Abstract
The factors necessary to maintain organ-specific progenitor cells are poorly understood and yet of extreme clinical importance. Here, we identify the transcription factor SOX9 as the first specific marker and maintenance factor of multipotential progenitors during pancreas organogenesis. In the developing pancreas, SOX9 expression is restricted to a mitotically active, Notch-responsive subset of PDX1+ pluripotent progenitors and is absent from committed endocrine precursors or differentiated cells. Similar to Notch mutations, organ-specific Sox9 inactivation in mice causes severe pancreatic hypoplasia resulting from depletion of the progenitor cell pool. We show that Sox9 maintains pancreatic progenitors by stimulating their proliferation, survival, and persistence in an undifferentiated state. Our finding that SOX9 regulates the Notch-effector HES1 suggests a Notch-dependent mechanism and establishes a possible genetic link between SOX factors and Notch. These findings will be of major significance for the development of in vitro protocols for cell replacement therapies.
Keywords: development, Hes1, Notch, Pdx1
Although the mechanisms of cell regeneration in the adult pancreas are still the subject of debate, neogenesis from a common pool of progenitor cells is the predominant mechanism of cell formation in the embryonic vertebrate pancreas. Progenitors in the early pancreatic anlagen are marked by the transcription factor PDX1 and provide the source for all differentiated pancreatic cells: the exocrine acinar and ductal cells and the four endocrine cell types of the islets of Langerhans, which include the insulin-producing beta cells (1). Those progenitor cells committed to an endocrine fate can be further identified by their expression of the transcription factor NGN3. Cell–cell signaling via the Notch pathway is required for maintaining cells in the progenitor state, in part by blocking the expression of Ngn3 and hence endocrine cell differentiation (2–4). Much progress has been made in the dissection of the transcriptional hierarchy governing pancreatic cell differentiation (5), but the cell-intrinsic determinants of progenitor cell maintenance remain largely elusive. Such knowledge, however, is vital for implementing cell-based therapies for diabetes because they require the expansion of tissue-specific precursors in vitro.
Members of the SOX transcription factor family have been implicated in maintaining cells in a stem cell-like state and inhibiting cell differentiation. In the CNS, Sox1, Sox2, and Sox3 universally mark neural stem/progenitor cells and prevent their exit from the cell cycle and the induction of neurogenesis (6, 7). Sox9 plays a similar role in stem/progenitor cells of the hair bulge, intestinal epithelium, and neural crest (8–10). Based on its expression in the emerging pancreatic rudiments, we identified Sox9 as a candidate Sox gene for pancreatic progenitors (11). Here, we show that SOX9 marks undifferentiated, pluripotent pancreatic progenitors, but it is excluded from lineage-committed progenitors or differentiated cells throughout organogenesis. Through pancreas-specific inactivation of Sox9 in mouse embryos, we show that Sox9 controls the maintenance of pluripotent progenitors by stimulating their proliferation and survival. SOX9-deficient progenitors have reduced expression of the Notch target HES1, thus establishing a possible link between SOX9 and the main conserved signal transduction pathway of stem cell maintenance.
Results
SOX9 Marks Pluripotent, Notch-Responsive Pancreatic Progenitors.
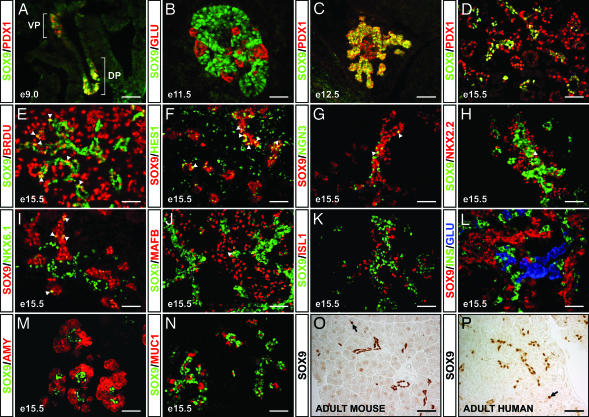
Using coimmunofluorescence, we characterized the domain(s) of SOX9 expression with respect to markers for pancreatic progenitors and differentiated cell types. At embryonic day (E) 9.0, SOX9 colocalized with PDX1 within the region of the gut endoderm that demarcates the future dorsal and ventral pancreatic buds (Fig. 1A). Almost complete overlap between the SOX9 and PDX1 domains persisted until E12.5 (Fig. 1C), thus demonstrating that SOX9 is expressed in pluripotent pancreatic progenitor cells. With the onset of major cell differentiation in the pancreas after E14, SOX9 became restricted to a subpopulation of PDX1+ cells (Fig. 1D). Although the PDX1+ domain includes newly differentiated endocrine and acinar cells (ref. 12; Fig. 1D), SOX9+ cells were confined to the centrally located epithelial cords, from which all pancreatic cell types are thought to arise (13). Consistent with the idea that SOX9 marks progenitor cells, 37.7 ± 2.5% of SOX9+ cells incorporated the mitotic marker BrdU (Fig. 1E).
