Abstract
Rolling-circle (RC) transposons, or Helitrons, are a newly recognized group of eukaryotic transposable elements abundant in the genomes of plants, invertebrates, and zebrafish. We provide evidence for the colonization of a mammalian genome by Helitrons, which has not been reported previously. We identified and characterized two families of Helitrons in the little brown bat Myotis lucifugus. The consensus sequence for the first family, HeliBat1, displays the hallmarks of an autonomous Helitron, including coding capacity for an ≈1,500-aa protein with an RC replication motif and a region related to the SF1 superfamily of DNA helicases. The HeliBatN1 family is a nonautonomous Helitron family that is only distantly related to HeliBat1. The two HeliBat families have attained high copy numbers (≈15,000 and > 100,000 copies, respectively) and make up at least ≈3% of the M. lucifugus genome. Sequence divergence and cross-species analyses indicate that both HeliBat families have amplified within the last ≈30–36 million years and are restricted to the lineage of vesper bats. We could not detect the presence of Helitrons in any other order of placental mammals, despite the broad representation of these taxa in the databases. We describe an instance of HeliBat-mediated transduction of a host gene fragment that was subsequently dispersed in ≈1,000 copies throughout the M. lucifugus genome. Given the demonstrated propensity of RC transposons to mediate the duplication and shuffling of host genes in bacteria and maize, it is tempting to speculate that the massive amplification of Helitrons in vesper bats has influenced the evolutionary trajectory of these mammals.
Keywords: Chiroptera, Helitron, horizontal transfer, mammalian genome, transposable elements
The largest fraction of most eukaryotic genomes is made up of interspersed repetitive DNA. Transposable elements (TEs) represent the major type of interspersed repeats and often constitute the single largest component of the genetic material. For example, approximately half of the human genome is made of TEs (1), and at least 60% of the maize genome is occupied by TEs (2). There is evidence that TEs have contributed profoundly to shaping eukaryotic genomes through their movement and amplification (for review, see refs. 3–5).
Mammalian TEs described so far fall within three types: non-long-terminal repeat (non-LTR) retrotransposons, retroviral-like (LTR) elements, and DNA transposons. The non-LTR retrotransposons are, by far, the most abundant type of TEs in the genomes of human, mouse, rat, and dog (1, 6–8). In humans, two predominant families of non-LTR retrotransposons (Alu and L1) account for more than one-fourth of the genome and have been major players in the structural genomic evolution of humans and other primates (9, 10). Much less is known about the nature and impact of TEs in the genome of other mammalian lineages.
We report on the discovery of rolling-circle (RC) transposons, also known as Helitrons, in the genome of the little brown bat, Myotis lucifugus. Although Helitrons and Helitron-related sequences have been identified in the genome of plants, fungi, invertebrates, and fish (11–14), these elements have not been described previously in a mammalian species. Helitrons are distinguished from other classes of TEs by their overall structure and by the putative enzymatic activities encoded by autonomous copies (11). Helitrons share structural and sequence similarities with a disparate group of prokaryotic mobile elements and single-stranded DNA (ssDNA) viruses that use RC replication, a process that involves the nicking, displacement, and ligation of an ssDNA intermediate (15–18). Helitrons are poorly characterized TEs in eukaryotes, but they make up ≈2% of the small genomes of Arabidopsis thaliana and Caenorhabidits elegans (11). Furthermore, recent genetic and sequence analyses have implicated Helitrons in large-scale duplication and exon shuffling of thousands of genes in the maize genome (19–22). We show here that Helitrons are a major component (at least 3%) of the M. lucifugus genome, and, as such, they have likely played an important role in shaping some mammalian genomes.
Results
Discovery of HeliBats.
Homology-based searches (tblastn) of the whole genome shotgun sequences (WGS) database using as a query a protein domain (≈160 residues) containing the RC motif of a sea urchin Helitron (E.J.P., unpublished data) yielded 94 hits with significant e values (<10−4) to M. lucifugus contigs. The first 75 hits span the entire RC domain and display 40–57% identity and 63–76% similarity to the query sequence. No significant hits were obtained with any other placental mammals, despite the current representation in the WGS database of 35 other species from most mammalian orders. Two significant hits were obtained with sequences from the platypus Ornithorhynchus anatinus (e values of 2 × 10−5 and 0.042). Closer inspection confirmed that these platypus contigs contain highly degenerated remnants of Helitron coding sequences (data not shown). Thus, a family of Helitrons also seems to have colonized the genome of an ancestral mammalian or monotreme species.
