Abstract
Ferrochelatase, the terminal enzyme in heme biosynthesis, catalyzes the insertion of ferrous iron into protoporphyrin IX to form protoheme IX. Human ferrochelatase is a homodimeric, inner mitochondrial membrane-associated enzyme that possesses an essential [2Fe-2S] cluster. In this work, we report the crystal structure of human ferrochelatase with the substrate protoporphyrin IX bound as well as a higher resolution structure of the R115L variant without bound substrate. The data presented reveal that the porphyrin substrate is bound deep within an enclosed pocket. When compared with the location of N-methylmesoporphyrin in the Bacillus subtilis ferrochelatase, the porphyrin is rotated by ≈100° and is buried an additional 4.5 Å deeper within the active site. The propionate groups of the substrate do not protrude into solvent and are bound in a manner similar to what has been observed in uroporphyrinogen decarboxylase. Furthermore, in the substrate-bound form, the jaws of the active site mouth are closed so that the porphyrin substrate is completely engulfed in the pocket. These data provide insights that will aid in the determination of the mechanism for ferrochelatase.
Keywords: heme biosynthesis, protoporphyrin IX, x-ray crystallography, metal insertion
Metallated tetrapyrroles are present in most organisms and participate in essential biochemical processes that include photosynthesis, oxygen transport, drug metabolism, transcriptional regulation, NO synthesis, and oxidative phosphorylation. Metallation of tetrapyrroles is catalyzed by a group of enzymes named chelatases. This group includes, but is not limited to, magnesium chelatase, which is essential for chlorophyll production (1), and ferrochelatase, which is essential for heme production (2). Because of the diverse functions of heme, the latter enzyme plays a critical role in human health. Human genetic defects affecting this enzyme have been identified and result in the disease erythropoietic protoporphyria (3). Human ferrochelatase, with its [2Fe-2S] cluster (4, 5), represents the convergence of tetrapyrrole synthesis with iron supply and must play a key role in overall body iron metabolism (6).
Ferrochelatases from a variety of sources have been cloned, expressed, and characterized to various extents (2, 4), but it is the Bacillus subtilis and mammalian ferrochelatases that have been studied most extensively. These two enzymes represent the broadest diversity among ferrochelatases examined to date with <10% sequence identity. The B. subtilis protein is a water-soluble, monomeric protein with no cofactors (7), whereas the human enzyme is an inner mitochondrial membrane-associated homodimer with a [2Fe-2S] cluster in each subunit (8). Nevertheless, there is clear structural similarity between these two enzymes. A comparison of the structures reveals a root-mean-square deviation of only 2.4 Å for the Cα atoms. The majority of the conserved residues are located in the active site pocket.
Since the initial work by DeMatteis' group that identified and characterized the “green pigment” in livers of 3,5-dicarbethoxy-1,4-dihydrocollidine-treated mice as N-methylprotoporphyrin, there has been considerable interest in N-alkylporphyrins as inhibitors of ferrochelatase (9–12). Because of the tight binding competitive inhibition of ferrochelatase by N-methylprotoporphyrin (13), the fact that nonenzymatic metal insertion into porphyrins is facilitated by macrocycle distortion, and the observation that an antibody raised against N-methylmesoporphyrin (N-MeMP) IX catalyzes the insertion of metal ions (14–17), the general hypothesis has been that N-alkyl porphyrins are transition-state analogs (18–20). The crystal structure of B. subtilis ferrochelatase with bound N-MeMP has served as the basis of mechanistic models for ferrochelatases (21). These models, as well as resonance Raman and site-directed mutagenesis studies, assume that the N-MeMP observed in the crystal structure is bound in the active site of ferrochelatase in an orientation that is identical to the spatial orientation of the natural substrate/product (22–24). Additionally, it has been assumed that the 36° macrocycle distortion observed in the N-MeMP-bound crystal structure represents a catalytic intermediate that occurs during normal turnover. Data presented herein provide evidence that these assumptions may not be valid.