Fig. 1.
SOX9 expression is restricted to uncommitted pancreatic progenitor cells. At E9.0, SOX9 colocalizes with PDX1 in the prepancreatic endoderm (A), persists in the PDX1+ pancreatic progenitors at E12.5 (C), and by E15.5 becomes restricted to a core subset of PDX1+ epithelial cords (D). At E15.5, ≈40% of SOX9+ cells incorporate the mitotic marker BrdU (E, arrowheads) and coexpress HES1 (F, arrowheads). Committed NGN3+ endocrine progenitors are intercalated within the SOX9+ epithelial cords but rarely express SOX9 (G, arrowheads). SOX9 rarely colocalizes with the endocrine differentiation factors NKX2.2 (H, arrowhead), NKX6.1 (I, arrowheads), and MAFB (J, arrowhead), and it is similarly excluded from differentiated endocrine cells expressing ISL1 (K), insulin or glucagon (B and L), differentiated amylase+ acinar cells (M), and MUC1+ ductal cells (N). In the adult, SOX9 expression is restricted to a subset of ductal epithelial and centroacinar (arrows in O and P) cells in both mouse (O) and human (P). VP, ventral; DP, dorsal prepancreatic endoderm; BRDU, bromodeoxyuridine; GLU, glucagon; INS, insulin; AMY, amylase; MUC1, mucin-1; e, embryonic day. [Scale bar, 50 μm (A, B, D–P); 100 μm (C).]
In the pancreas, Notch-mediated signaling is transduced through the intracellular Notch effector HES1 (3). We observed colocalization of SOX9 and HES1 in 38.3 ± 2.0% of SOX9+ cells (Fig. 1F) but almost no coexpression of SOX9 and NGN3 at E15.5 (Fig. 1G), therefore suggesting that SOX9 is enriched in Notch signal-transducing cells and absent from committed endocrine progenitors. Accordingly, SOX9 rarely colocalized with the endocrine differentiation factors NKX2.2, NKX6.1, or MAFB and was not coexpressed with ISL1 or endocrine hormones (Fig. 1 B and H–L). SOX9 was similarly excluded from amylase+ acini (Fig. 1M) and from the MUC1+ ductal epithelium (Fig. 1N). From E16.5 on, SOX9 expression was substantially down-regulated, and a greater divergence was observed in the spatial domains of SOX9 and PDX1. Although PDX1 becomes postnatally restricted to the islet beta and delta cells (12), SOX9 remained exclusive to a small subset of the ductal epithelial cells, including the centroacinar cells (Fig. 1O). In contrast to SOX9 protein, we previously reported detection of Sox9 transcripts in isolated adult mouse islets by using degenerate primers (11). Because we failed to amplify Sox9 mRNA from islets in subsequent analyses with intron-spanning Sox9-specific primers (data not shown), we conclude that residual genomic DNA or ductal cell contamination in the islet preparation led to a false-positive result. Significantly, we observed the same scattered ductal expression of SOX9 as seen in mice in the adult human pancreas (Fig. 1P). The expression in the adult pancreas is particularly intriguing because the adult pancreatic ductal epithelium has long been speculated to function as a reservoir of pancreatic progenitors (14).
SOX9 Regulation Confirms Its Role as a Progenitor Cell Marker.