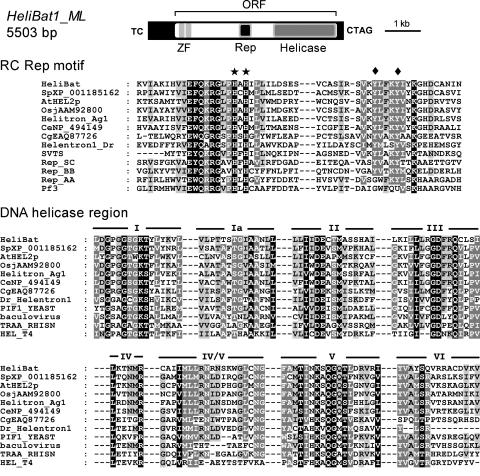
Using one of the M. lucifugus hits as a seed, we retrieved and aligned 15 closely related DNA sequences and their flanks. This multiple alignment was used to derive a consensus sequence of 5,503 bp that seems to represent a full-length member of a Helitron family that we named HeliBat1. The HeliBat1 consensus sequence possesses all of the hallmarks of a potential autonomous Helitron (11). First, the termini of the consensus and individual copies in the genome are defined by 5′-TC and CTAG-3′ motifs. Second, a short palindromic motif (18 bp with one mismatch and a single T nucleotide spacer) is located 11 nucleotides (nt) upstream of the 3′ terminus of the consensus (Fig. 1). These terminal features are consistent with those of Helitrons previously characterized in A. thaliana, rice, mosquito, and nematode (11, 12).
Fig. 1.
Terminal sequence features of HeliBat elements and other Helitrons. The 5′ and 3′ terminal sequences characteristic of Helitrons are shaded in black, and the 3′ palindromic motifs are underlined. The flanking A and T host nucleotides are in lowercase. Ml, M. lucifugus; Ce, C. elegans; Ag, Anopheles gambiae; Os, Oryza sativa; At, A. thaliana.
The HeliBat1 consensus sequence contains a long ORF that potentially encodes a 1,496-aa protein. This putative protein can be aligned over its entire length with the proteins encoded by potential autonomous Helitrons previously identified from the aforementioned species and other eukaryotic species [see supporting information (SI) Fig. 4]. The HeliBat1 protein is of similar length to other Helitron proteins (typically 1,300–1,700 aa) and contains the same domain organization (Fig. 2). The N-terminal region contains predicted zinc-finger-like motifs. The central region shares similarity with the Rep domain of other Helitron proteins (11) and contains a motif that can be confidently aligned with the so-called “two-His” replication initiator motifs of some prokaryotic mobile elements, plasmids, and ssDNA viruses (Fig. 2). The Rep motif is essential for the life cycle of these genetic elements, because it catalyzes the cleavage and ligation of DNA during RC replication (17, 18). The residues known to be critical for this activity are all conserved in the HeliBat1 putative protein (Fig. 2). The C-terminal half of the HeliBat1 protein has significant similarity with several eukaryotic and prokaryotic DNA helicases that bind to 5′ ssDNA and unwind the duplex DNA in a 5′ to 3′ direction (23). For example, it has 43% similarity over 418 aa with the TPR domain protein from the α-proteobacteria Oceanicaulis alexandrii (GenBank accession no. ZP_00957251). The strongest similarity is with members of the SF1 superfamily of DNA helicases and in particular with the Pif1p family (24). The seven conserved motifs that define the SF1 superfamily are present and well conserved in the HeliBat1 protein (Fig. 2).
Fig. 2.
Genetic organization and predicted functional protein domains of HeliBat1. (Top) A schematic representation of the genetic organization of HeliBat1 and domain structure of the putative encoded protein. ZF, zinc-finger-like motifs; Rep, RC replication initiator motif; Helicase, region similar to SF1 superfamily of DNA helicases. (Middle) An alignment of the REP motif of HeliBat1, representative Helitrons from seven other species [abbreviations as in Fig. 1, plus Sp, Strongylocentrotus purpuratus; Cg, Chaetomium globosum (a fungus); Dr, Danio rerio] and several RC viruses and plasmids (SVTS, Spiroplasma plectrovirus; Rep_SC, Streptomyces cyaneus plasmid; Rep_BB, Bacillus borstelensis plasmid; Rep_AA, Actinobacillus actinomycetemcomitans plasmid; TRAA_RHISN, Rhizobium sp.; NGR234Pf3, Pseudomonas aeruginosa bacteriophage). The positions of the two histidines and two tyrosines known to be critical for catalytic activity of the RC elements are highlighted above the alignment. (Bottom) An alignment of the seven conserved motifs of SF1 superfamily DNA helicases from yeast (P07271), baculovirus (T30397), bacteria (P55418), and T4 phage (P32270) with the corresponding regions of HeliBat1 and other Helitron proteins.