To provide insight into the interaction of the physiological substrate with ferrochelatase, we have determined the structure of human ferrochelatase with bound protoporphyrin IX. The human ferrochelatase enzyme used in this investigation was an E343K variant that has a higher affinity for protoporphyrin IX in comparison to the wild-type enzyme. We have also collected higher resolution data for the previously reported R115L variant of human ferrochelatase (25) without bound substrate. The position of the substrate in the E343K variant is distinctly different from the previously reported orientation of N-MeMP in the B. subtilis ferrochelatase (21). In addition to the spatial orientation of substrate within the active site of human ferrochelatase, this work also shows that the substrate-bound form of the enzyme possesses a “closed” active site conformation that is notably different from the structure of the inhibitor-bound B. subtilis ferrochelatase or the structure of the human enzyme without substrate. In both of the latter cases, the active sites are distinctly “open.” These observations provide insight into the ferrochelatase mechanism of catalysis and inhibition.
Results
General Description of the Overall Structure and Substrate Binding Sites.
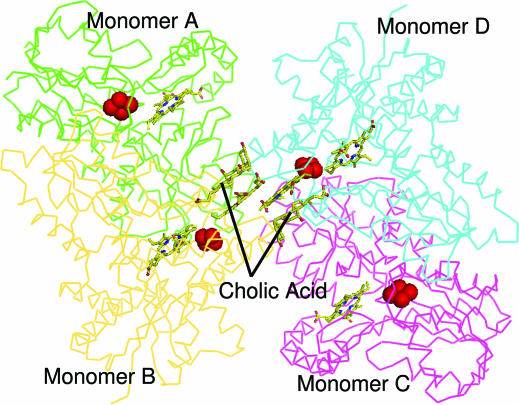
The crystals of the E343K variant of human ferrochelatase belong to the triclinic space group P1 (see Table 1), whereas the space group for the initial structure of human ferrochelatase (containing the amino acid substitution R115L) reported by Wu et al. (25) is orthorhombic (P212121). In the latter case, the asymmetric unit contained a single biological dimer. The different crystal symmetry between the substrate-bound form and the free enzyme suggests that substantial conformational differences exist between the two forms of the enzyme. Upon phasing and refinement, it was revealed that the asymmetric unit of the E343K variant of human ferrochelatase contains two copies of the biological dimer (Fig. 1), for a total of four peptide monomers and six protoporphyrin IX molecules. This finding is shown in Fig. 1, which highlights the relative positions of the four monomers, six porphyrin molecules, and two detergent molecules, as well as the [2Fe-2S] clusters. The fact that substrate was observed in the electron density is of interest because substrate was not added during the expression, isolation, or crystallization of the E343K variant. One protoporphyrin molecule is found in each of the four active sites, and two are located at a noncrystallographic 2-fold axis. Interestingly, the two protoporphyrin molecules that are located at the noncrystallographic 2-fold axis are positioned at an interface just outside the entrance to the active sites of monomers B and D. This is the same region that detergent molecules occupy in the original structure of the R115L human ferrochelatase variant (25). Consistent with previous work, we also see electron density for two detergent molecules bound at this interface (Fig. 1). All enzyme subunits in both the E343K variant and the higher resolution R115L variant possess [2Fe-2S] clusters, as was reported previously for human ferrochelatase.
Table 1.
Data collection and refinement statistics
| R115L | E343K | |
|---|---|---|
| Space group | P212121 | P1 |
| Wavelength | 0.98 | 1.54 |
| Resolution range, Å | 40.0–1.7 | 50.0–2.5 |
| Unique observations | 100,275 | 53,845 |
| Completeness | 99.6 (98.2)* | 96.7 (93.5) |
| Rsym, %† | 0.08 (14.7) | 0.08 (28.2) |
| I/σ | 29.1 (5.8) | 13.5 (3.4) |
| Unit cell (a, b, c) | 88.5, 92.9, 110.4 | 61.9, 88.3, 93.2 |
| Protein atoms | 5,782 | 11,564 |
| Solvent atoms | 601 | 338 |
| Resolution limits | 40.0–1.7 | 50.0–2.5 |
| Rcryst, % | 22.1 | 21.6 |
| Rfree, % | 24.2 | 27.8 |
| rmsd bonds, Å | 0.006 | 0.008 |
| rmsd angles, ° | 1.21 | 1.61 |
| average B factor, Å2 | 21.8 | 31.1 |
*Numbers in parentheses denote values for the outermost resolution shell.