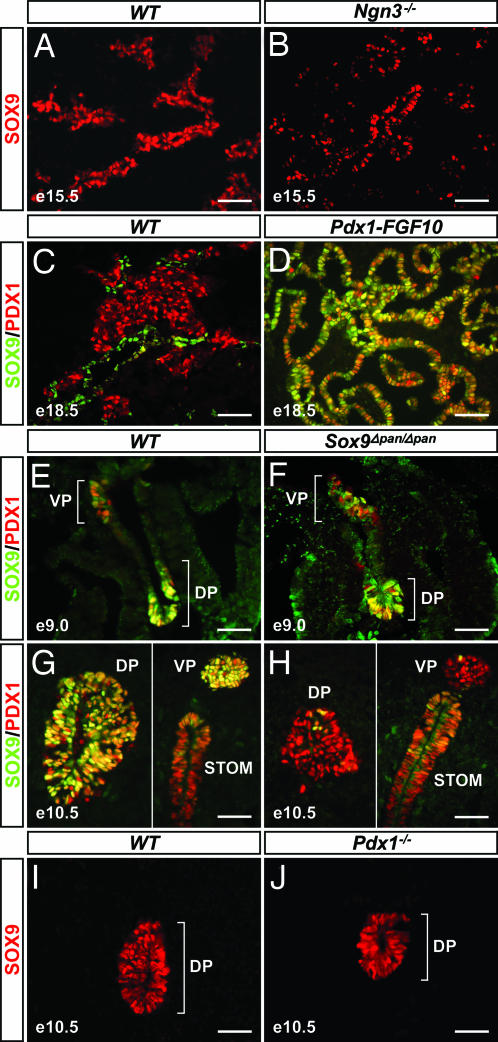
We reasoned that as a progenitor cell marker, Sox9 should not be a downstream target of genes that control endocrine differentiation or maturation, such as Ngn3 and Nkx6.1 (15, 16), and therefore we tested whether SOX9 is maintained in pancreata from Ngn3- and Nkx6.1-nullizygous embryos. As predicted, there was no difference in the number (wild-type, 334 ± 19 SOX9+ cells per field, n = 10, vs. Ngn3−/−, 318 ± 31, n = 6; P = 0.667 or vs. Nkx6.1−/−, 320 ± 46, n = 6; P = 0.790) or spatial distribution of SOX9+ cells between Ngn3−/− or Nkx6.1−/− and wild-type embryos [Fig. 2 A and B and supporting information (SI) Fig. 6]. Similarly, one would expect SOX9 to be maintained when differentiation is blocked and cells are arrested in the progenitor state. We therefore assayed for SOX9 in the pancreatic epithelium of Pdx1-FGF10 mice, in which ectopic FGF10 expression under control of the Pdx1 promoter completely abrogates pancreatic cell differentiation (17). We observed strong SOX9 expression throughout the tubular network of undifferentiated PDX1+ epithelial cells in Pdx1-FGF10 embryos at E18.5 (Fig. 2D), a time point at which PDX1 is normally confined to the endocrine compartment, whereas SOX9 persists in a subset of cells within the epithelial cords (Fig. 2C). Collectively, its spatiotemporal expression and genetic regulation suggest that SOX9 is a marker for uncommitted, pluripotent pancreatic progenitors.
Fig. 2.
Genetic regulation of SOX9 is consistent with a role as a pancreatic progenitor cell marker. The pattern of pancreatic SOX9 expression is identical in wild-type (A) and Ngn3-nullizygous embryos (B) at E15.5. Although SOX9 is restricted to a small subset of cells within the epithelial cords of E18.5 wild-type pancreas (C), SOX9 is maintained throughout the tubular network of undifferentiated PDX1+ epithelial cells in embryos that express an FGF10 transgene under the control of the Pdx1 promoter (Pdx1-FGF10) (D). In Sox9flox/flox;Pdx1-Cre (Sox9Δpan/Δpan) mice, SOX9 is robustly expressed throughout the PDX1+ prepancreatic endoderm at E9.0 (E and F). By E10.5, Pdx1-Cre has efficiently eliminated SOX9 from >95% of PDX1+ cells (G and H). PDX1 expression is maintained in the SOX9-deficient pancreatic epithelium (H). Likewise, SOX9 is expressed in pancreatic rudiments from PDX1-deficient embryos (J). DP, dorsal pancreas; VP, ventral pancreas; STOM, stomach; e, embryonic day. (Scale bar, 50 μm.)
SOX9-Deficient Progenitors Have Decreased Capacity to Contribute to Pancreas.
To study Sox9 function during pancreas development, we analyzed mice in which Sox9 was selectively deleted in pancreatic progenitors. Because neonatal lethality of heterozygous Sox9+/− mutant mice precluded the generation of SOX9-deficient embryos (18), we generated a pancreas-specific deletion through Pdx1-Cre-mediated recombination of the Sox9flox allele (1, 19). At E9.0, SOX9 was still detected throughout the dorsal and ventral PDX1+ prepancreatic endoderm of Sox9flox/flox;Pdx1-Cre/+ (Sox9Δpan/Δpan) embryos (Fig. 2F), thus indicating that inactivation of Sox9 occurs after endodermal progenitors have acquired a pancreatic fate (5). By E10.5, however, SOX9 was no longer detected in >95% of PDX1+ cells of the dorsal and ventral pancreatic buds (Fig. 2H). Interestingly, the robust expression of PDX1 in SOX9-deficient pancreatic epithelium (Fig. 2H) demonstrates that Sox9 is not required to maintain expression of Pdx1. To explore a possible reciprocal requirement for PDX1 in pancreatic Sox9 induction or maintenance, we examined whether PDX1 deficiency affects SOX9 expression. Because we observed no difference in the pattern and intensity of pancreatic SOX9 between Pdx1−/− and wild-type embryos (Fig. 2 I and J), we conclude that despite their coexpression in pluripotent pancreatic progenitors, PDX1 and SOX9 do not regulate each other.