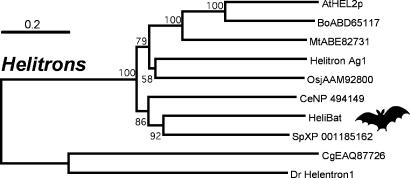
A neighbor-joining phylogenetic analysis based on the alignment of the complete HeliBat1 protein with representative Helitron proteins from other species reveals a strongly supported group consisting of all of the plant and most of the animal proteins (Fig. 3). Falling outside of this group is the Helentron protein from zebrafish (14) and a fungal Helitron-like protein. This topology is consistent with previous phylogenetic analyses (14) and suggests the existence of two distinct lineages of Helitrons in animals: the Helitron group per se and the Helentrons so far only identified in fish and distinguished by a C-terminal endonuclease domain (14). Within the Helitron group, the HeliBat1 protein appears most closely related to the sea urchin Helitron protein (Fig. 3).
Fig. 3.
Neighbor-joining phylogenetic analysis of HeliBat1 and other Helitron and Helitron-like proteins. The accession numbers for the Helitron putative proteins are preceded by the species name abbreviated as in Fig. 1 and 2. The Helitron2 and Helentron1 proteins are from refs. 11 and 14. The midpoint rooting option was used, and bootstrap scores > 50% were retained. Ag, Anopheles gambiae; Bo, Brassica oleracea; Mt, Medicago trunculata.
Nonautonomous HeliBats.
A common characteristic of Helitrons in plant and nematode genomes is the prevalence of nonautonomous elements relative to autonomous Helitron copies (11, 19, 20, 22). These elements are generally much shorter, have no compelling coding capacity, and group into various subfamilies with moderate to high copy numbers. It is thought that the proliferation of nonautonomous elements occurs through the recognition in trans of shared terminal sequence motifs by proteins encoded by autonomous elements residing within the same genome (11, 19, 25). We identified many nonautonomous Helitrons in the bat genome and characterized in more detail a major family called HeliBatN1.
We derived a 1,154-bp consensus HeliBatN1 sequence from an alignment of 13 closely related copies. HeliBatN1 contains no significant coding capacity but harbors terminal sequences typical of Helitrons (Fig. 1). Note however that the HeliBatN1 and HeliBat1 consensus do not share significant sequence similarity besides their very terminal nucleotides (Fig. 1). Even their 3′ palindromic motifs are considerably divergent, suggesting that HeliBatN1 likely has relied on transposition enzymes different from those encoded by HeliBat1. Regardless of the enzymatic source, the HeliBatN1 family has been markedly more successful at propagating than HeliBat1 (see below).
A consensus sequence for a distinct subfamily of HeliBatN1, called HeliBatN2, was constructed from the alignment of 15 copies. The HeliBatN2 consensus is 2,274 bp long and displays 88% and 75% similarity with the first 638 and last 107 bp, respectively, of HeliBatN1. Thus, HeliBatN1 and HeliBatN2 share very similar terminal sequences (Fig. 1) and therefore may have relied on the same machinery for amplification. Yet, the internal sequences of HeliBatN1 and HeliBatN2 appear to be of completely different origin. These data illustrate the considerable structural plasticity of HeliBats, a feature shared by plant Helitrons (19, 22, 26).
A third subfamily of nonautonomous Helitrons, HeliBatN3, was identified, and a consensus of 1,033-bp was reconstructed from the alignment of 10 copies. The termini of HeliBatN3 consensus shares strong similarity with those of HeliBat1, suggesting that HeliBatN3 are derived from HeliBat1 elements and borrowed their enzymatic machinery to propagate. However, the internal sequences are completely unrelated, and thus HeliBatN3 is a distinct subfamily. Based on a blastn search, we estimate that there are >1,000 elements in the M. lucifugus genome with 90–94% similarity over the entire HeliBatN3 consensus.