†Rsym = Σhkl [ΣI(Ihkl,I − 〈Ihkl〉)]/Σhkl,I〈Ihkl〉, where Ihkl is the intensity of an individual measurement of the reflection with indices hkl and 〈Ihkl〉 is the mean intensity of that reflection.
Fig. 1.
Overall backbone trace of the asymmetric unit for the E343K human ferrochelatase. The Cα trace for monomers A, B, C, and D are colored green, yellow, magenta, and cyan, respectively. The two detergent molecules and six protoporphyrin IX molecules are also shown with the carbon, oxygen, and nitrogen atoms colored yellow, red, and blue, respectively. Only the detergent molecules are labeled. The [2Fe-2S] clusters are represented as spheres and colored bright red.
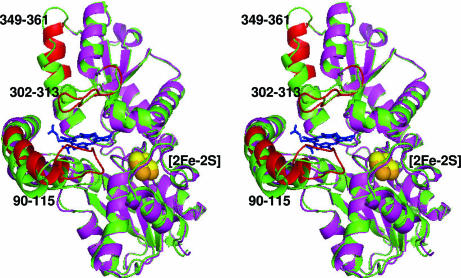
Comparison of the R115L variant that lacks bound substrate with the substrate-bound (E343K variant) form reveals that the enzyme with porphyrin bound possesses a significantly more “closed” active site conformation. The overall difference between these two conformations is illustrated in Fig. 2. There is an average deviation of 3.5 Å between the R115L and E343K variants for the backbone atoms of residues 90–115. Substantial differences in the torsion angles of the peptide backbone for the substrate-bound structure in comparison with the substrate-free model are also observed for residues 302–313 and 349–361 (Fig. 2). In addition, there is a reorientation of numerous amino acid side chains in the active site pocket. The overall result is an active site that completely engulfs the substrate.
Fig. 2.
Structural alignment of the substrate-bound (E343K) and substrate-free (R115L) forms of human ferrochelatase. The substrate-free form of human ferrochelatase is shown in green, and the substrate-bound form of human ferrochelatase is shown in magenta. Regions of significant movement in the substrate-bound form have been highlighted in red for clarity and include residues 90–115, 302–313, and 349–361. The [2Fe-2S] clusters for the substrate-free and substrate-bound forms are shown in yellow and orange, respectively.
Protoporphyrin IX Binding and Active Site Structure.
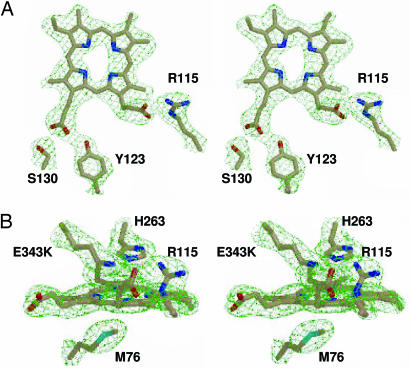
The binding of protoporphyrin IX is identical in all four of the ferrochelatase monomers in the asymmetric unit. A difference (Fo − Fc) composite omit map and the model for the active site are shown in Fig. 3. The omit map was generated by using the simulated annealing protocol with 7% of the model omitted during map generation. The planar and pseudosymmetrical nature of the protoporphyrin IX molecule makes it possible to model the substrate in either orientation with minimal impact on the R factor (<0.5%). However, if the porphyrin is rotated by 180°, then significant peaks appear in the difference map at the vinyl groups, suggesting that one orientation is preferred over the other.
Fig. 3.
Wall-eyed stereoview of the substrate and key residues in the active site of the human E343K ferrochelatase. (A) Top view of the substrate and residues R115, Y123, and S130. (B) Side view of the substrate and residues M76, R115, H263, and the point mutation E343K. Carbon, oxygen, nitrogen, and sulfur atoms are colored light brown, red, blue, and cyan, respectively. The light green cage represents a Fo − Fc composite omit map contoured at 3σ. The omit map was generated by using the simulated annealing protocol with 7% of the model being omitted per cycle.