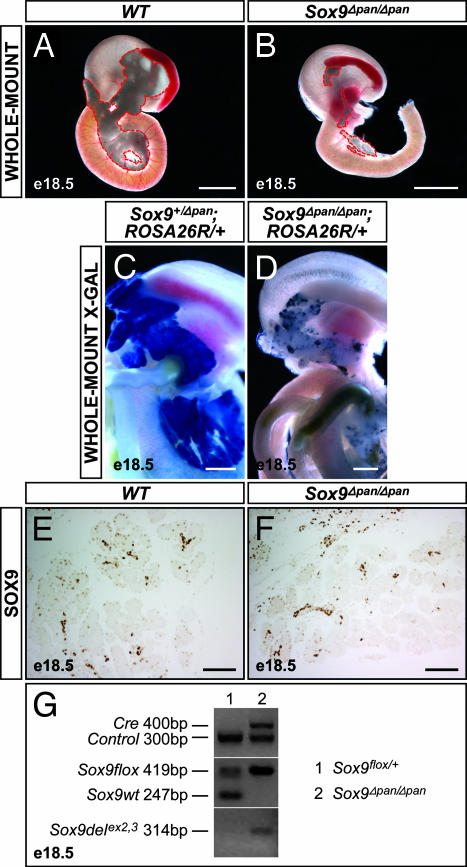
Shortly after birth, Sox9Δpan/Δpan pups manifested growth retardation and dehydration as well as dramatically elevated blood glucose levels (data not shown). All pups died within the first 4 days of life. In all cases where at least one wild-type allele for Sox9 was present, the pancreas was normal in appearance and weight (Fig. 3A and data not shown). By contrast, Sox9Δpan/Δpan embryos displayed a reduction of the pancreas to stunted rudiments in both the splenic and duodenal regions, indicating that the development of tissue from both pancreatic buds is abrogated (Fig. 3B). Although always reduced compared with wild-type pancreas, the degree of size reduction varied between individual mutant embryos. To test whether the rudimentary pancreatic tissue arose from a pool of cells that failed to undergo Cre-mediated recombination, we generated mice that carry the Sox9flox/flox, Pdx1-Cre, and Gt(ROSA)26Sortm1Sor (ROSA26R) alleles. In these mice, Pdx1-driven Cre recombination both inactivates the floxed Sox9 alleles and activates the heritable expression of β-gal, allowing all recombined cells and their progeny to be traced by X-Gal staining. Using this approach, we found that compared with the early stages, when pancreatic epithelial recombination was almost complete in Sox9Δpan/Δpan embryos (Fig. 2H), β-gal− unrecombined cells were overrepresented at E18.5. Although uniform β-gal staining was detected in the entire pancreas in Sox9Δpan/+ and wild-type backgrounds (Fig. 3C), the pancreatic remnant in Sox9Δpan/Δpan mice comprised a mosaic of β-gal+ recombined cells and β-gal− unrecombined cells (Fig. 3D). The percent contribution of unrecombined cells correlated with the size of the pancreatic remnants. These results indicate a partial repopulation of the Sox9Δpan/Δpan pancreas by presumably unrecombined SOX9+ progenitor cells. Consistent with this notion, we observed substantial numbers of SOX9+ cells in those embryos with relatively large remnants (Fig. 3 E and F). Furthermore, we detected both the unrecombined and the recombined Sox9 alleles in E18.5 Sox9Δpan/Δpan pancreatic rudiments (Fig. 3G). We conclude that SOX9-deficient cells are disadvantaged in their capacity to contribute to the forming pancreas, thus bestowing a selective advantage on progenitors that failed to undergo Cre-mediated recombination.
Fig. 3.
Decreased contribution of SOX9-deficient progenitors to cell neogenesis results in pancreatic hypoplasia. Conditional Sox9 deletion with a Pdx1-Cre transgene results in pancreatic hypoplasia at E18.5 (A and B; pancreas outlined by a red dashed line). Using the ROSA26R allele, progeny of cells that underwent Pdx1-Cre-mediated recombination are identified by X-Gal staining (C and D). Uniform blue X-Gal staining in Sox9flox/+;Pdx1-Cre;ROSA26R/+ (Sox9+/Δpan;ROSA26R/+) embryos at E18.5 shows that all pancreatic cells have arisen from Pdx1-Cre-expressing progenitors (C). By contrast, the pancreatic remnant of Sox9Δpan/Δpan;ROSA26R/+ littermates comprises a variable mosaic of β-gal+ recombined cells and β-gal− unrecombined cells (D), indicating that cells that failed to undergo recombination have a selective advantage to contribute to the pancreas. Consistent with the presence of unrecombined cells in Sox9Δpan/Δpan pancreas at E18.5, immunohistochemical staining reveals substantial numbers of SOX9-immunopositive pancreas cells in Sox9Δpan/Δpan embryos bearing relatively large remnants (E and F). In concordance, PCR of genomic DNA from E18.5 pancreatic rudiments of Sox9Δpan/Δpan embryos detects the Pdx1-Cre transgene, the unrecombined Sox9flox allele, as well as the recombined, deleted Sox9 allele, revealing only partial Cre-mediated recombination (G). In Sox9flox/+ controls that do not carry the Pdx1-Cre transgene, the wild-type Sox9 and floxed Sox9 alleles are amplified, but no deleted Sox9 allele is detected. e, embryonic day. [Scale bar, 200 μm (A and B); 100 μm (C–F).]
