Abstract
The polo-like kinase, Plk1, which is expressed and active in mitosis, is involved in regulation of mitotic entry, spindle pole assembly, mitotic exit, and cytokinesis [Donaldson MM, Tavares AA, Hagan IM, Nigg EA, Glover DM (2001) J Cell Sci 114:2357–2358]. In mammals, there are two other members of the polo-like kinase family that are less well understood, Plk2 and Plk3. Plk3 first was identified and cloned as an immediate early gene. Here, we report Plk3 localizes to the nucleolus and is involved in regulation of the G1/S phase transition. We demonstrate that the level of Plk3 protein is cell cycle regulated, peaking in G1. We have delivered Plk3-interfering RNA with lentivirus to serum-starved cells and found that, upon serum stimulation, Plk3 is required for cyclin E expression and entry into S phase. Plk3-interfering RNA-induced Plk3 depletion resulted in a large fraction of asynchronously proliferating cells to become quiescent. We propose the Plk3 requirement in the cell cycle is fulfilled in G1, and that once cells pass this point, they are able to complete cell division, whereas in the absence of Plk3, they fail to reenter the cell cycle. Additional data suggest that Plk3 may regulate entry into S phase in part through interaction with the phosphatase Cdc25A, because its depletion also resulted in attenuation of cyclin E expression.
Keywords: Cdc25A, cyclin E, nucleolus
In yeast and Drosophila, polo kinases are master mitotic regulators, involved in the regulation of mitotic entry, the metaphase-to-anaphase transition, and mitotic exit (1, 2). Whereas only one polo kinase is found in yeast and Drosophila, three polo-like kinases have been identified in mammals: Plk1, Plk2 (Snk), and Plk3 (Prk and Fnk). Polo kinases are characterized by an N-terminal serine/threonine kinase domain and a conserved C-terminal substrate-binding domain, termed the Polo box domain (Fig. 1). All three mammalian polo-like kinases have predicted nuclear localization signals (NLSs), suggesting that all three may pass through the nucleus. However, to date, only one NLS (in Plk1) has been shown to be functionally active (3). A fourth related mammalian kinase, Sak (Plk4) shows significant homology to Plk1, Plk2, and Plk3 in its kinase domain but lacks conservation of the typical bipartite polo box domain (4). Plk1 is the best-studied enzyme of this family. Like yeast and Drosophila polo kinases, Plk1 regulates multiple aspects of mitotic entry and exit (5). The functions of Plk2 and Plk3 are not well understood.
Fig. 1.
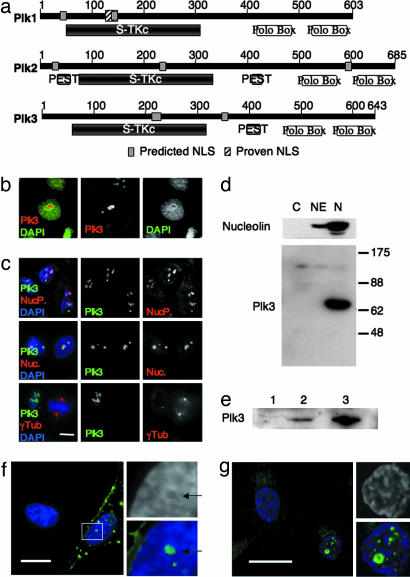
Plk3 localizes to the nucleolus. (a) Comparison of human polo-like kinases. Polo kinases are characterized by a conserved serine-threonine kinase domain (S-Tkc) and a bipartite polo box domain (polo box). All three mammalian polo-like kinases contain predicted nuclear localization signals (NLS), although, to date, only one has been experimentally tested and proven to be functional (proven NLS) (3). Plk2 and Plk3 both contain predicted proline-glutamate-serine-threonine (PEST) signals involved with targeting of proteins for degradation by the proteosome. (b) Endogenous Plk3 (red) appears as nuclear inclusions in interphase U2OS cells. Endogenous Plk3 was visualized by immunofluorescence microscopy using an affinity-purified rabbit polyclonal antibody (Fnk140; Santa Cruz Biotechnology). Nuclear DNA is visualized with DAPI (green). (c) Endogenous Plk3 (green) colocalizes with nucleolar marker proteins nucleophosmin (NucP., red; Top) and nucleolin (Nuc., red; Middle), in U2OS cells, but has no detectable colocalization with the centrosome and spindle pole marker γ-tubulin (γTub., red; Bottom). (Scale bar: 10 μm.) (d) Plk3 is enriched in nucleoli isolated from asynchronous HeLa cells. HeLa cells were fractionated and nucleoli isolated by the method of Andersen et al. (14). One hundred micrograms of protein from each fraction was run on a 10% polyacrylamide gel and analyzed by Western blot. The blot was probed for the nucleolar protein nucleolin and for Plk3, using the same antibodies used in b and c. C, cytoplasmic fraction; NE, soluble nuclear extract; N, nucleolar fraction. (e) Plk3 is resistant to detergent extraction. Asynchronously growing HeLa cells were extracted sequentially with equal volumes of STE (lane 1), RIPA (lane 2), and Hepes/glycerol/NaCl (lane 3) buffers. Equal volumes of each extract were analyzed by Western blot for Plk3. (f and g) Twenty hours after transfection, ectopically expressed strep-tagged mouse Plk3 (green) forms nuclear inclusions in HeLa cells (f) and in MCF10A cells (g). (Scale bars: 15 μm.)