Interestingly, most of the internal region of HeliBatN3 that distinguishes it from HeliBat1 aligns with the 5′ UTR and first exon of the human gene NUBPL (nucleotide binding protein-like; GenBank accession no. NP_079428) with 70% identity over 675 bp (see SI Fig. 5). This genomic region returns a single high-scoring blast hit in each of the complete genome sequences of human, dog, mouse, and rat. Indeed, NUBPL is a single-copy gene encoding a highly conserved protein in all these mammals (SI Fig. 5B). There is also a single high-scoring hit to the second exon of the human NUBPL gene in the M. lucifugus WGS database, supporting the presence of a single NUBPL homolog in M. lucifugus. Thus, it appears that the 5′ upstream region and first exon of the M. lucifugus NUBPL gene were captured by a HeliBat1-like element and subsequently amplified to >1,000 copies, forming the HeliBatN3 family. In support of this hypothesis, we identify what appears to be the remnant of the original 5′ terminus of a HeliBat1-like element, located at position 826 in the HeliBatN3 consensus, downstream of the NUBPL-derived region (SI Fig. 5A). It appears that the 5′ termination sequence of a different HeliBat copy was recognized instead of the legitimate 5′ terminus, resulting in the transduction of the intervening genomic sequence, including the promoter, first exon, and donor splice site of the first intron of the NUBPL gene (SI Fig. 5C). This event formed a new chimeric HeliBat element that subsequently propagated, giving rise to the HeliBatN3 family. These results indicate that HeliBat elements can transduce and replicate adjacent host sequences, including exons, akin to RC transposons in maize and bacteria (19–22, 27).
Insertion Specificity and Tandem Arrays of HeliBats.
Helitrons from plants and nematodes have been shown to insert between A and T nucleotides of the host chromosome without creating sequence alterations such as target site duplications commonly associated with the insertion of other types of TEs (11, 13, 20). The vast majority of HeliBat1 and HeliBatN1 elements examined were also immediately flanked by 5′-A and 3′-T nucleotides, respectively. To demonstrate that HeliBats indeed insert between A and T nucleotides, we searched for conserved paralogous HeliBat insertion sites that might be devoid of the insertion. We identified two such sites in the WGS database of M. lucifugus (see SI Fig. 6). These sites occur in multiple copies in the bat genome because they are part of other repeat families. In all three cases, comparison of the sites with and without the HeliBat element revealed that the insertion occurred precisely between A and T and did not induce duplication or alteration of the target site, consistent with the RC mechanism of transposition (SI Fig. 6). These data also provide evidence for the past mobility of HeliBats within the genome.
Another expected outcome of the RC replication mechanism is the occasional formation of tandem arrays of elements (17, 27, 28). Such arrangements have been observed for prokaryotic RC transposons (27), but have never been reported for eukaryotic Helitrons. To detect potential tandem arrays of HeliBat elements, we searched the M. lucifugus WGS database by using blastn with artificial queries where the last 50 bp of the HeliBat1 or HelibatN1 consensus sequence was fused to the first 50 bp of the respective consensus. These searches yielded 114 and 45 hits of perfect head-to-tail junctions of two HeliBat1 and HeliBatN1 elements, respectively (for examples, see SI Fig. 7). The short length of the contigs in the database (2.4 kb on average) precluded us from recovering tandem arrays of two complete HeliBats, and no such arrays could be unambiguously identified in the available BAC sequences. However, we were able to detect two contigs (AAPE01389610 and AAPE01505253) that each contains a full-length HeliBatN copy immediately flanked by the end and the beginning of another HeliBatN copy, suggesting that tandem arrays can contain more than two HeliBat copies. To our knowledge, tandem arrays of Helitrons have not been identified previously, and this finding serves as another line of evidence supporting an RC mechanism of replication for Helitrons.
How Many Helitrons Are in the M. lucifugus Genome?