It is important to note that the preparations of the E343K ferrochelatase protein generally contain slightly fewer than 0.5 protoporphyrin molecules per ferrochelatase dimer (26). Thus, the E343K amino acid substitution does not cause ferrochelatase to irreversibly bind porphyrin but only increases the affinity of the enzyme for porphyrin. In contrast, the crystallographic data confirm that the substrate occupancy is 100% and the conformation is “closed” for all of the ferrochelatase monomers in the asymmetric unit. Because the substrate propionates are buried inside the active site and do not contribute to surface charge or crystallographic contacts, it is reasonable to conclude that all ferrochelatase molecules in the closed conformation, independent of the presence or absence of enclosed substrate, would crystallize under identical conditions. Because there is porphyrin-free enzyme in the initial crystallization setup, the lack of porphyrin-free ferrochelatase in the crystal lattice suggests that the substrate-free E343K variant is not in a closed conformation and that substrate binding alone results in the closed conformation. These conclusions are consistent with the longstanding use of crystallization techniques to isolate enantiomerically pure compounds from racemic mixtures.
In the structural model for the E343K variant of human ferrochelatase, the protoporphyrin IX appears to be spatially positioned in the active site by a few key interactions. One of the protoporphyrin propionates forms a salt bridge with R115. This interaction is consistent with the published chemical modification data (27). The other propionate participates in a hydrogen-bonding network with the side chains of residues S130 and Y123 (Fig. 3A). These types of salt bridge and hydrogen-bonding interactions have been observed before with the propionates of bound substrate in the crystallographic models reported for S-adenosyl-l-methionine-dependent bismethylase dehydrogenase/ferrochelatase (CysG) (28) as well as uroporphyrinogen decarboxylase (29). In addition to the salt bridge and hydrogen-bonding interactions, there are other amino acids on either side of the tetrapyrrole that are within 3.5 Å of the macrocycle center. These residues are shown in Fig. 3B and include residues M76 and H263. Interestingly, W310 (W256 in mouse numbering), a residue that has been proposed to be involved in saddling of the porphyrin during catalysis (24), is farther from the porphyrin than predicted from the B. subtilis:N-MeMP structure, with a closest approach of 3.7 Å.
R115L Ferrochelatase.
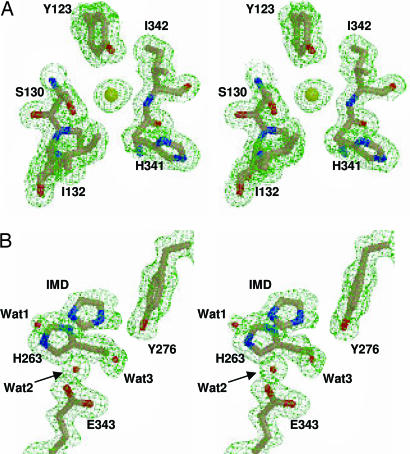
A higher resolution structure of the R115L human ferrochelatase has also been solved (Table 1). This variant form of human ferrochelatase has essentially identical catalytic properties as the wild-type recombinant human ferrochelatase. Two previously unreported features are revealed by our data (Fig. 4). First, a molecule with substantially larger electron density than water is bound in close proximity to the side chains of Y123 and S130 (an 8σ peak remained when modeled as a water molecule). When a chloride ion is placed in this position, there was no additional density observed in the difference map. The hydrogen bonding in this area is also consistent with the presence of a chloride ion (Fig. 4A). Specifically, the imide proton from the peptide bond between residues H341 and I342 points toward the chloride atom. The proton on the side chain of S130 must also point toward the chloride atom because the γO of S130 serves as a hydrogen bond acceptor for another backbone proton (the peptide bond between residues 131 and 132). These observations are consistent with the binding of chloride in other crystallographic models (30–32). Given the findings with the E343K variant, it is proposed that one of the substrate propionates displaces the chloride ion upon substrate binding.
