Abstract
Hemozoin (HZ) is an insoluble crystal formed in the food vacuole of malaria parasites. HZ has been reported to induce inflammation by directly engaging Toll-like receptor (TLR) 9, an endosomal receptor. “Synthetic” HZ (β-hematin), typically generated from partially purified extracts of bovine hemin, is structurally identical to natural HZ. When HPLC-purified hemin was used to synthesize the crystal, β-hematin had no inflammatory activity. In contrast, natural HZ from Plasmodium falciparum cultures was a potent TLR9 inducer. Natural HZ bound recombinant TLR9 ectodomain, but not TLR2. Both TLR9 stimulation and TLR9 binding of HZ were abolished by nuclease treatment. PCR analysis demonstrated that natural HZ is coated with malarial but not human DNA. Purified malarial DNA activated TLR9 but only when DNA was targeted directly to the endosome with a transfection reagent. Stimulatory quantities of natural HZ contain <1 μg of malarial DNA; its potency in activating immune responses was even greater than transfecting malarial DNA. Thus, although the malarial genome is extremely AT-rich, its DNA is highly proinflammatory, with the potential to induce cytokinemia and fever during disease. However, its activity depends on being bound to HZ, which we propose amplifies the biological responses to malaria DNA by targeting it to a TLR9+ intracellular compartment.
Keywords: fever, immunomodulator, parasitic diseases
Malaria infection affects hundreds of millions of people annually, and is an important cause of morbidity and mortality in tropical countries (1). No effective prophylactic vaccine is available, and chemotherapeutic treatment of large populations of infected individuals is, under current socioeconomic conditions, not possible. Little is known about the innate immune response to malaria infection (2). The life cycle of the Plasmodium parasite occurs primarily within erythrocytes. Typically, the rupture of parasitized erythrocytes is accompanied by the onset of symptoms like fever and rigors, which are due to the systemic release of proinflammatory cytokines (3, 4). However, it remains a contentious issue to define the component of the malaria-infected erythrocyte that is responsible for these events. Both host-derived and pathogen-derived molecules have been proposed as the cytokine-inducing “malaria toxin.”
Infected erythrocytes contain both the parasite and an abundant amount of malaria-derived debris. During the intraerythrocytic stage, parasites digest hemoglobin in the food vacuole, resulting in the production of potentially toxic heme metabolites (5). To protect itself from oxidative damage, the parasite has developed a detoxification process that converts heme into an insoluble crystal called hemozoin (HZ) (6). Synthetic HZ, hereafter referred to as β-hematin, is made from blood extracts enriched in hemin and forms rapidly at 60°C in acetate buffer. β-Hematin is chemically and structurally identical to HZ (7). Both natural HZ and β-hematin have been reported to induce inflammatory responses both in vitro and in vivo (8–11).
During infection, the concentration of HZ after erythrocyte rupture may be as high as 100μg/ml (12), but it is rapidly cleared from the circulation by the liver and spleen because of its particulate nature. As a result of its high concentration in immune tissue, HZ has been suggested to contribute to systemic inflammatory immune responses during malaria infection.
Toll-like receptors (TLRs) are central components of the innate immune system (13, 14). TLRs have been demonstrated to be involved in the recognition of constituents of protozoan parasites (15). The best studied parasitic molecules that engage TLRs include glycosylphosphatidylinositol (GPI) anchors, which activate host cells primarily through TLR2 (16). Indeed, GPI from Plasmodium falciparum have been reported to interact with immune cells through the activation of TLR2 and TLR4 (17). Blood-stage parasites have been shown to activate plasmacytoid dendritic cells (18). Furthermore, MyD88-null mice have a decreased production of IL-12 and less severe pathology than WT control mice (19). In human disease, polymorphisms in TLRs 2, 4, and 9 affect outcome (20). Finally, a Mal S180L polymorphism dramatically decreases mortality and morbidity (Luke O'Neill, personal communication). Thus, it is likely that malaria pathogenesis involves the engagement of TLRs.
In this study, we investigated whether activation of cells from the innate immune system by β-hematin (i.e., synthetic HZ) and natural HZ involves recognition by TLRs. We report that β-hematin is immunologically inert, whereas natural HZ activates cells of the innate immune system, similar to a report by Coban et al. (21). In sharp contrast to these authors, we found that the natural HZ crystal itself is not a ligand for TLRs. Rather, HZ functions as a carrier for malarial DNA, which, as a consequence of being bound to HZ, is targeted to TLR9. As a result of its association with HZ, malarial DNA changes in character from a benign, nonstimulatory molecule to a toxic, highly inflammatory one.