Polo kinases colocalize with their substrates through interactions with the polo box domain (6, 7). There are several conflicting reports of Plk3 localization in the literature. Plk3 has been reported to colocalize with the centrosome, spindle pole and spindle microtubules (8), with Golgi apparatus (9), and with actin-containing plaques (10). In contrast to previous reports, we demonstrate here that Plk3 localizes in the nucleolus, based on both immunofluorescence and subcellular fractionation.
We show that the level of Plk3 protein is cell cycle regulated; unlike Plk1, the level of Plk3 peaks in G1, concurrent with the reported peak in the level of Plk3 message (11–13). We further demonstrate that Plk3 is required for expression of cyclin E and for cells to pass from G1 to S phase.
Results
Plk3 Localizes to the Nucleolus.
Because Plks colocalize with their substrates, we reasoned that a clear understanding of Plk3 localization throughout the cell cycle should shed light on its functions. We initially examined its localization by using the antibody and conditions reported in refs. 8 and 9. However, in contrast to their results, we were unable to detect any specific immunofluorescent localization (data not shown). We next screened other commercially available antibodies for effective immunofluorescent staining. One affinity-purified rabbit polyclonal antibody, Fnk140 (Santa Cruz Biotechnology, Santa Cruz, CA), appeared effective. Unexpectedly, in U2OS cells, the staining of endogenous Plk3 appeared as discrete nuclear inclusions, morphologically similar to nucleoli (Fig. 1b). Identification of these nuclear compartments as nucleoli was confirmed by colocalization of Plk3 with nucleophosmin and nucleolin, proteins that are known to localize to the nucleolus (Fig. 1c). Similar colocalization was observed in HeLa, MCF10A, h-tert-RPE, T98G, and 293T cells (data not shown). Plk3 localization was detectable only within the nucleolus; we were unable to detect endogenous Plk3 at the centrosome, spindle pole, or any other cytoplasmic location (Fig. 1c).
Human Plk3 is predicted to migrate as a 72-kDa protein. To confirm that the observed immunolocalization represented endogenous Plk3 and not a cross-reactive protein, asynchronous HeLa cells were fractionated according to the method of Andersen et al. (14). The Fnk140 antibody recognized a band of approximately the expected size that was significantly enriched in nucleolar fractions but undetectable in cytoplasmic and soluble nuclear fractions (Fig. 1d). This ≈72-kDa band was specifically depleted by three different lentiviruses expressing Plk3-RNAi targeting different sequences in Plk3 mRNA, as discussed below (Figs. 3 and 4). These same RNAi vectors also significantly reduced the immunofluorescence signal at the nucleolus (see below). Moreover, we expressed full-length strep-tagged mouse Plk3 and evaluated immunolocalization by using anti-strep-tag antibodies. At 20–24 h after transfection, ectopically expressed Plk3 formed nuclear inclusions in both HeLa and MCF10A cells (Fig. 1 f and g). Taken together, these data show that Plk3 is a nucleolar protein.
Fig. 3.
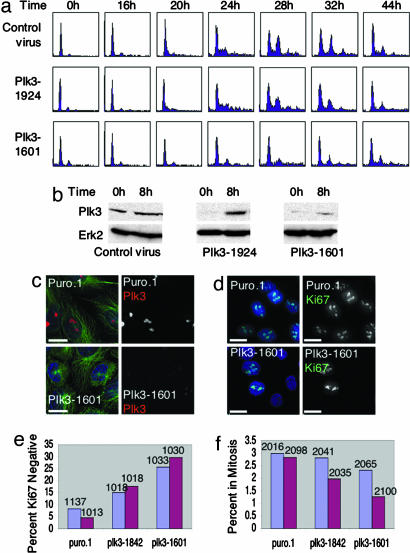
Plk3 is required for entry into S phase. (a and b) MCF10A cells were infected with lentivirus-expressing RNAi-targeting Plk3 at nucleotide 1601 or 1924 of mature message, or with control virus carrying the Plk0 puro.1 vector, then synchronized by 72 h of serum starvation. Cells were analyzed at the indicated times after addition of serum and hormones. (a) Cell cycle progression of treated cells was analyzed by FACS. Cells treated with the 1601 virus failed to enter S phase. Cells treated with the 1924 virus showed a modest reduction in S and G2/M phase entry relative to control cells. (b) Total cellular protein was analyzed from duplicate plates by Western blot to determine the effectiveness of the vectors. Both 1924 and 1601 showed a significant reduction in Plk3 levels at 0 h, but only 1601 maintained a low level of Plk3 throughout 8 h of induction. (c–f) Asynchronously growing MCF10A cells were treated with virus-targeting Plk3 or with plk0 puro.1 control virus, maintained at low density, and analyzed with time by immunofluorescence. (c) Cells treated with lentivirus-targeting Plk3 show a significant reduction in Plk3 (red) staining at the nucleolus and a normal cytoskeleton (α-tubulin, green; DNA, DAPI, blue). (Scale bars: 15 μm.) (d) The percentage of cycling cells was assessed by staining for cellular proliferation marker Ki67 (green), which is expressed in all phases of the cell cycle except G0. DNA was visualized by DAPI. (Scale bar: 20 μm.) (e) Asynchronous MCF10A cells treated with virus-targeting Plk3 showed a significant increase in percentage of Ki67-negative cells relative to puro.1 treated control at 3 days (blue bars) and 5 days (red bars) after infection. Number of cells counted for each treatment on each day appears above bar. (Plk3–1842: day 3, P = 3.8 × 10−38; day 5, P = 8.1 × 10−32), (Plk3–1601: day 3, P = 9.2 × 10−69; day 5, P = 2.7 × 10−62). (f) Plk3 virus-treated cells also showed a significant decrease in the percent of cells in mitosis on day 5 (Plk3–1842: day 3, P = 0.08; day 5, P = 7.4 × 10−6), (Plk3–1601: day 3, P = 0.0016; day 5, P = 4.8 × 10−9). The number of cells counted for each treatment appears above each bar. Blue bars, 3 days. Red bars, 5 days.
