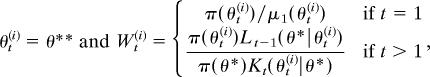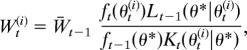Abstract
Recent new methods in Bayesian simulation have provided ways of evaluating posterior distributions in the presence of analytically or computationally intractable likelihood functions. Despite representing a substantial methodological advance, existing methods based on rejection sampling or Markov chain Monte Carlo can be highly inefficient and accordingly require far more iterations than may be practical to implement. Here we propose a sequential Monte Carlo sampler that convincingly overcomes these inefficiencies. We demonstrate its implementation through an epidemiological study of the transmission rate of tuberculosis.
Keywords: approximate Bayesian computation, Bayesian inference, importance sampling, intractable likelihoods, tuberculosis
Termed approximate Bayesian computation (ABC), recent new methods in Bayesian inference have provided ways of evaluating posterior distributions when the likelihood function is analytically or computationally intractable (1–4). ABC algorithms represent a substantial methodological advance because they now admit realistic inference on problems that were considered intractable only a few years ago. The rapidly increasing use of these methods has found application in a diverse range of fields, including molecular genetics (5), ecology (6), epidemiology (7), evolutionary biology (8, 9) and extreme value theory (1).
Given a likelihood function, f(x0|θ), and a prior distribution π(θ) on the parameter space, Θ, interest is in the posterior distribution f(θ|x0) ∝ f(x0|θ)π(θ), the probability distribution of the parameters having observed the data, x0 (10, 11).
To avoid directly evaluating the likelihood, all ABC algorithms incorporate the following procedure to obtain a random sample from the posterior distribution. For a candidate parameter vector θ* drawn from some density, a simulated data set x* is generated from the likelihood function f(x|θ*) conditioned on θ*. This vector is then accepted if simulated and observed data are sufficiently “close.” Here, closeness is achieved if a vector of summary statistics S(·) calculated for the simulated and observed data are within a fixed tolerance (ϵ) of each other according to a distance function ρ (e.g., Euclidean distance). In this manner, ABC methods sample from the joint distribution f(θ, x|ρ(S(x), S(x0)) ≤ ϵ), where interest is usually in the marginal f(θ|ρ(S(x), S(x0)) ≤ ϵ). The algorithms work by accepting a value θ with an average probability of Pr(ρ(S(x), S(x0)) ≤ ϵ|θ). If the summary statistics S(·) are near-sufficient and ϵ is small then f(θ|ρ(S(x), S(x0)) ≤ ϵ) should be a reasonable approximation to f(θ|x0).
Existing ABC methods for obtaining samples from the posterior distribution either involve rejection sampling (3, 4, 12) or Markov chain Monte Carlo (MCMC) simulation (1, 2). Both of these classes of methods can be inefficient. The ABC rejection sampler proceeds as follows
ABC-REJ Algorithm
REJ1. Generate a candidate value θ* ∼ π(θ) from the prior.
REJ2. Generate a data set x* ∼ f(x|θ*).
REJ3. Accept θ* if ρ(S(x*), S(x0)) ≤ ϵ.
REJ4. If rejected, go to REJ1.
Each accepted vector represents an independent draw from f(θ|ρ(S(x), S(x0)) ≤ ϵ). Acceptance rates for algorithm ABC-REJ can be very low as candidate parameter vectors are generated from the prior π(θ), which may be diffuse with respect to the posterior. Accordingly, Marjoram et al. (2) proposed to embed the likelihood-free simulation method within the well known MCMC framework. This algorithm proceeds as follows
ABC-MCMC Algorithm
MC1. Initialize θ1, i = 1.
MC2. Generate a candidate value θ* ∼ q(θ|θi), where q is some proposal density.
MC3. Generate a data set x* ∼ f(x|θ*).
MC5. If i < N, increment i = i + 1 and go to MC2.
Here 1(A) = 1 if A is true, and 0 otherwise. The candidate vector is generated from an arbitrary proposal density q(θ|·) and accepted with the usual Metropolis–Hastings acceptance probability. The (intractable) likelihood ratio is now coarsely approximated by 1 if simulated and observed data are sufficiently “close,” and 0 otherwise. Algorithm ABC-MCMC generates a sequence of serially and highly correlated samples from f(θ|ρ(S(x), S(x0)) ≤ ϵ). Determination of the chain length, N, is therefore obtained through a careful assessment of convergence (13) and consideration of the chain's ability to explore parameter space (i.e., chain mixing).