Results
Highly Pure β-Hematin Has No Intrinsic Stimulatory Activity.
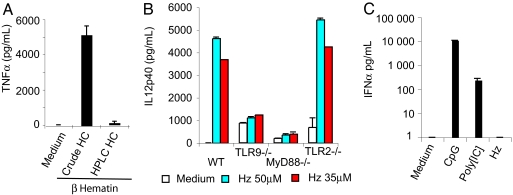
We synthesized β-hematin using two different sources of hemin. A crude bovine hemin preparation was guaranteed by the manufacturer to be 80% pure, although LC-mass spectroscopy analysis demonstrated that hemin accounted for only 65% of the mass. A second commercial preparation was HPLC-purified; LC-mass spectroscopic analyses failed to detect impurities (data not shown). We found that β-hematin from the first preparation of hemin was active at concentrations >100 μg/ml in HEK293/CD14/TLR2 cells. In contrast, β-hematin derived from HPLC-purified hemin had no activity at any concentration (data not shown). Similarly, FLT3-L-derived mouse dendritic cells (FL-DCs) responded to the β-hematin derived from the crude hemin. No significant responses were observed from cells stimulated with β-hematin made from HPLC-purified hemin (Fig. 1). We conclude that the “immune activity” of β-hematin was caused by contaminants from the crude bovine hemin.
Fig. 1.
Highly pure β-hematin has no stimulatory activity. (A) FL-DCs (2 × 105 cells per well) were exposed for 18 h to medium alone or 150 μM β-hematin (“synthetic” HZ) prepared from two different sources of hemin chloride. IL12p40 (data not shown) and TNFα were measured in the culture supernatant by ELISA. (B) Bone marrow-derived FL-DCs from WT and knockout mice were tested for their response by overnight stimulation with 50 μM natural HZ. The production of IL12p40 and TNFα (data not shown) was assessed by ELISA. Results are the mean level of triplicate determinations of released cytokines (±SD). (C) WT FL-DCs were stimulated for 24 h with medium alone or medium plus the indicated ligand. IFNα release into the medium was then measured by ELISA.
Natural HZ Activates Cells to Produce Proinflammatory Cytokines and Chemokines Through TLR9 and MyD88 Because of the Presence of DNA.
We purified HZ from P. falciparum cultures to determine whether natural HZ has inflammatory properties. This technique involves passing malaria-infected blood through a strong magnetic field. Once the blood has passed through, the column is washed extensively, and the retained HZ is eluted after removing the magnet. HZ “purified” in this way is not a pure naked crystal: when boiled in loading buffer and analyzed by SDS/PAGE, a ladder of Coomassie-stained proteins was observed (data not shown).
We used natural HZ to stimulate FL-DC. We observed strong stimulation of IL-12p40 (Fig. 1B) and Rantes (data not shown). Dendritic cells from TLR9 and MyD88 knockout mice failed to respond to HZ, whereas TLR2-null cells responded comparably to WT cells (Fig. 1B). These results confirm that natural HZ engages the TLR9/MyD88 pathway (21). We also confirmed that HZ, unlike CpG DNA (A-class oligonucleotides) and poly I:C, failed to induce the production of IFNα (Fig. 1C).
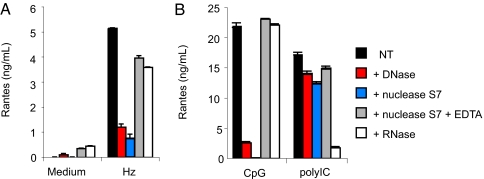
Unlike previous reports, we found that the activity of HZ was abolished by nuclease digestion (Fig. 2A), although the HZ crystal remained intact. When we inhibited the enzymatic activity of S7 nuclease with EDTA, we prevented the inactivation of HZ as a TLR ligand. RNase had no effect on HZ activity whatsoever. As additional controls, we tested the ability of DNase and RNase to inhibit the activity of A-class CpG oligonucleotides and poly I:C (Fig. 2B). Finally, we found that when HZ was mixed with glycerol and gently sonicated, associated genomic DNA could be observed on an ethidium-stained gel (data not shown). We conclude that the TLR9-inducing activity of HZ is due to contaminating DNA.
Fig. 2.