Fig. 4.
Wall-eyed stereoview of an anion-binding site (A) and a bound imidazole molecule in the active site of the human R115L ferrochelatase (B). Carbon, nitrogen, oxygen, and chloride atoms are colored tan, blue, red, and yellow, respectively. The green cage represents a Fo − Fc composite omit map contoured at 3σ. The omit map was generated by using the simulated annealing protocol with 7% of the model being omitted per cycle.
The second feature revealed by these data is the presence of an imidazole in the active site of the R115L variant (Fig. 4B). Of specific interest is the observation that the plane of the imidazole ring resides parallel to the imidazole ring from the side chain of residue H263. This position is strikingly similar to the position of the pyrrole ring of bound protoporphyrin substrate in the E343K variant described above. The arrangement of water molecules in the active site further supports our conclusion that this density is due to a bound imidazole molecule (Fig. 4B). Given that our purification procedure employs a metal affinity matrix for which imidazole is used to elute the protein, it is likely that the imidazole observed in the structure arises from the elution buffer.
Discussion
Although a variant form of the human enzyme was used in this study, these data provide significant insights into the binding mode of the physiological substrate. The data reveal that when protoporphyrin is bound in the active site of ferrochelatase, the enzyme surrounds the substrate in a snug pocket. The pyrrole nitrogen of the protoporphyrin IX “A” ring is positioned directly under and 3.2 Å distant from the ring nitrogen of the conserved and centrally located H263 side chain.
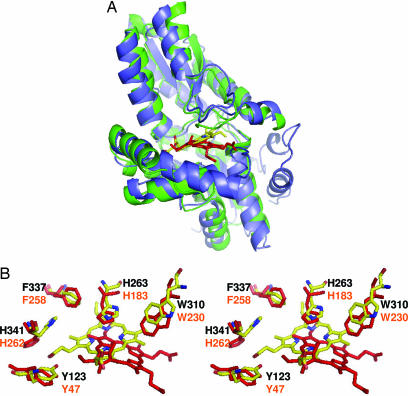
The spatial position of the protoporphyrin substrate in the active site of human ferrochelatase is significantly different from what has been reported for the position of the ferrochelatase inhibitor, N-MeMP, in the structure of the B. subtilis enzyme. Although these two enzymes have little sequence identity, they do share similar monomeric structures, positioning of key active site residues, and kinetic characteristics. In fact, a number of published spectroscopic and catalytic studies on mouse ferrochelatase base their conclusions on the B. subtilis structures (22–24). Thus, although possible, it seems unlikely that these two proteins would bind substrate and inhibitors in significantly different positions. The current data showing that the macrocycle is 4.5 Å “deeper” in the pocket and is rotated 100° in comparison to what was observed in the N-MeMP-bound ferrochelatase structure (Fig. 5) suggest that the previous assignment of specific catalytic roles to various active site residues needs to be reevaluated.
Fig. 5.
Comparison of the protoporphyrin IX and N-MeMP binding modes for human and B. subtilis ferrochelatase, respectively. (A) Structural alignment of the E343K human ferrochelatase model containing protoporphyrin IX (blue cartoon) with the model reported for B. subtilis ferrochelatase containing N-MeMP (green cartoon; PDB ID code 1C1H). (B) Wall-eyed stereoview showing the relative positions of protoporphyrin IX, N-MeMP, and the side chains of strictly conserved amino acids within the active site of the two structures.