Fig. 4.
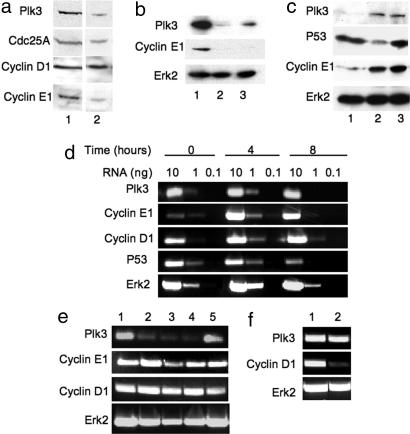
Plk3 regulates the level of cyclin E1 by a posttranscriptional mechanism. (a) Eight hours after serum addition, MCF10A cells infected with Plk3–1601 virus (lane 2) and synchronized by serum starvation show a significant reduction in cyclin E1 protein level relative to control cells 8 h after serum addition (lane 1). Levels of two other important G1 proteins, cyclin D1 and Cdc25A, were not significantly affected. (b) Cells treated with Plk3–1842 virus also showed a significant reduction in cyclin E1 at 8 h after release from serum starvation. Lanes: 1, control; 2, Plk3–1842; 3, Plk3–1601. (c) Eight hours after release from serum starvation, MCF10A cells treated with virus targeting P53 showed no effect on Plk3 or cyclin E1 levels relative to cells treated with the control virus PLK0 puro.1. Lanes: 1, Plk3–1601; 2, P53-virus; 3, control. (d) Uninfected MCF10A cells were synchronized by serum starvation, then relative message levels of Plk3 and the G1 cyclins D1 and E1 were evaluated by RT-PCR. P53 and Erk2 message levels were included for comparison. Plk3 and cyclin D1 message levels were high before serum addition, were higher at 4 h after release, and fell again by 8 h after serum addition. Cyclin E1 message was barely detectable before serum addition, peaked at 4 h, and was somewhat reduced by 8 h. P53 message showed no difference from 0–4 h but was significantly reduced at 8 h after serum addition. The message level of the constitutively expressed Erk2 is included for comparison. (e) Four hours after induction with serum, MCF10A cells treated with lentivirus-targeting Plk3 showed a significant reduction in message for Plk3 but no significant effect on message levels of cyclin E1 or cyclin D1. Erk2 message level is included for reference. Messages were amplified from 2 ng of RNA. Lanes: 1, control virus; 2, Plk3–1934; 3, Plk3–1845; 4, Plk3–1601; 5, P53-virus. (f) Plk3 message level was unaffected by virus-targeting cyclin D1, although cyclin D1 message levels were significantly reduced.
The Level of Plk3 Protein Is Cell Cycle Regulated.
Plk3 was initially cloned as an immediate early gene (11, 12). To shed further light on Plk3 function, we chose to investigate the protein level of Plk3 throughout the cell cycle. As is the case for other predominantly nucleolar proteins, endogenous Plk3 is resistant to detergent extraction and is not readily solubilized by standard isolation protocols for protein kinases. But it can be effectively solubilized from either isolated nucleoli (data not shown) or detergent-extracted cell pellets (Fig. 1e) with a buffer containing high salt, magnesium, and glycerol (15). Therefore, to follow the level of Plk3, we analyzed total protein by boiling the entire sample, not just the soluble fraction, in SDS sample buffer.
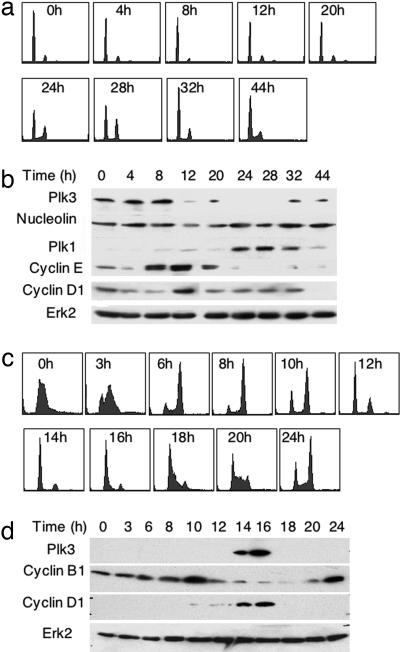
We analyzed the protein level of Plk3 throughout the cell cycle in MCF10A cells synchronized by serum starvation (Fig. 2 a and b) and in HeLa cells synchronized in early S phase by double thymidine block (Fig. 2 c and d). Plk3 expression was monitored by Western blot analysis; cell cycle progression was monitored by fluorescence-activated cell sorting (FACS). In MCF10A cells, Plk3 was detected in serum-deprived cells but increased and peaked in G1, ≈8 h after serum stimulation, before the level of cyclin E1 reached its peak. Mitosis occurred at 24–28 h, based on the proportion of cells with 4N DNA content, concurrent with peak expression of Plk1. This pattern of Plk3 protein expression is consistent with the expression pattern of Plk3 mRNA in serum-starved cells (11–13).