Natural HZ stimulates FL-DCs in a DNase-sensitive fashion. WT FL-DCs were treated with 50 μM natural HZ for 24 h with HZ before (black bars) and after (red bars) treatment with DNase I, S. aureus micrococcal nuclease S7 (blue bars), EDTA-inactivated S7 nuclease (gray bars), or RNase (white bars). The supernatants were collected and analyzed for Rantes and IL12p40 by ELISA. CpG oligonucleotide 2336 and poly[IC] were treated in the same way as the HZ. Medium controls represent tissue culture medium to which the final concentration of each nuclease was added. Each condition was assayed in triplicate; the results represent mean level of Rantes (±SD). Nearly identical results were observed when the release of IL12p40 was measured.
The Ectodomain of TLR9, but Not TLR2, Binds Directly to the Surface of HZ Through Its Interactions with DNA.
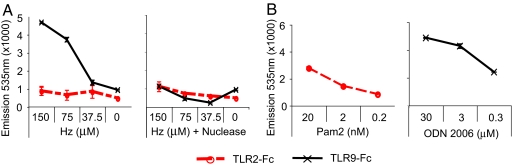
The activation of TLR9 by CpG DNA occurs as a result of the direct binding of DNA to the TLR9 ectodomain (22, 23). We devised an experimental protocol to determine whether TLR9 directly binds to HZ. HZ was used to coat 96-well fluorimeter dishes. The binding to HZ by TLR9 was measured by adding recombinant TLR-Fc fusion proteins containing the ectodomain of TLR9 or TLR2. Positive controls for TLR2:Fc and TLR9:Fc binding consisted of wells coated with lipopeptide and CpG-DNA, respectively. The wells were developed for the presence of bound chimeric molecules with an Alexa Fluor 488-conjugated anti-mouse IgG. This experiment demonstrated binding of TLR9, but not TLR2, to untreated HZ (Fig. 3A). TLR9 binding was entirely eliminated when HZ was pretreated with the nuclease S7, consistent with the concept that TLR9 binds surface DNA and not to the HZ crystal itself. Both TLR2:Fc and TLR9:Fc bound to their established ligands (Fig. 3B).
Fig. 3.
The ectodomain of TLR9 but not TLR2 binds to the DNA coating of natural HZ. Chimeric proteins composed of the extracellular domain of TLR9 or TLR2 and the Fc portion of mouse IgG2a were used to assess ligand binding. Titrated amounts (0–150 μM) of HZ or S7-nuclease treated-HZ (Hz plus nuclease) were absorbed to the plastic bottoms of 96-well black plates. The respective TLR2- or TLR9-Fc fusion proteins were added at 2 μg/ml. Bound Fc proteins were detected by using an anti-mouse IgG Alexa Fluor 488 pAb; fluorescent emissions were read at 535 nm. The binding of TLR2:Fc to PAM2CK4 (Pam2) and TLR9:Fc to CpG oligonucleotide 2006 (ODN 2006) was assessed as positive controls. Results representing triplicate determinations are shown as mean fluorescence emitted (±SD).
The DNA on the Surface of HZ Is Malarial in Origin.
To determine the source of the DNA on HZ, we used HZ as a template in a PCR, using established primer sets from the human, mouse, and P. falciparum genomes (24). Mouse CD14 primers and mouse genomic DNA were also examined because of the possibility that HZ might inhibit the PCR.
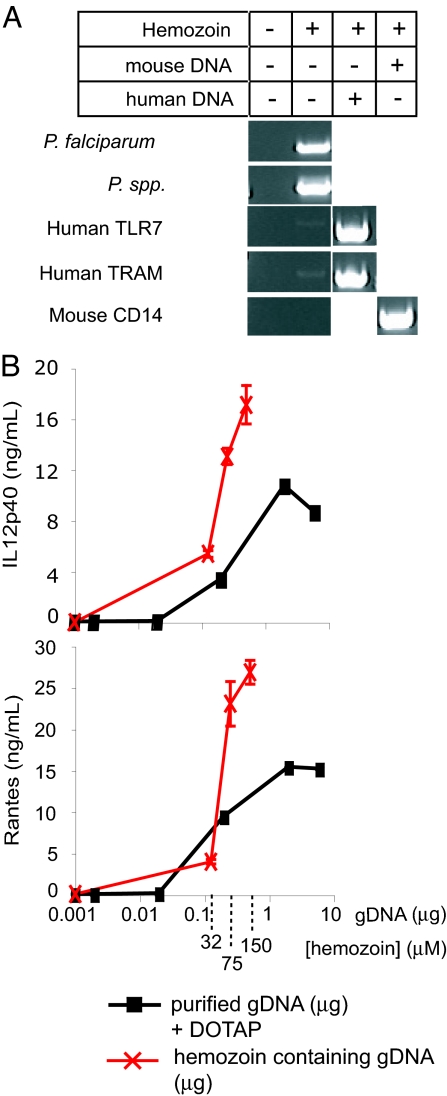
The results of this experiment, shown in Fig. 4A, show clearly that the DNA on the surface of HZ is almost exclusively from the parasite. Extremely small amounts of human DNA were amplified in comparison with malarial DNA, although the human primers were highly efficient at amplifying TLR7 and TRAM from purified human genomic DNA. HZ did not inhibit the ability of mouse CD14 primers to amplify mouse genomic CD14. Hence, the DNA on HZ appears to be malarial in origin.