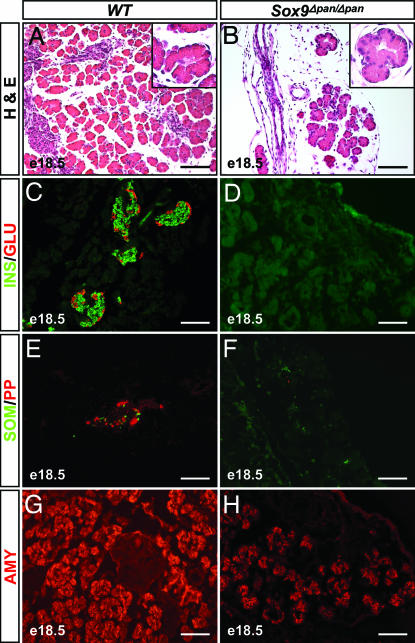
Histological examination of the pancreatic rudiments revealed that they comprised only a primitive ductular tree supplying isolated clusters of acini (Fig. 4B), of which some displayed a reduced cytoplasmic/nuclear ratio compared with wild-type (Fig. 4 A and B Insets). The majority of pancreatic acini in the Sox9Δpan/Δpan embryos exhibited consistently reduced intensity levels of amylase immunostaining (Fig. 4H). Throughout the Sox9Δpan/Δpan pancreatic rudiments we also observed abundant fibrous tissue as well as epithelial cysts of variant sizes (Fig. 4B), a phenotype reminiscent of the pancreatic to intestinal transformation seen in Pdx1-Shh mice (20). However, because we failed to detect intestinal markers in the epithelial cysts of Sox9Δpan/Δpan embryos (data not shown), they are unlikely to represent transformed intestinal cells. Consistent with the absence of recognizable islets (Fig. 4B), we did not detect any immunostaining for the four endocrine hormones in more than half of the examined Sox9Δpan/Δpan pancreatic rudiments (n = 4; Fig. 4 D and F). Isolated cells for each of the four hormones were occasionally found, but they never exceeded a total of five endocrine cells per section, and they could have arisen from progenitors that escaped Cre-mediated recombination. Together, these results demonstrate that pancreatic growth and cell differentiation require SOX9 activity.
Fig. 4.
Pancreatic growth and cell differentiation require SOX9 activity. Hematoxylin/eosin (H&E) staining of pancreatic sections from E18.5 embryos (A and B) shows that pancreatic rudiments from Sox9flox/flox;Pdx1-Cre (Sox9Δpan/Δpan) embryos comprise predominantly fibrous tissue and epithelial cysts surrounding isolated clusters of acini, some of which show densely packed nuclei (B Inset). Sox9Δpan/Δpan pancreatic rudiments display an almost complete absence of insulin+, glucagon+, somatostatin+, or pancreatic polypeptide+ endocrine cells (D and F) and scattered acinar cells that are weakly amylase+ (H). INS, insulin; GLU, glucagon; SOM, somatostatin; PP, pancreatic polypeptide; AMY, amylase; e, embryonic day. (Scale bar, 100 μm.)
SOX9 Stimulates Proliferation and Survival of Pluripotent Progenitors.
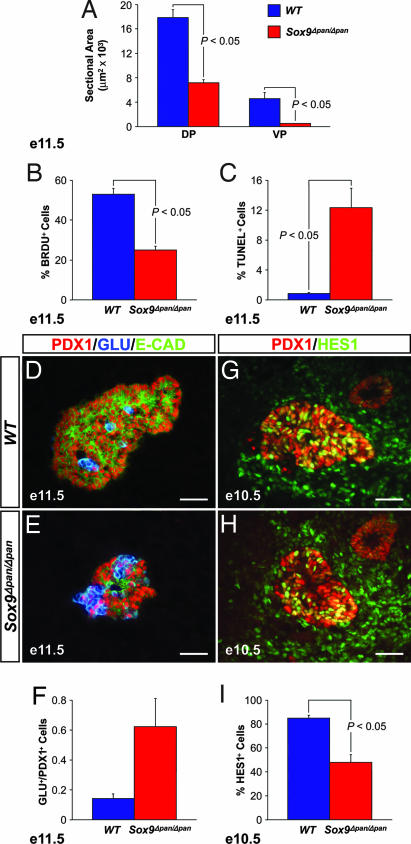
Using morphometry for the pancreatic epithelial area, we observed completely penetrant hypoplasia of both pancreatic buds as early as E11.5 (Fig. 5A). A slight size reduction was already detected at E10.5, which coincides with the earliest time point at which complete Pdx1-Cre-mediated recombination was observed (Fig. 2H). These findings indicate a requirement for Sox9 in pancreatic growth before the onset of major cell differentiation.