Fig. 2.
Plk3 is expressed in G1. (a and b) MCF10A cells were synchronized by 72 h of serum starvation. (a) Serum and hormones then were added to the cells, and the cells analyzed at 4-h intervals by FACS. (b) Total cellular protein was analyzed from duplicate plates by Western blot. (a) Under these conditions, G2/M occurred between 24–32 h. (b) The level of Plk3 protein peaked in G1 near 8 h, whereas the level of Plk1 peaked as expected during G2/M. Levels of the G1 cyclins, cyclin D1 and cyclin E1, were analyzed for comparison. Nucleolin and Erk2 loading controls are included. (c and d) HeLa cells were synchronized by double thymidine block. Cells were analyzed at 2-h intervals by FACS (c), and total cellular protein was analyzed from duplicate plates by Western blot (d). (c) Double thymidine block synchronizes cells in early S phase with slightly more than 2N DNA content (0 h). M phase cyclin B1 expression peaked at 10 h after release and again at 24 h. The level of Plk3 protein peaked in G1 at 14–16 h (c and d), concurrent with the peak expression of the G1 phase cyclin D1. Erk2 loading control is also shown.
In HeLa cells synchronized by double thymidine block, the cells arrest in early S phase, with slightly more than 2N DNA content (Fig. 2c, 0 h). Mitosis occurred at 8–10 h after release, concurrent with peak expression of cyclin B1, and Plk3 peaked in G1 at 14–16 h, concurrent with peak cyclin D1 expression, just before the onset of the next S phase (Fig. 2 c and d). Taken together, these results demonstrate that Plk3 expression is cell cycle regulated and Plk3 is expressed primarily in G1. This suggests a possible role for Plk3 in regulating G1 events.
Plk3 Is Required for Progression from G1 to S Phase.
To investigate the function of Plk3 in vivo, we designed lentivirus vectors expressing RNAi, specifically targeting Plk3. The lentivirus system has several advantages over siRNA treatments, including efficient infection of difficult-to-transfect cells, and the ability to select infected cells with puromycin, ensuring that all cells in a study are, in fact, expressing RNAi. MCF10A cells were infected, synchronized by serum starvation, and analyzed after addition of serum (Fig. 3a and b). Cells that were efficiently depleted of Plk3 failed to enter S phase (Fig. 3 a and b; Plk3–1601), whereas cells that were less efficiently depleted (Plk3–1924) showed only modest reduction is S phase progression, and cells infected with control virus entered S and G2/M phases normally.
We also evaluated the effect of Plk3 depletion on logarithmically growing MCF10A cells (Fig. 3 c–f). Cells treated with lentivirus targeting Plk3 maintained a normal cellular morphology, even 5 days after infection, although the level of Plk3 was visibly reduced (Fig. 3c). We used immunofluorescence microscopy to assess the percentage of cells expressing the proliferation marker Ki67, which is normally expressed in cells in G1, S, G2, and M phases but not in G0 (16) (Fig. 3 d and e). Cells treated with lentivirus-targeting Plk3 showed a significant increase in the percentage of cells not expressing Ki67 at 3 and 5 days (P < 10−5), indicating that a significant percentage of Plk3-depleted cells had exited the cell cycle. By 5 days after infection, the percentage of depleted cells reaching mitosis was significantly reduced (Fig. 3f). Those cells that did reach mitosis appeared to pass through mitosis normally, no mitotic defects were observed in these cells. The results of Plk3 depletion in synchronized cells (Fig. 3 a and b) and asynchronous cells (Fig. 3f) suggest that Plk3 is required in G1; after passage of that restriction point, cells complete the cell cycle normally but are unable to initiate a new cycle.
Plk3 Is Required for Cyclin E Expression.
Because Plk3-depleted cells arrested in G1/G0, we examined MCF10A cells depleted of Plk3, synchronized by serum starvation, and analyzed by Western blot 8 h after serum addition for levels of known major regulators of entry into S phase. Cells with reduced Plk3 also had a significantly reduced level of cyclin E protein, although levels of cyclin D and Cdc25A appeared unaffected (Fig. 4 a and b). This suggests that Plk3 functions upstream of cyclin E in S phase entry. RNAi sequences may produce off-target effects; therefore, it is important to demonstrate target specificity by demonstrating the same effect with multiple sequences targeting the same message of interest. The attenuation of cyclin E protein was observed for virus-targeting Plk3 at different sites (Fig. 4b), suggesting that the effect was not due to off-target effects from a single RNAi site. As an additional control to rule out off-target effects from viral infection, we analyzed the effect of lentivirus-based RNAi of P53 on cyclin E and Plk3 levels 8 h after serum stimulation (Fig. 4c). The level of P53 was significantly reduced, but Plk3 and cyclin E levels were unaffected, further indicating that the reduction in cyclin E protein is a specific effect caused by depletion of Plk3.