When the prior and posterior are dissimilar, algorithm ABC-MCMC delivers substantial increases in acceptance rates over algorithm ABC-REJ [Marjoram et al. (2) report 0.2% acceptance rates over 0.0008% in a simple coalescent tree analysis], although at the price of generating dependent samples. However, because acceptance rates for ABC samplers are directly proportional to the likelihood; if the ABC-MCMC sampler enters an area of relatively low probability with a poor proposal mechanism, the efficiency of the algorithm is strongly reduced because it then becomes difficult to move anywhere with a reasonable chance of acceptance, and so the sampler “sticks” in that part of the state space for long periods of time. This is illustrated in the following toy example.
Toy Example
As an illustration, suppose that the posterior of interest is given by the mixture of two normal distributions
where φ(μ, σ2) is the density function of a N(μ, σ2) random variable. Here, the second component implies large regions of relatively low probability with respect to the lower variance first component (Figure 1Lower). In the ABC setting, this posterior may be realized by drawing x = (x1,…, x100), xi ∼ N(θ, 1) and by specifying
where x̄ = 1/100 Σ xi denotes the sample mean.
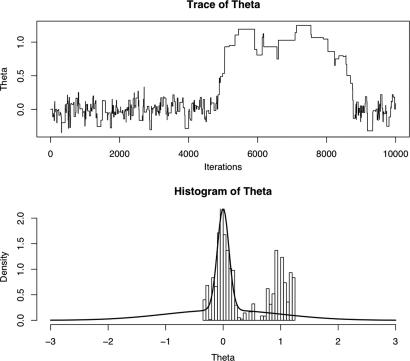
Fig. 1.
Trace and histogram of θ. (Upper) Trace of 10,000 ABC-MCMC sampler iterations. (Lower) Target mixture distribution (solid line) and histogram of ABC-MCMC sampler output.
With a prior of π(θ) ∼ U(−10, 10) and ϵ = 0.025, the ABC-REJ algorithm required a mean of 400.806 data-generation steps (simulations from the likelihood) for each accepted realization based on 1,000 accepted realizations. With an acceptance rate of <0.25%, this is highly inefficient.
In contrast, Fig. 1 shows the result of implementing the ABC-MCMC algorithm initialized at θ0 = 0, with N = 10,000 iterations and with proposals generated via the random walk q(θ|θt) ∼ N(θt, 0.152). When the sampler is within the high-density region, transitions between different parameter values are frequent (acceptance rate ≈5%). However, when the sampler moves outside of this region, the frequency of transitions drops markedly, especially so for the extended period at ≈5,000–9,000 iterations. Of course, the samples should visit the distributional tails and will do so for a number of iterations proportional to the posterior probability. However, as is evident from the histogram of the sampler output, the number of further iterations required so that the realizations in the tail are in proportion to the target distribution will be far in excess of the initial 10,000.
In the search for improved ABC methods, poor Markov chain performance may be improved by embedding the target distribution as a marginal distribution within a larger family of related distributions among which it is far easier for the sampler to move around. This was essentially the approach adapted for ABC by Bortot et al. (1), although such algorithms are wasteful by construction in that samples from the auxiliary distributions are not used in the final analysis.
As an alternative, we propose to improve upon simple rejection sampling by adopting a sequential Monte Carlo (SMC)-based simulation approach. Here, a full population of parameters θ(1),…, θ(N) (termed particles) is propagated from an initial, user-specified distribution, through a sequence of intermediary distributions, until it ultimately represents a sample from the target distribution. SMC methods can be considered an extension of importance sampling. We will demonstrate that the SMC approach can substantially outperform both MCMC and rejection sampling in the ABC framework.
Advantages of the SMC approach are
Like rejection sampling, the sampler will never become “stuck” in regions of low probability.
Unlike rejection sampling, severe inefficiencies generated by mismatch of (initial) sampling and target distributions are avoided.
Particles that represent the target posterior poorly are eliminated in favor of those particles that represent the posterior well.
The population-based nature of the sampler means that complex (e.g., multimodal) posteriors may be explored more efficiently.
Samples are obtained from a number of distributions with differing tolerances (ϵ). This permits an a posteriori, or dynamic, examination of the robustness of the posterior to this choice.
Unlike MCMC, SMC particles are uncorrelated and do not require the determination of a burn-in period or assessment of convergence.
Disadvantages of the SMC approach are considered in Discussion.
Here, we propose an ABC sampler based on SMC simulation. We initially outline a generic SMC algorithm before exploiting ideas based on PRC to derive a more efficient algorithm for the ABC setting. Finally, we demonstrate its utility with regard to the toy example considered above and in a reexamination of a previously implemented analysis of the transmission rate of tuberculosis.
Methods
SMC Without Likelihoods.
We wish to sample N particles θ(1),…, θ(N) from the posterior f(θ|ρ(S(x), S(x0)) ≤ ϵ), for observed data x0, and for unknown parameter vector θ ∈ Θ. We assume that the summary statistics S(·), the tolerance ϵ, and the distance function ρ are known.