Fig. 4.
Malarial DNA is coated with HZ; the activity of HZ as an inducer of TLR9 is due to malaria DNA. (A) PCR was performed by using 3.5 nmol of natural HZ or 25 ng of genomic DNA as template. Plasmodium-specific primers (P. falciparum, P. spp), human-specific primers (TLR7, TRAM), or mouse-specific primers (CD14) were tested in the absence or presence of template as indicated in the table. Note that the presentation of the data required reorganization of digital images; the original gels can be seen as SI Fig. 5. (B) FL-DCs from WT mice were stimulated with 32, 75, and 150 μM natural HZ (red X symbols) or purified genomic DNA (black squares) mixed with DOTAP. The x axis of the HZ data has been aligned to also indicate its content of genomic DNA. After 24 h, supernatants were analyzed by ELISA.
Malarial DNA Activates TLR9, and HZ Effectively Traffics Malarial DNA into a TLR9-Positive Compartment.
Typically, stimulatory DNAs are rich in CpG motifs (25). The malaria genome is one of the most AT-rich genomes sequenced to date (26). Thus, the concept that HZ stimulated cells by presenting malaria DNA to TLR9 seemed potentially flawed, and it was important to determine whether malaria DNA truly had the capacity to activate innate immune responses.
We purified malaria DNA from cultures of P. falciparum grown in human blood. These preparations of malaria DNA were endotoxin-free, as assessed by the lack of an effect of the LPS inhibitor E5531 on HZ stimulation; furthermore, FL-DCs from TLR4-null mice responded similarly when compared with WT mice (data not shown). In addition, we used a commercial kit designed to look for the presence of bacterial DNA by PCR; none was found (data not shown).
We first tested the effects of malarial DNA as a potential stimulant of FL-DCs. Purified malaria DNA failed to detectably activate Flt-3-derived dendritic cells at concentrations as high as 50 μg/ml (data not shown). This finding led us to consider our previous finding that TLR9 is localized in an endocytic compartment when DNA is internalized by plasmacytoid dendritic cells (23). In addition, our older unpublished work with Escherichia coli DNA suggested that genomic DNA is not efficiently internalized into cells. One possibility for the failure to observe TLR9 activation by malarial DNA might have been that it was not being properly internalized into an intracellular compartment that had access to TLR9. The monocationic transfection reagent DOTAP has been used to target nucleotides into an endosomal compartment (27). Hence, we mixed malaria DNA with DOTAP and “transfected” the DNA into mouse dendritic cells. Under these conditions, the DNA strongly activated cells to secrete IL12p40 and RANTES (Fig. 4B, black line). The ability of HZ/DOTAP to activate FL-DCs was completely absent in cells from the TLR9−/− mouse (data not shown). When we measured the quantity of DNA present on the surface of HZ, and determined the quantity of DNA per mole of HZ, we found that malarial DNA complexed to the surface of HZ was as potent as malaria DNA mixed with DOTAP, suggesting that HZ is as effective as DOTAP at targeting malaria DNA to an endosomal compartment (Fig. 4B, red line).
Discussion
Very few topics related to the pathogenesis of a common clinical disorder are as poorly established as the biological basis of fever in malaria. On the one hand, there is widespread agreement that the basic rules of fever apply: cytokines with pyrogenic activity are secreted from phagocytes, circulate and alter temperature-regulatory systems in preoptic areas within the hypothalamus. But two basic questions are unresolved. First, what molecule (if any), from the parasite induces cytokines? Second, what receptors recognize this molecule?
Fever in malaria is temporally associated with the rupture of infected RBCs and the release of merozoites. Hence, it has long been assumed that the release of merozoites or some malarial waste product represent a pyrogenic malarial “toxin.” Because the parasite is coated with GPI anchors, many have long thought that this family of molecules represents the malarial toxin. Although we do not dispute such claims, we note that GPI anchors are an established TLR2 ligand (16). We and others have found that the role of TLR2 in rodent malaria appears to be minor [(19), D.T.G. and R.T.G., data not shown]. Furthermore, we and others have also found that purified late trophozoites and schizonts have no obvious TLR2-inducing activity [(18) and data not shown]. These data suggest that the influence of the GPI anchor on cytokine production is limited. Hence, although we believe that GPI anchors have importance in malarial pathogenesis, we have looked to other molecules as the primary inducer of fever.
