we obtain the condition
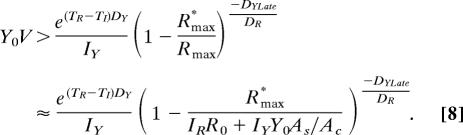 |
The minimum initial number of Y→A←L cells required for CoSMO viability is obtained after setting R0 in inequality 8 to the saturation density 5 × 107 cells per ml:
Footnotes
The authors declare no conflict of interest.
See Commentary on page 1741.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610575104/DC1.
References
- 1.Hanahan D, Folkman J. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 2.Boucher DH. The Biology of Mutualism: Ecology and Evolution. New York: Oxford Univ Press; 1985. [Google Scholar]
- 3.Bergstrom CT, Bronstein JL, Bshary R, Connor RC, Daly M, Frank SA, Gintis H, Keller L, Leimar O, Noe R, Queller DC. In: Genetic and Cultural Evolution of Cooperation. Hammerstein P, editor. Boston: MIT Press; 2002. [Google Scholar]
- 4.Rainey PB, Rainey K. Nature. 2003;425:72–74. doi: 10.1038/nature01906. [DOI] [PubMed] [Google Scholar]
- 5.Buchsbaum R, Buchsbaum M. Science. 1934;80:408–409. doi: 10.1126/science.80.2079.408. [DOI] [PubMed] [Google Scholar]
- 6.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 7.Jeon KW. Science. 1972;176:1122–1123. doi: 10.1126/science.176.4039.1122. [DOI] [PubMed] [Google Scholar]
- 8.Fiegna F, Yu YTN, Kadam SV, Velicer GJ. Nature. 2006;441:310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- 9.Hata H, Kato M. Biol Lett. 2006;2:593–596. doi: 10.1098/rsbl.2006.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowan R, Knowlton N, Baker A, Jara J. Nature. 1997;388:265–269. doi: 10.1038/40843. [DOI] [PubMed] [Google Scholar]
- 11.Cook JM, Rasplus JY. Trends Ecol Evol. 2003;18:325–325. and erratum (2003) 18:241–248. [Google Scholar]
- 12.Wernegreen JJ. Nat Rev Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 13.Pellmyr O, Leebens-Mack J. Proc Natl Acad Sci USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zientz E, Dandekar T, Gross R. Microbiol Mol Biol Rev. 2004;68:745–770. doi: 10.1128/MMBR.68.4.745-770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellmyr O, Thompson JN, Brown JM, Harrison RG. Am Nat. 1996;148:827–847. [Google Scholar]
- 16.Fleming TH, Holland JN. Oecologia. 1998;114:368–375. doi: 10.1007/s004420050459. [DOI] [PubMed] [Google Scholar]
- 17.Stams AJM, de Bok FAM, Plugge CM, van Eekert MHA, Dolfing J, Schraa G. Environ Microbiol. 2006;8:371–382. doi: 10.1111/j.1462-2920.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 18.Dean-Raymond D, Alexander M. Appl Environ Microbiol. 1977;33:1037–1041. doi: 10.1128/aem.33.5.1037-1041.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurmikko V. Experientia. 1956;12:245–249. doi: 10.1007/BF02157327. [DOI] [PubMed] [Google Scholar]
- 20.Yeoh HT, Bungay HR, Krieg NR. Can J Microbiol. 1968;14:491–492. doi: 10.1139/m68-083. [DOI] [PubMed] [Google Scholar]
- 21.Levin SA. Ecology. 1992;73:1943–1967. [Google Scholar]
- 22.Gause GF. The Struggle for Existence. Baltimore: Williams and Wilkins; 1934. [Google Scholar]
- 23.Messenguy F, Colin D, ten Have JP. Eur J Biochem. 1980;108:439–447. doi: 10.1111/j.1432-1033.1980.tb04740.x. [DOI] [PubMed] [Google Scholar]
- 24.Nagy M. Biochim Biophys Acta. 1979;558:221–232. doi: 10.1016/0005-2736(79)90062-2. [DOI] [PubMed] [Google Scholar]
- 25.Feller A, Ramos F, Pierard A, Dubois E. Eur J Biochem. 1999;261:163–170. doi: 10.1046/j.1432-1327.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- 26.Armitt S, Woods RA. Genet Res. 1970;15:7–17. doi: 10.1017/s0016672300001324. [DOI] [PubMed] [Google Scholar]
- 27.Pellmyr O, Huth CJ. Nature. 1994;372:257–260. [Google Scholar]
- 28.Kroon AGM, van Ginkel CG. Environ Microbiol. 2001;3:131–136. doi: 10.1046/j.1462-2920.2001.00170.x. [DOI] [PubMed] [Google Scholar]
- 29.Rai AN, Soderback E, Bergman B. New Phytologist. 2000;147:449–481. doi: 10.1046/j.1469-8137.2000.00720.x. [DOI] [PubMed] [Google Scholar]
- 30.Mohanty S, Firtel RA. Semin Cell Dev Biol. 1999;10:597–607. doi: 10.1006/scdb.1999.0343. [DOI] [PubMed] [Google Scholar]
- 31.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Tomlin GC, Wixon JL, Bolotin-Fukuhara M, Oliver SG. Yeast. 2001;18:563–575. doi: 10.1002/yea.703. [DOI] [PubMed] [Google Scholar]
- 33.Hagen DC, McCaffrey G, Sprague GF., Jr Proc Natl Acad Sci USA. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray GS, Bhattacharjee JK. J Gen Microbiol. 1976;97:117–120. doi: 10.1099/00221287-97-1-117. [DOI] [PubMed] [Google Scholar]
- 35.Stade K, Ford CS, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 36.Bevis BJ, Glick BS. Nat Biotechnol. 2002;20:83–87. [Google Scholar]
- 37.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic; 1991. [Google Scholar]
- 38.Meyer JS, Tsuchiya HM. Biotechnol Bioeng. 1975;17:1065–1081. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.