The structure of human ferrochelatase with bound protoporphyrin has significant differences in the position of several loop regions (residues 90–115, 302–313, and 349–361) when compared with the structure of human ferrochelatase without bound substrate. The positions of residues 90–115 are displaced an average of 3.5 Å, and the spatial positions of residues 302–313 and 349–361 are also substantially different from what is seen in the structure without bound substrate. In addition to the difference in the positions of atoms, several side chains are also reoriented within the active site pocket. In particular, the guanidino side chain of R164 is flipped 180° out of the pocket, the benzyl ring of F337 is flipped 90° and rotated into the pocket, and the side chains of H341 and M76 are rotated ≈90°. The overall effect is that the “mouth” of the active site pocket is closed in the presence of porphyrin and forms a snug fit around the porphyrin macrocycle. The two porphyrin propionates interact with active site residues via hydrogen and ionic bonds to ensure a highly specific spatial orientation of the porphyrin in the active site. This observation is consistent with the high degree of porphyrin substrate specificity (20, 33) and spectroscopic studies (34) on the enzyme. Similar observations have been made for another heme biosynthetic enzyme, uroporphyrinogen decarboxylase (28). In this case, the propionate groups are also held in place through ionic interactions with arginine residues and hydrogen- bonding interactions with tyrosine and serine residues. In comparison, the published B. subtilis ferrochelatase structures with and without bound N-MeMP do not show any difference in active site amino acid side chain orientations or overall active site pocket shape (21).
One somewhat surprising finding is that the enzyme-bound porphyrin macrocycle is only modestly distorted (≈11.5° bend). This distortion is significantly less than the 36° bend found with N-MeMP bound to the B. subtilis enzyme (21). The bound porphyrin in the E343K variant has a modest saddle conformation that is consistent with theoretical calculations made by Sigfridsson and Ryde (35) for a B. subtilis ferrochelatase:porphyrin complex. Their theoretical model proposes a tilt of all four rings, with one pyrrole ring being 13–15° out of plane and the other three being 1–10° out of plane. However, because their model was based on the position of N-MeMP in the B. subtilis enzyme structure rather than what is reported here for the physiological substrate, protoporphyrin IX, some differences may be expected between the theoretical model and the crystallographic data for human ferrochelatase.
Previous resonance Raman studies have examined conformational changes that occur to porphyrins and metalloporphyrins on binding to ferrochelatase (22–24). Although these data are consistent with some form of macrocycle distortion, the conclusions drawn by the authors were necessarily influenced by use of the B. subtilis:N-MeMP structure as the reference model. With the current structural data, new assignments for the spectral shifts can be made that may provide further insight into the ferrochelatase mechanism. Of additional interest is the crystallographic observation that human ferrochelatase not only binds porphyrin within the active site but also has associated “free” porphyrin nearby, but outside, the active site. This observation may account for the published resonance Raman data that reported spectra characteristic of both planar and ruffled macrocycle conformations (23). These data were obtained for the mouse equivalent of the human E343Q variant enzyme. This variant form of mouse ferrochelatase, and the human E343K and E343H variants (26), have significant amounts of bound protoporphyrin when purified.
Before the current data, catalytic models for ferrochelatase were largely influenced by the proposal that N-alkylporphyrins were transition-state analogs. Thus, when the structure of B. subtilis ferrochelatase with bound N-MeMP became available (21), the assumption was made that the N-MeMP was bound in the active site in an orientation similar to what would occur with the natural substrate protoporphyrin IX. This assumption has resulted in a number of studies that proposed specific roles for active site residues and has even served as the basis for a recently proposed model for metal substrate specificity (36). On the basis of the data presented above, it would seem appropriate to reevaluate these current models.
Materials and Methods
Enzyme Expression, Isolation, and Crystallization.
Mutagenesis, expression, and purification of human ferrochelatase were performed as described (8, 26, 37). Concentration buffer for variants consisted of 0.045 M Tris-Mops (pH 8.1), 0.09 M potassium chloride, 0.9% sodium cholate, 0.225 M imidazole, and 10% glycerol. Crystallization was carried out by using the hanging drop method in the EasyXtal crystallization tool (Qiagen, Valencia, CA). Mother liquor for the R115L crystals consisted of 0.05 M calcium chloride, 0.1 M Bistris (pH 6.5), and 30% vol/vol poly(ethylene glycol) methyl ether 550. Mother liquor for the E343K ferrochelatase crystals consisted of 0.1 M Bistris (pH 5.5), 0.2 M magnesium chloride, and 25% wt/vol PEG 3350. Crystals typically took a minimum of 1 week at 18°C to grow. For both variants, the glycerol concentration was gradually increased to 20% before flash freezing the crystals in liquid nitrogen.