Fig. 5.
Sox9 ensures maintenance of pancreatic progenitors through regulation of the Notch-effector HES1. At E11.5, Sox9flox/flox;Pdx1-Cre (Sox9Δpan/Δpan) embryos exhibit a significantly smaller mean sectional area of both dorsal and ventral pancreatic buds than wild-type littermates (A). The SOX9-deficient pancreatic epithelium shows reduced numbers of BrdU-incorporating cells among the PDX1+ pancreatic progenitor pool at E11.5 (B) and an increased number of TUNEL+ cells per total epithelial cells (C). At E11.5, the ratio of glucagon+ endocrine cells to the total number of PDX1+ progenitors is 4.4-fold increased in the pancreatic epithelium of Sox9Δpan/Δpan compared with wild-type embryos (D–F). SOX9 deficiency is associated with decreased HES1 staining within the PDX1+ progenitor pool at E10.5 (G–I). (A–C) n = 3. (F and I) n = 4. DP, dorsal pancreas; VP, ventral pancreas; GLU, glucagon; E-CAD, E-cadherin; e, embryonic day. (Scale bar, 50 μm.)
When examining the mechanism(s) responsible for the pancreatic hypoplasia, we considered three main possibilities: (i) a reduction in pancreatic progenitor cell proliferation; (ii) an increased incidence of apoptosis; and (iii) precocious cell cycle exit and differentiation of pancreatic progenitors, all three leading to a depletion of the progenitor pool. Quantification of immunohistochemical markers in the ventral pancreatic bud of Sox9Δpan/Δpan embryos was difficult because of its severe size reduction, so we focused our analysis on the dorsal pancreas. First, we examined whether loss of Sox9 affects proliferation and/or survival of the PDX1+ progenitors by assaying for BrdU incorporation and apoptosis. We observed a 53% reduction in cell proliferation of the PDX1+ cell population as well as a 14-fold increase in the number of apoptotic cells in Sox9Δpan/Δpan embryos (Fig. 5 B and C and SI Fig. 7). Cell death appeared to affect mainly the progenitor cell pool because the domain of TUNEL+ cells coincided with the domain of PDX1+ cells and not with the domain of newly differentiated glucagon+ cells on adjacent sections (data not shown). Thus, in the absence of SOX9, both decreased proliferation and increased cell death contribute to a reduction of the PDX1+ progenitor cell pool.
Beginning at E9.5, glucagon-expressing cells are the first cells to differentiate (5). Given the markedly diminished size of the progenitor cell pool in Sox9Δpan/Δpan embryos, one would expect fewer glucagon+ cells to emerge from the undifferentiated epithelium. Contrary to this prediction, we observed a relative increase in the number of glucagon+ cells in the pancreatic buds from SOX9-deficient embryos at E11.5 (Fig. 5 D–F; the merged images are separated for clarity in SI Fig. 8 A–F and SI Table 1). Although wild-type embryos displayed an average ratio of glucagon+ to PDX1+ cells of 0.14 ± 0.06, this ratio was increased 4.4-fold in Sox9Δpan/Δpan embryos. We observed a similar relative increase in the number of cells expressing the pan-endocrine marker ISL1, but we rarely detected insulin+ cells (data not shown), which is consistent with their normal occurrence after E13 (5). In both wild-type and SOX9-deficient pancreatic epithelium, very few cells were negative for both PDX1 and glucagon, and conversely, few cells coexpressed PDX1 and glucagon (Fig. 5 D and E and SI Fig. 8 A–F). As in wild-type embryos, cell differentiation in Sox9Δpan/Δpan pancreas was associated with cell cycle exit, as inferred by an absence of BrdU incorporation by glucagon+ endocrine cells (data not shown), thus confirming that endocrine maturation occurs in the absence of SOX9. Together, these findings suggest a possible role for SOX9 in preventing precocious cell cycle exit and differentiation of progenitors.
SOX9 Regulates HES1 Expression.