HZ was the next most attractive molecule that appeared to be a cytokine inducer. Ironically, we initially focused on β-hematin, i.e., “synthetic HZ,” because it seemed less likely to be contaminated with endotoxin and other biologically active molecules than the natural product. This judgment was erroneous, because both β-hematin that is produced from a high-quality source of hemin chloride and the natural crystal that has been “cleaned” with nuclease have no apparent immunomodulatory activity. In contrast to what appears to be the currently prevailing opinion (if not the current dogma), we conclude that the pure HZ crystal cannot, by itself, activate a TLR.
This conclusion does not mean, however, that HZ lacks importance as a cause of fever in malaria. Our study suggests that HZ functions to internalize malaria DNA into an intracellular compartment where it may be sensed by TLR9. In other words, HZ has carrier properties, similar to DOTAP, and transforms malaria DNA from an otherwise harmless molecule to one that has the ability to potently generate cytokines.
On first glance, malaria DNA appeared to be an unusual DNA-based ligand for TLR9, because the P. falciparum genome is highly AT-rich (26). Thus malaria DNA is a poor candidate for a TLR9 inducer. Mouse cells, for example, are optimally activated by oligonucleotides containing CpG sequences flanked by two 5′ purines and two 3′ pyrimidines, such as the motif GACGTT; human TLR9 is optimally triggered by the motif GTCGTT and/or TCGTA (28). Over the last few years, DNA ligands for TLR9 have been categorized in three classes, A, B, and C. A-class oligonucleotides, are strong inducers of Type I interferons (29), in contrast to B-class oligonucleotides, whose IFN-inducing activity is virtually nonexistent. C-class oligonucleotides have features of both classes.
Despite the AT-rich nature of the malaria genome, we found several “classic” CpG motifs in the malaria genome by genome-wide scanning. We identified 269 sequences resembling the CpG B-class motif, whereas only two A-class and three C-class CpG motifs were found. Almost two thirds of the 20 most abundant CpG B-class sequences are located in subtelomeric regions and include some very important var genes, such as P. falciparum erythrocyte membrane protein 1 (PfEMP1). We have found that oligonucleotides based on malaria CpG-rich motifs are highly immunostimulatory [supporting information (SI) Table 1 and SI Fig. 6). In addition to classic CpG motifs, recent reports have also highlighted the potential for certain AT motifs, particularly those flanking a run of 4–5 Ts, to be TLR9 activators (30). We have also identified several regions of the malaria genome that contain such motifs. Like the CpG-containing genes, over half of the 20 most abundant sequences were located in subtelomeric regions.
The larger question that remains unanswered in these studies is whether or not HZ is, in fact, the “malaria toxin” in humans. Such a question is difficult to answer in a compelling manner, although HZ crystal is often detected within the circulating phagocytes of septic patients. The coexpression of pyrogenic cytokines in such cells might be considered good evidence for a role for HZ in fever in human malaria, although this would not be conclusive evidence. TLR9 inhibitors are currently being developed for a variety of conditions, including immune disorders such as systemic lupus erythematosis. Ultimately, the administration of this class of drugs may be the best way we will have to determine the true significance of malarial DNA and TLR9 in malarial septicemia.
Materials and Methods
All of the experiments presented in this report were repeated on three occasions, except for that Fig. 4A, which was repeated twice.
Reagents.
Unless otherwise stated, reagents were purchased from Sigma-Aldrich (St. Louis, MO). LPS from E. coli strain 0111:B4 was reextracted by phenol chloroform (31) to eliminate lipoprotein contamination. Two sources of hemin chloride were used. The first was Sigma-brand hemin chloride, which was extracted from cow blood and was guaranteed to be >80% pure; the second brand was Fluka-brand hemin chloride (Fluka, Buchs, Switzerland), purified by HPLC and guaranteed to be >98% pure. The purity of these preparations, as assessed by LC-mass spectrometry at the University of Massachusetts core facility, was ≈65% and >98%, respectively (data not shown). Pam2CysK4 was from EMC Microcollection (Tubingen, Germany), and human CpG 2336 ODN and CpG 2006 ODN were from Coley Pharmaceutical Group (Wellesley, MA). Poly[IC] was from Amersham (Piscataway, NJ). All ELISA kits were from R & D Systems (Minneapolis, MN), except for mouse IFN-α (Bender MedSystems, Burlingame, CA). C57BL/6 WT mice were obtained from The Jackson Laboratory (Bar Harbor, ME). TLR knockout mice were provided by S. Akira (Osaka University, Osaka, Japan) and analyzed for genetic background by microsatellite analysis (Charles River Laboratories, Wilmington, MA). All mice were subsequently bred to ≥10 generations onto a C57BL/6 background. Antibodies for FACS were from BD Biosciences (San Jose, CA). DNaseI was from Invitrogen (Carlsbad, CA), RNase was from Ambion (Austin, TX), and Staphylococcus aureus S7 nuclease was from USB (Cleveland, OH). DOTAP was from Roche (Indianapolis, IN).