Because cyclin E accumulation in G1 reflects increased transcription of cyclin E message in normal cells (17), we sought to determine whether Plk3 regulates cyclin E1 transcription. We evaluated the relative cyclin E message level by RT-PCR in cells depleted of Plk3. In untreated MCF10A cells synchronized by serum starvation, Plk3, cyclin E1, and cyclin D1 message levels were all elevated 4 h after serum addition (Fig. 4d). We therefore chose the 4-h time point for lentivirus studies. Depletion of Plk3 had no significant effect on induction of message for cyclin E1 (Fig. 4e), suggesting that Plk3 must regulate the level of cyclin E1 by a posttranscriptional mechanism. Cyclin D1 message was also unaffected (Fig. 4e). Depletion of cyclin D1 had no effect on the levels of Plk3 message (Fig. 4f) or Plk3 protein (data not shown), suggesting that Plk3 expression is not regulated by cyclin D1.
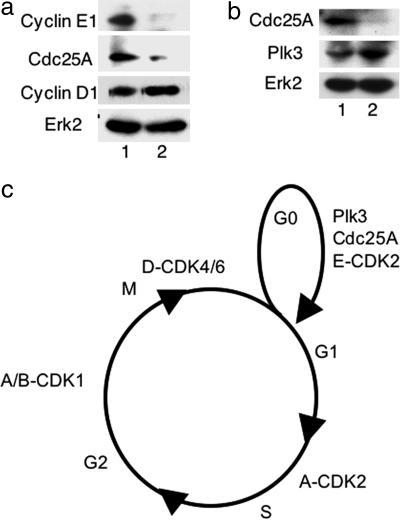
Cdc25A has been reported to be a substrate of Plk3 in vitro (18), and Cdc25A regulates the activity of the cyclin E/Cdk2 complex (17, 19, 20). This suggested the possibility that Plk3 may influence cyclin E stability indirectly through interaction with or activation of Cdc25A. To investigate this question, we designed lentivirus-based RNAi to deplete Cdc25A. MCF10A cells were depleted of Cdc25A, synchronized by serum starvation, and analyzed at 8 h after serum addition (Fig. 5a). Importantly, although the level of cyclin D was unaffected, Cdc25A was depleted and cyclin E expression was attenuated, indicating that Cdc25A is required for cyclin E accumulation. Plk3 was expressed in cells depleted of Cdc25A, showing that the loss of cyclin E expression was not caused by loss of Plk3 (Fig. 5b).
Fig. 5.
Depletion of Cdc25A causes loss of cyclin E. (a) Loss of Cdc25A by lentivirus-based RNAi resulted in a significant reduction in cyclin E expression without significantly altering the level of cyclin D. MCF10A cells were infected with lentivirus-targeting Cdc25A (lane 2) or with control virus (lane 1) and synchronized by serum starvation. The levels of Cdc25A, cyclin E1, and cyclin D1 were analyzed by Western blot 8 h after addition of serum. The level of Erk2 is included as a loading control. (b) Loss of cyclin E in MCF10A cells treated with lentivirus-based RNAi targeting Cdc25A is not caused by loss of Plk3. MCF10A cells were treated with lentivirus-targeting Cdc25A (lane 2) or with control virus (lane 1) and synchronized by serum starvation as above. The levels of Cdc25A and Plk3 were analyzed by Western blot 8 h after addition of serum. The level of Erk2 is included as a loading control. (c) Cyclin-dependent kinase complexes and Plk3 promote cell cycle progression. Cyclin E is required to restart the cell cycle after serum starvation (21). Depletion of Plk3 or Cdc25A attenuates expression of cyclin E and blocks cell cycle progression.
Discussion
We have demonstrated that Plk3 expression is cell cycle regulated. The level of Plk3 protein peaks in G1; Plk3 expression becomes undetectable as cells enter mitosis. We have shown that Plk3 is required for progression from G1 to S phase in cells synchronized by serum starvation. We have demonstrated that loss of Plk3 attenuated cyclin E expression and that Plk3 may influence cyclin E expression indirectly through regulation of Cdc25A. Plk3 is also required for continued cycling in asynchronous cells. We have demonstrated that Plk3 localizes to the nucleolus. The data presented here do not agree with previous reports indicating that Plk3 localized to the centrosome, spindle microtubules, Golgi apparatus, and actin-containing plaques (8–10). We are unable to account for the differences, but as shown in Results, the antibody used here indeed recognizes Plk3 based on the loss of immunoflourescence and Western blot signals upon down-regulation and specific recognition of protein expressed after transfection with cloned Plk3 cDNA.
A recent study involving knockout mice suggests that cyclin E is required for reinitiation of the cell cycle after serum starvation but is dispensable for continued cycling under permissive conditions (21). Herein we show that Plk3 attenuates cyclin E expression, and by doing so, may regulate the G1 restriction point, start. We have also found that Plk3 is required for continued cycling under permissive conditions. This difference in phenotype between the cyclin E knockout and Plk3 depletion with RNAi indicates that Plk3 regulates targets in addition to cyclin E. Taken together, our data on cell cycle progression in Plk3-depleted cells indicates that Plk3 must be a critical regulator of G1 events.