To measure the abundance of the two HeliBat families in M. lucifugus, we used RepeatMasker (A. F. A. Smit, R. Hubley, and P. Green; http://repeatmasker.org) to identify all nonoverlapping segments matching either the HeliBat1 or HeliBatN1 consensus in the current WGS database of M. lucifugus. This database includes a total of 1,674 Mb of genomic sequence covering ≈73% of the M. lucifugus haploid genome size (see Materials and Methods). Out of this data set, 57 Mb (or 3.4%) were masked as either HeliBat1 or HeliBatN1 sequences. A total of 24,556 segments (spanning 4.2 Mb) and 205,663 segments (spanning 32.8 Mb) were annotated as HeliBat1 and HeliBatN1, respectively. For both families, there is a large excess (≈6-fold) of segments matching at or near the 3′ terminus compared with the 5′ terminus of the respective consensus sequences. This discrepancy may be attributed to (i) various degrees of 5′ truncation during integration, (ii) deletion and rearrangements following integration of full-length copies, or (iii) a higher level of sequence variation at the 5′ end of the elements. Thus, we consider that the number of segments containing the 3′ terminus can be used as a proxy of HeliBat copy numbers. There were 10,813 and 79,357 segments matching the last 200 bp (more or less 6 bp) of the HeliBat1 and HeliBatN1 consensus, respectively. Considering each of these segments as one HeliBat copy, these figures suggest conservative copy numbers of ≈15,000 and ≈110,000 for the HeliBat1 and HeliBatN1 families, respectively, in the M. lucifugus haploid genome. Note however that the vast majority of the elements detected were much smaller in size than their respective consensus sequences and often fragmented by DNA of unknown origin. For example, the RepeatMasker analysis detects only 307 full-length and uninterrupted copies of HeliBatN1. We did not use the HeliBatN2 consensus for repeat masking, because HeliBatN2 elements share strong similarity with HeliBatN1. Thus, HeliBatN2 elements are partially masked as HeliBatN1 in our analysis. blastn searches of the WGS suggest the presence of ≈500 bona fide copies of HeliBatN2 in the M. lucifugus genome.
As an independent measurement of HeliBat1 and HeliBatN1 abundance, we used the two consensus sequences to mask a second data set of 46 high-quality BAC sequences generated by the National Institutes of Health National Intramural Sequencing Center for the ENCODE project (29) (see Materials and Methods). Approximately 200 kb (or 2.5%) of the M. lucifugus BAC sequences were masked as HeliBat elements, and most of these (181 kb) were identified as being derived from the HeliBatN1 family. The observation that a lesser amount of sequences were masked as HeliBat elements in this data set could be due to the relatively small size of the database (≈0.3% of the genome) and to the relative enrichment of genes in this data set (see Materials and Methods). Indeed, in A. thaliana and C. elegans, Helitrons are thought to accumulate preferentially in gene-poor heterochromatic regions (11). In any case, the RepeatMasker analysis of BAC and WGS data sets are congruent in revealing that HeliBats occupy a significant fraction of the M. lucifugus genome.
Age of the HeliBat Families.
To estimate the age of the HeliBat1 and HeliBatN1 families, we used the sequence divergence of each element to their consensus as given by the milliDiv field of the RepeatMasker output for the M. lucifugus WGS database. The average sequence divergence of segments >2,000 bp masked as HeliBat1 (n = 46) was 7.9% (±2.9 SD). The average divergence of segments masked as HeliBatN1 and >75% of the consensus length for HeliBatN1 (that is, >859 bp, n = 1,535) was 8.2% (± 4.0). Furthermore, a set of 307 full-length HeliBatN1 copies (segments covering >99% of the consensus length) displays an average of 6.8% (±4.1) divergence to the consensus. These values suggest that the bulk of HeliBat1 and HeliBatN1 elements transposed around the same evolutionary time, ≈30–36 million years ago (MYA), assuming an average mammalian neutral substitution rate of 2.2 × 10−9 per bp per year (30). These age estimates should be regarded cautiously for the following reasons. First, there is currently no reliable estimate of the neutral substitution rate in the bat lineage (M. Springer, personal communication), and wide variations are known to occur among mammalian orders (30, 31). For example, if one used the faster substitution rate of the murid lineage (6), HeliBat1/N1 families would appear twice as young. Considering the short generation time, small body weight, and high metabolic rate of bats, all of which are thought to result in increased mutation rates (31), it is possible that we overestimated the age of the HeliBat1/N1 families. Second, we used the raw sequence divergence given by the RepeatMasker output without any correction, such as to account for nucleotide composition.
As an alternative approach to date the HeliBats, we used the ENCODE comparative data (29) to assess for the presence/absence of HeliBat elements masked in the M. lucifugus BAC data set at orthologous genomic positions in other mammalian species. None of the elements examined were present at orthologous positions in any of the 23 other mammalian species represented, including two other bat species, the Seba's short-tailed bat, Carollia perspicillata, and the great horseshoe bat, Rhinolophus ferrumequinum. Closer inspection of sequence alignments of orthologous regions confirmed that HeliBat elements were inserted between A and T nucleotides in M. lucifugus and were precisely missing in the other species (an example is shown in SI Fig. 6).