Synthetic HZ (β-Hematin) Preparation.
Synthetic HZ was prepared as described (32). The dried pigment was suspended in endotoxin-free PBS (Cellgro, Herndon, VA) at a final concentration of 2.5 mg/ml and stored at −20°C.
Culture of Parasite and Preparation of Natural HZ.
Erythrocytic stages of P. falciparum 3D7 isolate were cultured as described (33, 34). The culture was checked monthly for Mycoplasma contamination (PCR Mycoplasma Test kit; MDB, St. Paul, MN). HZ was isolated by exploiting its metallic properties. The P. falciparum culture was washed once in PBS and suspended at a hematocrit of ≈2%. The RBC suspension was loaded onto an LS column (Miltenyi Biotec, Auburn, CA) and placed in the MACS separator. The magnetic field allowed the free HZ to be purified because the flow rate was too fast for the retention of infected RBCs. The column was washed with PBS and removed from the magnetic field, and the HZ was eluted, quantified, and frozen at a concentration between 2.5 and 3.7 mM. The quantity of DNA associated with HZ was estimated by OD reading after sequential phenol-chloroform extraction and used for the calculations in Fig. 4B.
Heme Quantitation.
Total heme content was determined by depolymerizing HZ in 6 ml of 20 mM NaOH/2% SDS and incubating the suspension at room temperature for 3 h. An OD reading at 400 nm was then obtained on a Spectronic 20 (Milton Roy, Rochester, NY); 25 μg of β-hematin or natural HZ contains 26–29 nmol of heme (35).
P. falciparum Genomic DNA Preparation.
DNA from P. falciparum culture was purified by using three consecutive phenol-chloroform extractions followed by ethanol precipitation. The DNA was washed six times in 70% ethanol and subjected to RNase treatment. DNA was analyzed for the presence of contaminating bacterial DNA by using a commercial PCR kit designed for this purpose (Onar EUB; Minerva Biolabs, Berlin, Germany).
PCR Analysis of HZ.
PCR was performed by using a stock solution of HZ (3.5 mM) or genomic DNA from human or mouse cells as template (25 ng of DNA per microliter). PCRs included the following: 17.3 μl of H20, 2.5 μl of 10× Hotmaster Taq buffer (Eppendorf, Hamburg, Germany), 1.25 μl each of 10 μM primers, 0.5 μl of dNTP mix (10 mM), 0.2 μl Hotmaster Taq, and 1 μl of template with and without water. PCR conditions were: one cycle at 95°C for 1 min; 35 cycles at 95°C for 45 sec, 58°C for 45 sec, 72°C for 1 min; and one cycle at 72°C for 8 min. The P. falciparum and Plasmodium spp. PCR primers are described in ref. 24. The sequences of the human and mouse primers used are as follows: hTLR7: 5′-C CCCAGCGTCCTTTCACAGA-3 and 5′-CGAGGGCAATTTCCACTTAGGTC-3′; hTRAM: 5′-TCAGAGCGTGGAAGAGATGT-3′ and 5′-CCGCATGGGTATAACAGAGT-3′; mCD14: 5′-CCAAGTTTTAGCGCTGCGTAAC-3′ and 5′-GCCAGCCAAGGATACATAGCC-3′. PCR products were analyzed on a 3% agarose gel.
Cells.
The B16 cell line (36) stably expressing a retrovirus coding FLT3 ligand (FLT3-L) was the source of FLT3-L; levels of FLT3-L in culture supernatants were quantified by ELISA. FL-DCs were generated by culturing bone marrow cells with 100 ng of FLT3-L per milliliter for 7 days in RPMI medium 1640 (Invitrogen) containing 0.1 mM MEM nonessential amino acid (Invitrogen), 10% heat-inactivated FBS (HyClone, Logan, UT), 1 mM sodium pyruvate, 2 mM l-glutamine (Cellgro), 50 mM β-mercaptoethanol, and 10 μg of ciprofloxacin per milliliter (Bayer, West Haven, CT). Flow cytometry demonstrated that 92–96% of the cells were CD11c+; CD11c+ cells were approximately half mDC (CD11b+/B220−) and half plasmacytoid dendritic cells (CD11b−/B220+).