Cyclin D/cdk4/6 regulates G1/S primarily by phosphorylating and inhibiting transcriptional repressors (17). Plk3 expression is not affected by cyclin D1 depletion; it appears Plk3 functions in a pathway parallel to be unregulated by cyclin D.
Cyclin E is an unstable protein with a half-life of <30 min (22, 23). Moreover, its mode of degradation has been reported to depend on whether it is associated with Cdk2 (24). Two distinct pathways of ubiquitin-dependent proteolysis have been described for cyclin E. Cyclin E unassociated with Cdk2 is polyubiquitinated and targeted for proteolysis through binding to cullin-3 (24). Perhaps the events in these processes are regulated by Plk3 and/or Cdc25A. Cyclin E in a complex with Cdk2 is targeted for degradation by Fbw7 in a phosphorylation-dependent manner (25, 26). Thus, Plk3 and Cdc25A may impact the accumulation of cyclin E by modulating its capacity to associate with Cdk2 or with cullin-3 or Fbw7 ubiquitin ligases. Our observations also may reflect unexplored mechanisms of cyclin E regulation.
Localization of Plk3 to the nucleolus places it in position to participate in normal nucleolar activities. The nucleolus is well known as the site of ribosome biogenesis. Recently it has become apparent that the nucleolus also functions in the regulation of mitotic exit and the DNA-damage response, primarily through sequestration of regulators, such as P53, MDM2, CDC14b, pRB, pRB2, E2F4, p107, and p14ARF (27–34). Several major regulators of the G1/S transition, including cyclin D, cyclin E, and Cdc25A, are nuclear and may transit through the nucleolus (35–38). Cyclin E, in particular, has been reported to relocate from the nuclear matrix to the nucleolus at the onset of S phase (36). Taken together with our findings on Plk3, these data suggest that the nucleolus may also play a hitherto unappreciated role in regulating the G1/S transition.
Materials and Methods
Cell Culture and Synchronization.
HeLa, hTert-RPE, U2OS, and 293T cell lines were maintained in DMEM supplemented with 10% FCS supplemented with penicillin and streptomycin (Pen/Strep). Cells were synchronized in early S phase by double thymidine block; asynchronous cells were treated for 16 h with 2 mM thymidine, released for 8 h into fresh medium, and then treated for an additional 16 h with 2 mM thymidine. Human mammary epithelial cells (MCF-10A) were maintained in DMEM/F12 (50/50) supplemented with 5% horse serum/20 ng/ml EGF/0.5 μg/ml hydrocortisone/100 ng/ml cholera toxin/10 μg/ml insulin/Pen/Strep as described in ref. 39. These cells were synchronized by serum starvation for 72 h in DMEM/F12 with Pen/Strep, then stimulated by addition of 20% horse serum/20 ng/ml EGF/0.5 μg/ml hydrocortisone/100 ng/ml cholera toxin/10 μg/ml insulin.
Immunofluorescence.
For immunofluorescence, cells were fixed with −20°C methanol and stained as described (42). Data were collected on an Applied Precision Deconvolution Microscope as a Z series with 0.3 μm between planes. Images were subjected to a single round of deconvolution by using Deltavision software. All images were rendered two-dimensional by simple projection.
Antibodies.
Anti-Plk3 (Fnk140), anti-cyclin E, anti-cyclin D1, anti-Cdc25A, and anti-Erk2 antibodies were purchased from Santa Cruz Biotechnology. Anti-γ-tubulin GTU88 antibody was obtained from Sigma (St. Louis, MO). Anti-Plk1 and anti-nucleophosmin antibodies were purchased from Zymed (Carlsbad, CA). Anti-nucleolin antibody was obtained from Research Diagnostics (Flanders, NJ). Anti-Ki67 monoclonal antibody and anti-PRK monoclonal antibody were obtained from BD Biosciences (San Jose, CA). Anti-P53 monoclonal antibody was obtained from Imagenex (San Diego, CA). Anti-strep-tag antibody was obtained from IBA (Goettingen, Germany).
Molecular Cloning and Transfections.
Full-length mouse PLK3 was cloned by PCR as described (11) into the IBA45 mammalian expression vector (IBA) by using the forward and reverse primers CCAAAACAGCGAATTCTTCCTATTGGCTGACAGGGC and CCAAAACAGCCTCGAGTTCCTATTGGCT, respectively. The product was cut with EcoRI and XhoI and inserted into the EcoRI and XhoI sites in the IBA45 expression vector. The clone was fully sequenced before use. The clone was transfected into HeLa cells with GenePorter reagents, and into MCF10A cells with calcium phosphate.
Lentivirus-Based RNAi Plasmid Preparation and Virus Production.