Let θ 1(1),…, θ1(N) ∼ μ1(θ) be an initial population from a known density, μ1, from which direct sampling is possible, and by fT(θ) = f(θ|ρ(S(x), S(x0)) ≤ ϵ) the target distribution (this notation will become clear shortly). Standard importance sampling would then indicate how well each particle θ1(i) adheres to fT(θ) by specifying the importance weight, WT(i) = fT(θ 1(i))/μ1(θ1(i)), it should receive in the full population of N particles. The effectiveness of importance sampling is sensitive to the choice of sampling distribution, μ1. The prior π(θ) is often used for this purpose. Importance sampling can be highly inefficient if μ1 is diffuse relative to fT and can fail completely in the case of sampling and target distribution mismatch.
The idea behind sequential sampling methods is to avoid the potential disparity between μ1 and fT by specifying a sequence of intermediary distributions f1,…, fT−1, such that they evolve gradually from the initial distribution towards the target distribution. For example, one can choose a geometric path specification where ft(θ) = fT(θ)φt μ1(θ)1−φt with 0 < φ1 < … < φT = 1 (14, 15). Hence, it is possible to move smoothly and effectively in sequence from μ1 to fT using repeated importance sampling, generating a series of particle populations {θt(i)} = {θt(i) : i = 1,…, N}, for t = 1, … T. That is, sequential methods proceed by moving and reweighting the particles according to how well they adhere to each successive distribution, ft.
In the ABC setting, we may naturally define the sequence of distributions f1…, fT as
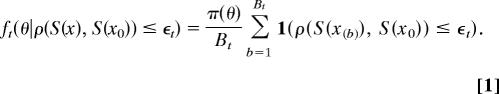 |
Here, x(1),…, x(Bt) are Bt data sets generated under a fixed parameter vector, x(b) ∼ f(x|θ), and {ϵt} is a strictly decreasing sequence of tolerances. The nested family of distributions generated by varying ϵ (continuously) was considered by Bortot et al. (1) in their augmented state space ABC algorithm. By specifying ϵt < ϵt−1, we ensure that the likely range of parameter values in each progressive distribution is a subset of the one it precedes. This is a desirable property for our sampling distributions. By specifying ϵT = ϵ, we realize the final particle population {θT(i)} as a weighted sample from the target distribution. Setting Bt = B and ϵt = ϵ for all t reduces to the likelihood specified by Marjoram et al. (2), and further B = 1 to the likelihood adopted in algorithm ABC-MCMC. The target distribution, fT, is specified by ϵT = ϵ.
PRC.
In ABC samplers, Bt = 1 is the most computationally efficient specification, in that some action occurs (e.g., a realization or move proposal is accepted) each time a nonzero likelihood is generated. However, because the particle weight under SMC methods is directly proportional to the likelihood, there is a large probability that the likelihood, and therefore the particle weight, will be zero, thereby rendering the particle useless. Fortunately, the idea of PRC (see chapters 2 and 3 of ref. 16) permits the repeated resampling (and moving) of particles from the previous population to replace those particles with zero weight. PRC continues until N particles with nonzero weight are obtained. See Appendix for further details.
At each step, SMC methods move each particle according to a Markov kernel Kt to improve particle dispersion. This induces a particle approximation to the importance sampling distribution μt(θt) = ∫Θ μt−1(θt−1)Kt(θt|θt−1)dθt−1, for populations t = 2,…, T. Choices of Kt include a standard smoothing kernel (e.g., Gaussian) or a Metropolis–Hastings accept/reject step. The kernel accordingly enters the particle weight calculation.
A recent innovation in SMC methods has been the introduction of a backward Markov kernel Lt−1 with density Lt−1(θt−1|θt) into the weight calculation (17). The backward kernel relates to a time-reversed SMC sampler with the same target marginal distribution as the (forward-time) SMC sampler induced by Kt. Because only specification of Kt is required in order to implement an SMC sampler, the backward kernel is essentially arbitrary. The kernel Lt−1 may therefore be optimized to minimize the variance of the weights induced by the importance sampling distribution μt (through Kt). This is difficult in general, however, so simplified forms are often adopted. See ref. 17 for further discussion.
SMC algorithms measure the degree of sample degeneracy within each population through the effective sample size (ESS). ESS calculates the equivalent number of random samples required to obtain an estimate, such that its Monte Carlo variation is equal to that of the N weighted particles. This may be estimated as 1 ≤ [Σi=1N (Wt(i))2]−1 ≤ N for each t (16, 18). Sample degeneracy can occur through sampling and target distribution mismatch when a small number of particles have very large weights. Through a resampling step, particles with larger weights become better represented in the resampled population than those with smaller weights. Those particles with sufficiently small weights, which poorly approximate ft, may be eliminated. The resampling threshold, E, is commonly taken to be N/2.