Data Collection, Structure Determination, and Refinement.
Data for the R115L variant were collected on Beamline 8.2.2. at the Advanced Light Source (ALS) in Berkeley, CA, by using a single crystal rotated a full 360°. Data for the E343K variant were collected at the University of Georgia on a Rigaku (Tokyo, Japan) RU-200 rotating anode equipped with Osmic focusing mirrors and an R-axisIIc image plate detector. A single crystal was used, and a full 360° of data were collected. In all cases, molecular replacement, model building, and refinement were performed by using the programs O (38) and CNS (39). All graphic representations were rendered by using PyMOL (40), Molscript (41), Bobscript (42), and Raster3D (43).
Acknowledgments
This work was supported by American Heart Association Grant 0465228B (to W.N.L.) and National Institutes of Health Grant DK32303 (to H.A.D.).
Abbreviation
- N-MeMP
N-methylmesoporphyrin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates for the R115L and E343K human ferrochelatase structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2HRC and 2HRE, respectively).
References
- 1.Willows RD, Hansson M. In: The Porphyrin Handbook. Kadish KM, Smith KM, Guilard R, editors. New York: Academic; 2003. pp. 1–48. [Google Scholar]
- 2.Dailey HA, Dailey TA. In: The Porphyrin Handbook. Kadish KM, Smith KM, Guilard R, editors. New York: Academic; 2003. pp. 93–121. [Google Scholar]
- 3.Todd DJ. Br J Dermatol. 1994;131:751–766. doi: 10.1111/j.1365-2133.1994.tb08577.x. [DOI] [PubMed] [Google Scholar]
- 4.Dailey HA, Dailey TA, Wu CK, Medlock AE, Wang KF, Rose JP, Wang BC. Cell Mol Life Sci. 2000;57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd M, Dailey TA, Dailey HA. Biochem J. 2006;397:47–52. doi: 10.1042/BJ20051967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, et al. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 7.Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hederstedt L. Structure (London) 1997;5:1501–1510. doi: 10.1016/s0969-2126(97)00299-2. [DOI] [PubMed] [Google Scholar]
- 8.Burden AE, Wu C, Dailey TA, Busch JL, Dhawan IK, Rose JP, Wang B, Dailey HA. Biochim Biophys Acta. 1999;1435:191–197. doi: 10.1016/s0167-4838(99)00196-x. [DOI] [PubMed] [Google Scholar]
- 9.De Matteis F, Gibbs AH, Smith AG. Biochem J. 1980;189:645–648. doi: 10.1042/bj1890645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Matteis F, Gibbs AH, Tephly TR. Biochem J. 1980;188:145–152. doi: 10.1042/bj1880145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tephly TR, Wagner G, Sedman R, Piper W. Fed Proc. 1978;37:35–39. [PubMed] [Google Scholar]
- 12.Tephly TR, Gibbs AH, Ingall G, De Matteis F. Int J Biochem. 1980;12:993–998. doi: 10.1016/0020-711x(80)90200-1. [DOI] [PubMed] [Google Scholar]
- 13.Dailey HA, Fleming JE. J Biol Chem. 1983;258:11453–11459. [PubMed] [Google Scholar]
- 14.Cochran AG, Schultz PG. Science. 1990;249:781–783. doi: 10.1126/science.2389144. [DOI] [PubMed] [Google Scholar]
- 15.Venkateshrao S, Yin J, Jarzecki AA, Schultz PG, Spiro TG. J Am Chem Soc. 2004;126:16361–16367. doi: 10.1021/ja0465395. [DOI] [PubMed] [Google Scholar]
- 16.Blackwood ME, Rush TS, Romesberg F, Schultz PG, Spiro TG. Biochemistry. 1998;37:779–782. doi: 10.1021/bi972616f. [DOI] [PubMed] [Google Scholar]