Three lentivirus RNAi transfer plasmids (pLKO.1-Plk3) were constructed as described in ref. 40. The human Plk3 sequences targeted were AAGAAAGACTGTGCACTACAA starting at position 1601, CTGTCCAGGTGAACTTCTA starting at position 1842, and AAATCGTAGTGCTTGTACTTA starting at position 1924 (GenBank accession no. NM_004073). Human cyclin D1 was targeted with the sequence CAAACAGATCATCCGCAAACA, which starts at position 730 (GenBank accession no. NM_053056). Cdc25A was targeted with the sequence GCACCACGAGGACTTTAAAGA, which starts at position 1711 (GenBank accession no. NM_201567). P53 was depleted with a vector that targets the P53 sequence from 1019–1039 (41).
To generate lentivirus, 293T cells in 100-mm dishes were transfected with 12 μg of pLKO.1-Plk3 (or control pLKO.1 vector), 12 μg of pHR′-CMV-ΔR8.20 vpr and 6 μg of pHR′-CMV-VSV-G by using calcium phosphate. Virus was harvested every 12 h from 24 to 72 h after transfection. Virus was passed through a. 45-μm filter to remove contaminating cell debris, then concentrated at 16,700 rpm for 90 min in a SW27 rotor at 5°C. The viral pellet was resuspended in 500 μl of TNE buffer (50 mM Tris·HCl, pH 7.8/130 mM NaCl/1 mM EDTA) per 100-mm dish. Infections were carried out in the presence of 10 μg/ml polybrene/10 mM Hepes. Selection for infected cells was initiated 24 h after infection by addition of 2 μg/ml puromycin.
Biochemical Techniques.
Nucleoli were isolated by the method of Andersen et al. (14). Briefly, HeLa cells were lysed in a dounce homogenizer in hypotonic buffer containing 10 mM Hepes, pH 7.9/10 mM KCl/1.5 mM MgCl2/0.5 mM DTT and complete protease inhibitors (catalog no. 1-873-580; Roche) supplemented with 10 μg/ml PMSF. Intact nuclei were sedimented by centrifugation at 200 × g for 5 min, and the soluble cytoplasmic extract was collected. The nuclear pellet was resuspended in a solution of 0.25 M sucrose, 10 mM MgCl2, and complete protease inhibitors with PMSF and then sedimented through a cushion of 0.35 M sucrose/0.5 mM MgCl2/protease inhibitors with PMSF at 1,500 × g for 5 min. Sedimented nuclei were resuspended in 0.35 M sucrose/0.5 mM MgCl2/protease inhibitors with PMSF and lysed by sonication. The released nucleoli were sedimented through a cushion of 0.88 M sucrose/0.5 mM MgCl2/protease inhibitors with PMSF at 3,000 × g for 10 min and resuspended in 0.35 M sucrose/0.5 mM MgCl2/protease inhibitors with PMSF. The cytoplasmic extract, the soluble nuclear extract, and the nucleolar suspension were boiled in SDS sample buffer and then analyzed by Western blot.
Preparation of Sequential Cell Extracts.
HeLa cells were sedimented in a microcentrifuge tube, then lysed on ice with STE extraction buffer (150 mM NaCl/50 mM Tris, pH 7.2/1.5 mM EDTA/1% Tween 20/2.5 mM sodium vanadate/protease inhibitor mixture with PMSF). The extract was clarified in a microcentrifuge for 15 min at high speed. The pellet was resuspended in RIPA (150 mM NaCl/10 mM Tris, pH 7.2/1% sodium deoxycholate/1% Triton X-100/0.1% SDS/2.5 mM sodium vanadate/protease inhibitor mixture with PMSF). Plk3 in the pellet after RIPA extraction was solubilized in a buffer containing 20 mM Hepes, 25% glycerol, 400 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 2.5 mM sodium vanadate, and protease inhibitor mixture with 0.5 mM PMSF.
RT-PCR.
RNA was extracted by use of an RNeasy kit from Qiagen (Valencia, CA). Message sequences were amplified by RT-PCR with a OneStep RT-PCR kit, also from Qiagen. Plk3 message was amplified with the primer pair TTGTGCTGGTGGGATTGTAGTGCACAG and GTGGCCACAGTAGTGGAGTCAGCCC. Cyclin E1 message was amplified with the primer pair CCTTGGGACAATAATGCAGTCTGTGC and CCATCTGTCACATACGCAAACTGGTGC. Cyclin D1, Erk2, and P53 messages were amplified with the primer pairs CACACTTGATCACTCTGGAGAGGAAGC, CGAGGAGCTGCTGCAAATGGAGCTGC; GCTACACCAACCTCTCGTACATCGGC, GAACCCTGTGATCATGGTCTGGATCTG; and GCTGTGACTGCTTGTAGATGGCCATGG, GTGCAGCTGTGGGTTGATTCCACACC, respectively.