Finally, we could identify sequences related to HeliBatN1 elements in three other microbat species, M. myotis (AF203644), Kerivoula papillosa (AM157686), and Pipistrellus abramus (GenBank accession no. AB258749), that diverged from M. lucifugus less than ≈16–25 MYA (32, 33). These sequences display ≈85% identity with the HeliBatN1 consensus over 239, 480, and 80 bp, respectively. Furthermore, a reciprocal blastn search of the WGS database with the HeliBatN1 element and its flanking sequence in the M. myotis GenBank accession no. AF203644 revealed that this element is present at orthologous position in M. lucifugus (contig AAPE01059303). Therefore, this HeliBatN1 copy must have inserted before the divergence of these two species.
Together, the cross-species analyses indicate that the HeliBat1 and HeliBatN1 families were active after the divergence of M. lucifugus, C. perspicillata, and R. ferrumequinum, estimated at 54 MYA, but before the divergence of the vesper bats, ≈16–25 MYA (32, 33). This evolutionary window is consistent with our age estimate based on sequence divergence (≈30–36 million years). However, it should be emphasized that we only examined two HeliBat families, so it cannot be excluded that the bat genome contains more recent families and still actively transposing Helitrons.
Discussion
The mammalian TE landscape has been finely delineated for the genomes of human, mouse, rat, and dog (1, 6–8). Although these genomes are rich in varied types of TEs, no Helitrons have been identified. Furthermore, only retrotransposons are known to be recently active in these lineages, and there has been no evidence that any mammalian DNA transposons have been active within the last 50 million years (1, 6, 9). Our results provide evidence for the relatively recent amplification of Helitrons, an atypical class of DNA transposons, in a mammalian lineage.
Reiterative searches of all current National Center for Biotechnology Information (NCBI) databases with the REP/Helicase domains of HeliBat1 (or Helentrons) revealed no evidence of Helitrons in any other placental or marsupial species. This result is surprising because complete or partial WGS are presently available for 36 species from 15 different mammalian orders (www.ncbi.nlm.nih.gov/genomes/leuks.cgi). In addition, we could not detect any evidence of Helitrons among the substantial amount of genomic sequences (≈1% of each genome) generated by the ENCODE project for 22 placental species besides M. lucifugus, including two other bat species, C. perspicillata and R. ferrumequinum. The presence of a handful of Helitron relics in the platypus genome most likely results from the (moderate) activity of Helitrons in a monotreme ancestor, because this lineage was separated from the other mammals ≈250 MYA. It also might be indicative of the presence of Helitrons in the mammalian common ancestor. However, given the overall slow rate of sequence decay in mammals (30, 34, 35), the absence of detectable Helitron coding sequences in any other placental or marsupial species suggests that, even if Helitrons were present in the common ancestor of mammals, they were most likely already extinct in the common ancestor of placental mammals or at least quiescent during the early placental radiation, from ≈65 to ≈105 MYA (36). Because the genome sequences currently available for closely related bat species are limited, further experiments will be necessary to decipher the intriguing evolutionary history of HeliBats.
Our preliminary analyses of HeliBat copy numbers reveal that a substantial fraction of the M. lucifugus genome is composed of Helitrons; ≈3.4% of the WGS data set is masked with either the HeliBat1 or HeliBatN1 consensus sequences. We estimate that the HeliBatN1 family alone is reiterated in >100,000 copies per haploid genome. It is the largest quantity of Helitron sequences ever reported in any species. Yet it is certainly still an underestimate of the amount of sequences generated by the activity of Helitrons in the vesper bat genomes. As with Helitrons characterized in other species, we found that nonautonomous Helitrons greatly outnumber autonomous elements in M. lucifugus. Nonautonomous elements are less readily detected by standard homology-based searches, and most of them probably await discovery in the M. lucifugus genome. For example, there were a total of 125,596 segments in the WGS data set matching the last 46 bp of HeliBatN1 consensus. As mentioned earlier, only approximately half of these hits extended significantly toward the internal region of the HeliBatN1 consensus. Most of the remaining 3′-terminal sequences probably derive from other subfamilies or families of HeliBat yet to be characterized. Considering this observation, it is likely that much more than 3.4% of the bat genome results from the activity of Helitrons. Characterizing additional HeliBat families will help ascertain the amount of genetic material derived from RC transposition in vesper bats.