The phenotypic alterations in the SOX9-deficient pancreas show a striking resemblance to the pancreatic defects associated with mutations in components of the Notch signaling pathway. Hes1−/− mutant embryos display a reduction in pancreas size resulting from depletion of the pancreatic progenitor cell pool because of reduced proliferation of PDX1+ progenitors and precocious differentiation into glucagon+ cells (3, 21). To test whether Sox9 functions as a modulator of Notch activity in pancreatic progenitors, we examined the expression of HES1 in Sox9Δpan/Δpan embryos. To avoid potential skewing of the results because of the relative increase in glucagon+ cells, we quantified the number of HES1+ cells as a percentage of the PDX1+ population at E10.5. We found that the proportion of the PDX1+ cells expressing HES1 was reduced by 43% in Sox9Δpan/Δpan embryos (Fig. 5 G–I; merged images are separated for clarity in SI Fig. 8 G–J), thus raising the possibility that Sox9 controls the activity of Notch signal transduction.
Discussion
The Notch pathway and Sox genes have been implicated as “molecular gatekeepers” of the pluripotent state in an evolutionarily conserved manner in many tissues (6, 8–10). This work demonstrates that SOX9 fulfills such a role in the embryonic pancreas by stimulating proliferation and preventing apoptosis of pluripotent progenitors. Similar functions for SOX9 have been found in the nervous system, hair bulge, testis, and notochord (10, 22–24). Our observation that HES1 expression is severely reduced in the absence of SOX9 activity raises the possibility that SOX9 controls pancreatic progenitor cell maintenance by modulating Notch signal transduction. Regulation of Notch signaling by SOX proteins has been previously demonstrated in neurogenesis, where SOX1–SOX3 maintain neural progenitors in an undifferentiated state by inducing the expression of Notch effectors and repressing NGN (6, 25). Consistent with the idea that a similar mechanism of SOX/Notch-mediated lateral inhibition may operate in the pancreas, SOX9/HES1-coexpressing cells showed an intercalated arrangement with NGN3+/SOX9-negative cells within a continuous cell layer of epithelial progenitors. However, although the effect on progenitor cell proliferation in SOX9-deficient pancreas mirrors the defects in Hes1 mutant embryos (21), there are also phenotypic differences that suggest Notch-independent functions of SOX9. Such Notch-independent functions of SOX9 could explain why despite only a 43% reduction of HES1+ cells, Sox9Δpan/Δpan mice show more severe pancreatic hypoplasia than do Hes1−/− mice. Notably, in contrast to Sox9Δpan/Δpan embryos, cell death was not increased in early pancreatic progenitors from Hes1-nullizygous embryos (3). Furthermore, although HES1-deficient embryos show an absolute increase in the number of glucagon+ cells in the pancreatic epithelium, we observed only a relative increase in the ratio of glucagon cell numbers to PDX1+ progenitors in SOX9-deficient pancreas. This difference could be a mere consequence of the delayed SOX9 inactivation by the Pdx1-Cre transgene compared with the null allele that was examined in the case of Hes1. However, it is also possible that SOX9 functions only in the maintenance of progenitors for mature pancreatic cells and that the “first wave” of endocrine differentiation in the early pancreas proceeds at a normal rate despite an overall reduction of the progenitor cell pool. Because similar genetic programs underlie the differentiation of the early and mature endocrine cells (15, 26), we consider it more likely that SOX9 functions to prevent premature differentiation of progenitors.
In support of a role for SOX9 as a marker for stem/progenitor cells, lineage-tracing studies have recently demonstrated that Sox9+ cells contribute to the formation of all cell types in a variety of tissues (22). The persistent expression of Sox9 in adult pancreatic ductal cells, specifically in centroacinar cells, raises the possibility that Sox9 continues to define a population of facultative stem/progenitor cells in adult pancreas. It has been hypothesized that centroacinar cells and ductal epithelial cells can transiently dedifferentiate and serve as multipotent progenitor cells, providing a capacity for a regenerative response to injury (14, 27). Future work will resolve whether Sox9 serves a role in progenitor cell maintenance of the adult pancreas similar to that demonstrated in the embryo.
Materials and Methods
Mice.
Sox9flox, Pdx1-Cre, Pdx1−/−, Ngn3−/−, Nkx6.1−/−, Pdx1-FGF10, and ROSA26R mice have been described previously (1, 15–17, 19, 28, 29). In Sox9flox × Pdx1-Cre crosses, age-matched Sox9flox/+ or Sox9flox/flox littermates without the Pdx1-Cre transgene were regarded as wild-type. For BrdU labeling, pregnant females were injected i.p. with 50 μg of BrdU per g of body weight, and embryos were harvested 45 min after injection.
Histology.