Acknowledgments
We thank Eleanor Erikson, Ming Lei, Tianhua Zhou, and Xiaoqi Liu for critical comments. This work was supported by National Institutes of Health Grant GM59172. R.L.E. is the John F. Drum American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lee KS, Park JE, Asano S, Park CJ. Oncogene. 2005;24:217–229. doi: 10.1038/sj.onc.1208271. [DOI] [PubMed] [Google Scholar]
- 2.Glover DM. Oncogene. 2005;24:230–237. doi: 10.1038/sj.onc.1208279. [DOI] [PubMed] [Google Scholar]
- 3.Taniguchi E, Toyoshima-Morimoto F, Nishida E. J Biol Chem. 2002;277:48884–48888. doi: 10.1074/jbc.M206307200. [DOI] [PubMed] [Google Scholar]
- 4.Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. Nat Struct Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- 5.Barr FA, Sillje HH, Nigg EA. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 6.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elia AE, Cantley LC, Yaffe MB. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Xie S, Chen J, Fukasawa K, Naik U, Traganos F, Darzynkiewicz Z, Jhanwar-Uniyal M, Dai W. Mol Cell Biol. 2002;22:3450–3459. doi: 10.1128/MCB.22.10.3450-3459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan Q, Wang Q, Xie S, Fang Y, Darzynkiewicz Z, Guan K, Jhanwar-Uniyal M, Dai W. Exp Cell Res. 2004;294:51–59. doi: 10.1016/j.yexcr.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Holtrich U, Wolf G, Yuan J, Bereiter-Hahn J, Karn T, Weiler M, Kauselmann G, Rehli M, Andreesen R, Kaufmann M, et al. Oncogene. 2000;19:4832–4839. doi: 10.1038/sj.onc.1203845. [DOI] [PubMed] [Google Scholar]
- 11.Donohue PJ, Alberts GF, Guo Y, Winkles JA. J Biol Chem. 1995;270:10351–10357. doi: 10.1074/jbc.270.17.10351. [DOI] [PubMed] [Google Scholar]
- 12.Chase D, Feng Y, Hanshew B, Winkles JA, Longo DL, Ferris DK. Biochem J. 1998;333:655–660. doi: 10.1042/bj3330655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anger M, Kues WA, Klima J, Mielenz M, Kubelka M, Motlik J, Esner M, Dvorak P, Carnwath JW, Niemann H. Mol Reprod Dev. 2003;65:245–253. doi: 10.1002/mrd.10289. [DOI] [PubMed] [Google Scholar]
- 14.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 15.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes J, Lemke H, Baish H, Wacker H, Schwab U, Stein H. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 17.Bartek J, Lukas C. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 18.Myer DL, Bahassi el M, Stambrook PJ. Oncogene. 2005;24:299–305. doi: 10.1038/sj.onc.1208278. [DOI] [PubMed] [Google Scholar]
- 19.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann I, Draetta G, Karsenti E. EMBO J. 1994;13:4302–4310. doi: 10.1002/j.1460-2075.1994.tb06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng Y, Yu Q, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 22.Clurman BE, Sheaf RJ, Thress K, Groudine M, Roberts JM. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 23.Won KA, Reed SJ. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 24.Singer JD, Gurian-West M, Clurman B, Roberts JM. Genes Dev. 2006;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minella AC, Welcker M, Clurman B. Proc Natl Acad Sci USA. 2005;102:9649–9654. doi: 10.1073/pnas.0503677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Drogen F, Sangfelt O, Malyukova A, Matskova L, Yeh E, Means AR, Reed SI. Mol Cell. 2006;23:37–48. doi: 10.1016/j.molcel.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 27.Wsierska-Gadek J, Horky M. Ann NY Acad Sci. 2003;1010:266–272. doi: 10.1196/annals.1299.046. [DOI] [PubMed] [Google Scholar]
- 28.Cerutti L, Simanis V. Curr Opin Genet Dev. 2000;10:65–69. doi: 10.1016/s0959-437x(99)00044-1. [DOI] [PubMed] [Google Scholar]
- 29.Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. J Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argentini M, Barboule N, Wasylyk B. Oncogene. 2000;19:3849–3857. doi: 10.1038/sj.onc.1203737. [DOI] [PubMed] [Google Scholar]
- 31.Rogalsky V, Todorov G, Moran D. Biochem Biophys Res Commun. 1993;192:1139–1146. doi: 10.1006/bbrc.1993.1535. [DOI] [PubMed] [Google Scholar]
- 32.Zini N, Trimarchi C, Claudio PP, Stiegler P, Marinelli F, Maltarello MC, La Sala D, De Falco G, Russo G, Ammirati G, et al. J Cell Physiol. 2001;189:34–44. doi: 10.1002/jcp.1135. [DOI] [PubMed] [Google Scholar]
- 33.Green C, Chatterjee R, McGarrigle HH, Ahmed F, Thomas NS. J Mol Endocrinol. 2000;25:275–286. doi: 10.1677/jme.0.0250275. [DOI] [PubMed] [Google Scholar]
- 34.Rizos H, Darmanian AP, Mann GJ, Kefford RF. Oncogene. 2000;19:2978–2985. doi: 10.1038/sj.onc.1203629. [DOI] [PubMed] [Google Scholar]
- 35.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juan G, Cordon-Cardo C. Cancer Res. 2001;61:1220–1226. [PubMed] [Google Scholar]
- 37.Cazzalini O, Perucca P, Valsecchi F, Stivala LA, Bianchi L, Vannini V, Prosperi E. Histochem Cell Biol. 2004;121:377–381. doi: 10.1007/s00418-004-0650-8. [DOI] [PubMed] [Google Scholar]
- 38.Kim SH, Li C, Maller JL. Dev Biol. 1999;212:381–391. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- 39.Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 40.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Lei M, Erikson RL. Mol Cell Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purohit A, Tynan SH, Valtee R, Doxsey SJ. J Cell Biol. 1999;147:481–491. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]