TE activity can lead to tremendous change in genome organization and also has an evolutionarily important impact on gene function, in part as a result of transposition, but also through processes such as illegitimate recombination between closely related repeated DNA sequences (3–5, 9). Helitron amplification may further lead to genome rearrangement through the transduction of genome fragments that occurs, possibly as a by-product of their mobilization through RC intermediates (25). The transduction capacity of Helitrons was highlighted recently in maize, where it was estimated that thousands of cellular gene fragments were captured, duplicated, and rearranged by Helitrons (19–21). We describe a HeliBat-mediated transduction event that resulted in the capture of the first exon of a well conserved mammalian gene and its dispersion in ≈1,000 copies throughout the M. lucifugus genome. The evolutionary implications of this amplification are unknown, but we note that the transduced segment also likely contained a donor splice site and proximal promoter elements (see SI Fig. 5), which could potentially be reused to assemble new chimeric genes (akin to ref. 37). Some of the maize Helitrons were found to produce chimeric transcripts that fused fragments of genes transduced from different loci, suggesting that they could mediate the formation of new genes through exon shuffling (21, 22). Transduction of adjacent sequences by bacterial RC insertion sequences has been shown to occur experimentally at frequencies ranging from 1% to 10% (16). Furthermore, bacterial RC transposons are associated with a plethora of virulence and antibiotic resistance genes and are thought to be involved in the mobilization of these factors among strains and species (17, 38). These data highlight the tremendous potential of RC transposition for genome evolution.
Bats constitute >20% of living mammal species (≈1,000 species), which make them the most speciose group of mammals after the rodents (32). Among bats, the lineage of M. lucifugus (vesper bats) has the most species, as well as the broadest geographic distribution. It is intriguing that the early expansive radiation of the vesper bats roughly coincides with the explosive amplification of the HeliBat families characterized in the present study. The impact that HeliBats may have played in altering the genome organization and contributing to the diversification of vesper bats is a fascinating question that warrants further investigation.
Materials and Methods
Sequence Data.
Two sequence data sets specific for M. lucifugus were used in this study. The first data set consists of 640,786 contigs generated by WGS by the Broad Institute (GenBank accession no. AAPE00000000). The average of the contigs is ≈2.4 kb and the entire data set sum is up to ≈1,674 Mb of genomic sequences. Assuming a haploid genome size of ≈2,300 Mb for M. lucifugus (T. R. Gregory, Animal Genome Size Database; www.genomesize.com), the WGS data cover ≈73% of the genome.
The second data set consists of 46 BAC sequences (7.9 Mb) generated for the ENCODE project by the National Institutes of Health National Intramural Sequencing Center (www.nisc.nih.gov). These BACs map to six different human reference genomic regions manually selected for the ENCODE pilot project: ENm001, ENm005, ENm008, ENm009, ENm013, and ENm014. Note that these regions were selected in part based on the presence of well studied genes (e.g., CFTR, globin), and therefore it may represent a more gene-rich data set relative to the WGS database and to the entire genome. GenBank accession nos. and additional information for these sequences are available at www.nisc.nih.gov/projects/encode/index.cgi?all_grid = 1.
Consensus sequences for the HeliBat families described in this study were deposited in Repbase (www.girinst.org/repbase). The accession and coordinates of the individual sequences used to reconstruct the consensus are listed in SI Table 1.
Sequence Analyses.
Data mining.
Homology searches (BLASTN, BLASTX, and TBLASTN) of the NCBI databases were undertaken up to October 24, 2006. Initial searches were performed with a query sequence identified from the purple sea urchin S. purpuratus representing the REP domain of a putative Helitron protein identified from that species (GenBank accession no. XP_001185162; E.J.P., unpublished data).
Phylogenetic analysis.
Sequence alignments were constructed and edited by using T-Coffee (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi) and GENEDOC (www.psc.edu/biomed/genedoc). Phylogenetic analysis was carried out with Mega (Version 3; www.megasoftware.net) by using the neighbor-joining method and the equal input model, which allows for varying substitution rates at the same site and between sites. Bootstrap analysis was performed for 1,000 replicates.
Repeatmasking.