Generation of Chimeric TLR2:Fc and TLR9:Fc Fusion Proteins.
The soluble extracellular domain of human TLR2 was produced as an Fc fusion protein in a retroviral shuttle vector, pCLNCX4, and expressed as a secreted protein in HEK293 cells (37). In the case of TLR9, the ectodomain was cloned into pcDNA3 (Invitrogen) and expressed as an intracellular protein in HEK293 cells, exactly as for full-length TLR9 (23). Cells were grown in protein-free medium (293CD; Invitrogen).
The TLR fusion proteins consisted of the extracellular domains of either human TLR2 (amino acids 1–587) or hTLR9 (amino acids 26–819) fused in-frame with the C-terminal 233-aa Fc portion of mouse IgG2a; in the case of TLR2, the protein was modified by the addition of the linker sequence GAAGGG, which is necessary for efficient secretion. Secreted Fc chimeric proteins were purified from culture supernatants (TLR2:Fc) or cell lysates (TLR9:Fc) by protein A affinity chromatography (GE Healthcare Bio-Sciences, Piscataway, NJ).
TLR Binding Assay.
To study the interaction between HZ and TLRs, an ELISA-like binding assay was developed. Natural HZ, poly[IC], or CpG 2006 were immobilized onto black nontreated 96-well plates (Costar, Corning, NY) in Reacti-Bind DNA coating solution (Pierce, Rockford, IL) overnight at room temperature (TLR9 binding assay). Alternatively, HZ (37.5–150 μM), LPS (10–1,000 ng/ml), or Pam2CysK4 (0.2–20 nM) were immobilized onto black Optiplate 96-well High Binding (PerkinElmer, Wellesley, MA) in PBS overnight at room temperature (TLR2-Fc binding assay). Plates were then washed three times in PBS–0.05% Tween 20, blocked with Superblock (Pierce) for 1 h, and incubated with 2 μg of TLR fusion protein per milliliter at room temperature for 2 h. After three washes, Fc-TLRs bound to their ligands were detected by using Alexa Fluor 488 anti-mouse IgG (Molecular Probes, Eugene, OR). The fluorescence of each sample was measured at 535 nm after excitation at 485 nm by an Envision plate reader (PerkinElmer). The results are presented as the average of triplicate determinations (±SD).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM54060, R01 AI065483, and R21 AI060737 (to D.T.G. and F.N.L.) and a grant from the Netherlands Organization for Scientific Research (to F.N.L.). R.T.G. was supported by the John Simon Guggenheim Memorial Foundation, the Millenium Institute/CNPq, and the World Health Organization.
Abbreviations
- FL-DC
FLT3-L-derived mouse dendritic cell
- GPI
glycosylphosphatidylinositol
- HZ
hemozoin
- TLR
Toll-like receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 1743.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608745104/DC1.
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson MM, Riley EM. Nat Rev Immunol. 2004;4:169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 3.Clark IA, Schofield L. Parasitol Today. 2000;16:451–454. doi: 10.1016/s0169-4758(00)01757-9. [DOI] [PubMed] [Google Scholar]
- 4.Miller LH, Good MF, Milon G. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 5.Francis SE, Sullivan DJ, Jr, Goldberg DE. Annu Rev Microbiol. 1997;51:97–123. doi: 10.1146/annurev.micro.51.1.97. [DOI] [PubMed] [Google Scholar]
- 6.Arese P, Schwarzer E. Ann Trop Med Parasitol. 1997;91:501–516. doi: 10.1080/00034989760879. [DOI] [PubMed] [Google Scholar]
- 7.Pagola S, Stephens PW, Bohle DS, Kosar AD, Madsen SK. Nature. 2000;404:307–310. doi: 10.1038/35005132. [DOI] [PubMed] [Google Scholar]
- 8.Jaramillo M, Godbout M, Olivier M. J Immunol. 2005;174:475–484. doi: 10.4049/jimmunol.174.1.475. [DOI] [PubMed] [Google Scholar]
- 9.Jaramillo M, Plante I, Ouellet N, Vandal K, Tessier PA, Olivier M. J Immunol. 2004;172:3101–3110. doi: 10.4049/jimmunol.172.5.3101. [DOI] [PubMed] [Google Scholar]
- 10.Pichyangkul S, Saengkrai P, Webster HK. Am J Trop Med Hyg. 1994;51:430–435. [PubMed] [Google Scholar]