- 17.Lavallee DK. The Chemistry and Biochemistry of N-Substituted Porphyrins. New York: Wiley; 1987. [Google Scholar]
- 18.Lavallee DK. In: Mechanistic Principles of Enzyme Activity. Liebman JF, Greenberg A, editors. New York: VCH; 1988. pp. 279–314. [Google Scholar]
- 19.Dailey HA. In: Biosynthesis of Heme and Chlorophylls. Dailey HA, editor. New York: McGraw–Hill; 1990. pp. 123–162. [Google Scholar]
- 20.Dailey HA, Jones CS, Karr SW. Biochim Biophys Acta. 1989;999:7–11. doi: 10.1016/0167-4838(89)90021-6. [DOI] [PubMed] [Google Scholar]
- 21.Lecerof D, Fodje M, Hansson A, Hansson M, Al-Karadaghi S. J Mol Biol. 2000;297:221–232. doi: 10.1006/jmbi.2000.3569. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y, Sousa A, Franco R, Mangravita A, Ferreira GC, Moura I, Shelnutt JA. Biochemistry. 2002;41:8253–8262. doi: 10.1021/bi025569m. [DOI] [PubMed] [Google Scholar]
- 23.Franco R, Ma JG, Lu Y, Ferreira GC, Shelnutt JA. Biochemistry. 2000;39:2517–2529. doi: 10.1021/bi991346t. [DOI] [PubMed] [Google Scholar]
- 24.Shi Z, Franco R, Haddad R, Shelnutt JA, Ferreira GC. Biochemistry. 2006;45:2904–2912. doi: 10.1021/bi051907i. [DOI] [PubMed] [Google Scholar]
- 25.Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. Nat Struct Biol. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 26.Sellers VM, Wu CK, Dailey TA, Dailey HA. Biochemistry. 2001;40:9821–9827. doi: 10.1021/bi010012c. [DOI] [PubMed] [Google Scholar]
- 27.Dailey HA, Fleming JE. J Biol Chem. 1986;261:7902–7905. [PubMed] [Google Scholar]
- 28.Stroupe ME, Leech HK, Daniels DS, Warren MJ, Getzoff ED. Nat Struct Biol. 2003;10:1064–1073. doi: 10.1038/nsb1007. [DOI] [PubMed] [Google Scholar]
- 29.Phillips JD, Whitby FG, Kushner JP, Hill CP. EMBO J. 2003;22:6225–6233. doi: 10.1093/emboj/cdg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park C, Schultz LW, Raines RT. Biochemistry. 2001;40:4949–4956. doi: 10.1021/bi0100182. [DOI] [PubMed] [Google Scholar]
- 31.Li SJ. Biopolmers. 2006;81:74–80. doi: 10.1002/bip.20367. [DOI] [PubMed] [Google Scholar]
- 32.Natesh R, Schwager SL, Evans HR, Sturrock ED, Acharya KR. Biochemistry. 2004;43:8718–8724. doi: 10.1021/bi049480n. [DOI] [PubMed] [Google Scholar]
- 33.Honeybourne CL, Jackson JT, Jones OT. FEBS Lett. 1979;98:207–210. doi: 10.1016/0014-5793(79)80185-4. [DOI] [PubMed] [Google Scholar]
- 34.Dailey HA. Biochemistry. 1985;24:1287–1291. doi: 10.1021/bi00327a003. [DOI] [PubMed] [Google Scholar]
- 35.Sigfridsson E, Ryde U. J Biol Inorg Chem. 2003;8:273–282. doi: 10.1007/s00775-002-0413-8. [DOI] [PubMed] [Google Scholar]
- 36.Al-Karadaghi S, Franco R, Hansson M, Shelnutt JA, Isaya G, Ferreira GC. Trends Biochem Sci. 2006;31:135–142. doi: 10.1016/j.tibs.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer MR, Dailey TA, Baucom CM, Supernak JL, Grady MC, Hawk HE, Dailey HA. J Struct Funct Genomics. 2004;5:159–165. doi: 10.1023/B:JSFG.0000029202.77832.34. [DOI] [PubMed] [Google Scholar]
- 38.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. PyMOL v0.99. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 41.Kraulis PJ. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 42.Gouet P, Robert X, Courcelle E. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]