We used a local copy of RepeatMasker (Version 3.1.5; A. F. A. Smit, R. Hubley, and P. Green, www.repeatmasker.org) to mask the M. lucifugus WGS and BAC sequences with the consensus HeliBat1 and HeliBatN1. Sequence comparisons were performed locally with WU-BLAST (http://blast.wustl.edu). The RepeatMasker output was copied into and analyzed with Microsoft Excel (BAC) or a mySQL database (WGS). The output is available from the authors upon request.
Supplementary Material
Acknowledgments
We thank John Pace for outstanding technical assistance with RepeatMasker, Claudio Casola for helpful comments on the manuscript, and Mark Springer and Emma Teeling for information about the neutral substitution rates of bats. This work was supported by start-up funds from the University of Texas, Arlington.
Abbreviations
- MYA
million years ago
- RC
rolling circle
- TE
transposable element
- WGS
whole genome shotgun sequences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data desposition: Consensus sequences for the HeliBat families described in this study were deposited in Repbase, www.girinst.org/repbase (accession numbers and coordinates of the individual sequences used to reconstruct the consensus are listed in SI Table 1).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609601104/DC1.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KF, Wing RA. Proc Natl Acad Sci USA. 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidwell MG, Lisch DR. Evol Int J Org Evol. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 4.Eichler EE, Sankoff D. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- 5.Biemont C, Vieira C. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 6.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 8.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ III, Zody MC, et al. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 9.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Hedges DJ, Batzer MA. BioEssays. 2005;27:785–794. doi: 10.1002/bies.20268. [DOI] [PubMed] [Google Scholar]
- 11.Kapitonov VV, Jurka J. Proc Natl Acad Sci USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapitonov VV, Jurka J. Proc Natl Acad Sci USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC. Plant Cell. 2003;15:381–391. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulter RT, Goodwin TJ, Butler MI. Gene. 2003;313:201–212. doi: 10.1016/s0378-1119(03)00679-6. [DOI] [PubMed] [Google Scholar]
- 15.Novick RP. Trends Biochem Sci. 1998;23:434–438. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 16.Tavakoli N, Comanducci A, Dodd HM, Lett MC, Albiger B, Bennett P. Plasmid. 2000;44:66–84. doi: 10.1006/plas.1999.1460. [DOI] [PubMed] [Google Scholar]
- 17.Garcillan-Barcia M, Bernales I, Mendiola V, De La Cruz F. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: Am Soc Microbiol; 2002. pp. 891–904. [Google Scholar]
- 18.Khan SA. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S, Gallavotti A, Stryker GA, Schmidt RJ, Lal SK. Plant Mol Biol. 2005;57:115–127. doi: 10.1007/s11103-004-6636-z. [DOI] [PubMed] [Google Scholar]
- 20.Lai J, Li Y, Messing J, Dooner HK. Proc Natl Acad Sci USA. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 22.Brunner S, Pea G, Rafalski A. Plant J. 2005;43:799–810. doi: 10.1111/j.1365-313X.2005.02497.x. [DOI] [PubMed] [Google Scholar]
- 23.Tuteja N, Tuteja R. Eur J Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- 24.Boule JB, Zakian VA. Nucleic Acids Res. 2006;34:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feschotte C, Wessler SR. Proc Natl Acad Sci USA. 2001;98:8923–8924. doi: 10.1073/pnas.171326198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tempel S, Giraud M, Lavenier D, Lerman IC, Valin AS, Couee I, Amrani AE, Nicolas J. Bioinformatics. 2006;22:1948–1954. doi: 10.1093/bioinformatics/btl337. [DOI] [PubMed] [Google Scholar]
- 27.Mendiola MV, Bernales I, de la Cruz F. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rochaix JD, Bird A, Barkken A. J Mol Biol. 1974;87:473–487. doi: 10.1016/0022-2836(74)90098-9. [DOI] [PubMed] [Google Scholar]
- 29.Consortium TEP. Science. 2004;306:636–640. [Google Scholar]
- 30.Kumar S, Subramanian S. Proc Natl Acad Sci USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang DG, Green P. Proc Natl Acad Sci USA. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 33.Jones KE, Bininda-Emonds OR, Gittleman JL. Evol Int J Org Evol. 2005;59:2243–2255. [PubMed] [Google Scholar]
- 34.Petrov DA, Lozovskaya ER, Hartl DL. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- 35.Yi S, Ellsworth DL, Li WH. Mol Biol Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
- 36.Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordaux R, Udit S, Batzer MA, Feschotte C. Proc Natl Acad Sci USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toleman MA, Bennett PM, Walsh TR. Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.