- 11.Sherry BA, Alava G, Tracey KJ, Martiney J, Cerami A, Slater AF. J Inflamm. 1995;45:85–96. [PubMed] [Google Scholar]
- 12.Sullivan DJ, Jr, Gluzman IY, Russell DG, Goldberg DE. Proc Natl Acad Sci USA. 1996;93:11865–11870. doi: 10.1073/pnas.93.21.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medzhitov R, Janeway C., Jr N Engl J Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 14.Takeda K, Kaisho T, Akira S. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 15.Gazzinelli RT, Ropert C, Campos MA. Immunol Rev. 2004;201:9–25. doi: 10.1111/j.0105-2896.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- 16.Campos MA, Almeida IC, Takeuchi O, Akira S, Valente EP, Procopio DO, Travassos LR, Smith JA, Golenbock DT, Gazzinelli RT. J Immunol. 2001;167:416–423. doi: 10.4049/jimmunol.167.1.416. [DOI] [PubMed] [Google Scholar]
- 17.Krishnegowda G, Hajjar AM, Zhu J, Douglass EJ, Uematsu S, Akira S, Woods AS, Gowda DC. J Biol Chem. 2005;280:8606–8616. doi: 10.1074/jbc.M413541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pichyangkul S, Yongvanitchit K, Kum-arb U, Hemmi H, Akira S, Krieg AM, Heppner DG, Stewart VA, Hasegawa H, Looareesuwan S, et al. J Immunol. 2004;172:4926–4933. doi: 10.4049/jimmunol.172.8.4926. [DOI] [PubMed] [Google Scholar]
- 19.Adachi K, Tsutsui H, Kashiwamura S, Seki E, Nakano H, Takeuchi O, Takeda K, Okumura K, Van Kaer L, Okamura H, et al. J Immunol. 2001;167:5928–5934. doi: 10.4049/jimmunol.167.10.5928. [DOI] [PubMed] [Google Scholar]
- 20.Mockenhaupt FP, Cramer JP, Hamann L, Stegemann MS, Eckert J, Oh NR, Otchwemah RN, Dietz E, Ehrhardt S, Schroder NW, et al. Proc Natl Acad Sci USA. 2006;103:177–182. doi: 10.1073/pnas.0506803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coban C, Ishii KJ, Kawai T, Hemmi H, Sato S, Uematsu S, Yamamoto M, Takeuchi O, Itagaki S, Kumar N, et al. J Exp Med. 2005;201:19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 23.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 24.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 25.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 26.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuda K, Yu P, Kirschning CJ, Schlatter B, Schmitz F, Heit A, Bauer S, Hochrein H, Wagner H. J Immunol. 2005;174:6129–6136. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 28.Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB. Proc Natl Acad Sci USA. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krug A, Towarowski A, Britsch S, Rothenfusser S, Hornung V, Bals R, Giese T, Engelmann H, Endres S, Krieg AM, Hartmann G. Eur J Immunol. 2001;31:3026–3037. doi: 10.1002/1521-4141(2001010)31:10<3026::aid-immu3026>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Shimosato T, Kimura T, Tohno M, Iliev ID, Katoh S, Ito Y, Kawai Y, Sasaki T, Saito T, Kitazawa H. Cell Microbiol. 2006;8:485–495. doi: 10.1111/j.1462-5822.2005.00640.x. [DOI] [PubMed] [Google Scholar]
- 31.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 32.Egan TJ, Ross DC, Adams PA. FEBS Lett. 1994;352:54–57. doi: 10.1016/0014-5793(94)00921-x. [DOI] [PubMed] [Google Scholar]
- 33.Cranmer SL, Magowan C, Liang J, Coppel RL, Cooke BM. Trans R Soc Trop Med Hyg. 1997;91:363–365. doi: 10.1016/s0035-9203(97)90110-3. [DOI] [PubMed] [Google Scholar]
- 34.Trager W, Jensen JB. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan AD, Ittarat I, Meshnick SR. Parasitology. 1996;112(Pt 3):285–294. doi: 10.1017/s003118200006580x. [DOI] [PubMed] [Google Scholar]
- 36.Mach N, Gillessen S, Wilson SB, Sheehan C, Mihm M, Dranoff G. Cancer Res. 2000;60:3239–3246. [PubMed] [Google Scholar]
- 37.Wang RF, Wang X, Johnston SL, Zeng G, Robbins PF, Rosenberg SA. Cancer Res. 1998;58:3519–3525. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.