For histology and immunostaining, tissues were fixed, sectioned, and stained as described previously (30). The following primary antibodies were used: rabbit anti-SOX9, 1:2,000 (31); guinea pig anti-PDX1, 1:10,000 (provided by C. Wright, Vanderbilt University, Nashville, TN); guinea pig anti-insulin, 1:5,000 (Linco Research, St. Charles, MO); mouse anti-glucagon, 1:5,000 (Sigma, St. Louis, MO); rabbit anti-amylase, 1:500 (Sigma); mouse anti-BrdU, 1:200 (Chemicon, Temecula, CA); rabbit anti-HES1, 1:5,000 (provided by T. Sudo, Toray Industries, Inc., Tokyo, Japan); guinea pig anti-NGN3, 1:1,000 and guinea pig anti-NKX6.1, 1:1,000 (both ref. 32); mouse anti-ISL1, 1:20 [kindly provided by T. Jessell (Columbia University, New York, NY)/Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA]; Armenian hamster anti-MUC-1, 1:500 (Lab Vision Corporation, Fremont, CA); mouse anti-NKX2.2, 1:50 (T. Jessell/DSHB); goat anti-MAFB, 1:10,000 (33), and rat anti-E-cadherin, 1:2,000 (Sigma). X-Gal staining was performed as described previously (34).
PCR.
The allele-specific PCRs for the Sox9flox and the Sox9Δex2/3 alleles were performed as described previously (19).
More detailed descriptions of the methods are available in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to G. Gradwohl (INSERM U682, Strasbourg, France), D. A. Melton (Howard Hughes Medical Institute, Harvard University, Cambridge, MA), M. Wegner (Universität Erlangen, Germany), C. Wright, R. Stein (Vanderbilt University, Nashville, TN), T. Sudo, and T. Jessell for mice and antibodies; and R. MacDonald for technical advice. We thank Jeannie Chui for technical assistance and A. Lander and members of the M.S. laboratory for reading of the manuscript. This work was supported by the Juvenile Diabetes Research Foundation International Grants CDA 2-2001-728 (to M.S.) and 3-2004-608 (to P.A.S.), National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health Grant R01-DK06847 (to M.S.), and American Diabetes Association Grant 35306 (to M.S.).
Abbreviation
- E
embryonic day.
Footnotes
Author contributions: P.A.S. and M.S. designed research; P.A.S., K.K.F., M.N.T., and E.E.M. performed research; J.J., R.K., and G.S. contributed new reagents/analytic tools; P.A.S., K.K.F., M.N.T., and M.S. analyzed data; and P.A.S. and M.S. wrote the paper.
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains suppoting information online at www.pnas.org/cgi/content/full/0609217104/DC1.
References
- 1.Gu G, Dubauskaite J, Melton DA. Development (Cambridge, UK) 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 2.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 3.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 4.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen J. Dev Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- 6.Bylund M, Andersson E, Novitch BG, Muhr J. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 7.Graham V, Khudyakov J, Ellis P, Pevny L. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 8.Cheung M, Briscoe J. Development (Cambridge, UK) 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 9.Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidal VP, Chaboissier MC, Lutzkendorf S, Cotsarelis G, Mill P, Hui CC, Ortonne N, Ortonne JP, Schedl A. Curr Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 11.Lioubinski O, Muller M, Wegner M, Sander M. Dev Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 12.Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Development (Cambridge, UK) 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 13.Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma A, Zangen DH, Reitz P, Taneja M, Lissauer ME, Miller CP, Weir GC, Habener JF, Bonner-Weir S. Diabetes. 1999;48:507–513. doi: 10.2337/diabetes.48.3.507. [DOI] [PubMed] [Google Scholar]
- 15.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Proc Natl Acad Sci USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Development (Cambridge, UK) 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 17.Norgaard GA, Jensen JN, Jensen J. Dev Biol. 2003;264:323–338. doi: 10.1016/j.ydbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 19.Kist R, Schrewe H, Balling R, Scherer G. Genesis. 2002;32:121–123. doi: 10.1002/gene.10050. [DOI] [PubMed] [Google Scholar]
- 20.Apelqvist A, Ahlgren U, Edlund H. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- 21.Georgia S, Soliz R, Li M, Zhang P, Bhushan A. Dev Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 22.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Proc Natl Acad Sci USA. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Barrionuevo F, Taketo MM, Scherer G, Kispert A. Dev Biol. 2006;295:128–140. doi: 10.1016/j.ydbio.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 25.Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, Sikorska M. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Development (Cambridge, UK) 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 27.Jensen JN, Cameron E, Garay MV, Starkey TW, Gianani R, Jensen J. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. Development (Cambridge, UK) 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 29.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 30.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 31.Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henseleit KD, Nelson SB, Kuhlbrodt K, Hennings JC, Ericson J, Sander M. Development (Cambridge, UK) 2005;132:3139–3149. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- 33.Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 34.Seymour PA, Bennett WR, Slack JM. J Anat. 2004;204:103–116. doi: 10.1111/j.1469-7580.2004.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.