Combining PRC with SMC, we obtain the following (ABC-PRC) algorithm
ABC-PRC Algorithm
-
PRC1. Initialize ϵ1,…, ϵT, and specify initial sampling distribution μ1.
Set population indicator t = 1.
PRC2. Set particle indicator i = 1.
-
PRC2.1. If t = 1, sample θ** ∼ μ1(θ) independently from μ1.
If t > 1, sample θ* from the previous population {θt−1(i)} with weights {Wt−1(i)}, and perturb the particle to θ** ∼
-
Kt(θ|θ*) according to a Markov transition kernel Kt. Generate a data set x** ∼ f(x|θ**).
If ρ(S(x**), S(x0)) ≥ ϵt, then go to PRC2.1.
If i < N, increment i = i + 1 and go to PRC2.1.
-
PRC3. Normalize the weights so that Σi=1N Wt(i) = 1.
If ESS = [Σi=1N(Wt(i))2]−1 < E then resample with replacement, the particles {θt(i)} with weights {Wt(i)} to obtain a new population {θt(i)}, and set weights {Wt(i) = 1/N}.
PRC4. If t < T, increment t = t + 1 and go to PRC2.
Samples {θT(i)} are weighted samples from the posterior distribution f(θ|ρ(S(x), S(x0)) ≤ ϵ). The validity of this algorithm is derived by construction from the validity of the combination of general SMC methods and the PRC process (see Appendix). Algorithm ABC-PRC corresponds to algorithm ABC-REJ for the special case when T = 1 and μ1(θ) = π(θ).
For the remainder of this article, we consider Kt(θt|θt−1) = Lt−1(θt−1|θt) as a Gaussian kernel with common variance (following ref. 19), which we have found to perform adequately. For discussions on closer to optimal choices of Lt−1, see ref. 17, and for applications of SMC and more technical proofs of the SMC algorithm's validity, see refs. 16, 17, and 20–24. Finally, we note that if Kt(θt|θt−1) = Lt−1(θt−1|θt), μ1(θ) = π(θ) and the prior π(θ) ∝ 1 over Θ, then all weights are equal throughout the sampling process and may safely be ignored [in addition to ignoring all population (PRC3) resampling steps].
Results
Toy Example (Revisited).
We now implement algorithm ABC-PRC in the mixture of normals posterior considered earlier. We generate a sample of N = 1,000 particles by considering a sequence of three distributions f1, f2, and f3 defined by Eq. 1 with ϵ1 = 2, ϵ2 = 0.5 and ϵ3 = 0.025, and with prior distribution π(θ) ∼ U(−10, 10). We specify μ1(θ) = π(θ) and Kt(θt|θt−1) = Lt−1(θt−1|θt) as a Gaussian random walk so that all weights are equal.
The initial (μ1) population and histograms of f1 to f3 are illustrated in Figure 2. The movement in distribution towards the target distribution is a clear progression, with the final sample adhering remarkably well to the target distribution, especially in the tails where the ABC-MCMC algorithm performed particularly poorly (Figure 1).
Fig. 2.
(Upper Left to Lower Right) Particle distributions μ1, f1, f2, and f3 defined with ϵ1 = 2, ϵ2 = 0.5, and ϵ3 = 0.025 using ABC-PRC algorithm. Dashed line denotes π(θ). The mixture of normals target distribution is superimposed.
ABC algorithms may be intuitively compared through the number of likelihood “evaluations,” that is, the number of data-generation steps. Table 1 enumerates the mean number of data-generation steps required to move a particle between two successive populations. As the tolerance reduces with each successive distribution, ft, the number of data-generation steps we expect increases. This effect is partially offset by the degree of similarity between population distribution ft and its induced sampling distribution μt. The total number of data-simulation steps in the ABC-PRC algorithm was 75,895. This is more than the illustrated 10,000 for the ABC-MCMC algorithm (Figure 1), but this latter simulation requires a substantially longer run before we can be satisfied that a representative sample has been drawn. Accordingly, the ABC-PRC algorithm is far more efficient for this case.
Table 1.
Mean number of data-generation steps per final particle for each population, based on 1,000 particles, under algorithms ABC-PRC and ABC-REJ
| t | ϵt | ABC-PRC | ABC-REJ |
|
|---|---|---|---|---|
| Prior | Posterior | |||
| 1 | 2.000 | 4.907 | – | – |
| 2 | 0.500 | 4.899 | – | – |
| 3 | 0.025 | 66.089 | 400.806 | 21.338 |
| Total | 75.895 | 400.806 | 21.338 | |
Final two columns use U(−10, 10) prior and known posterior mixture of normals as sampling distributions.
In contrast, using the ABC-REJ algorithm results in utilizing >400 data-simulation steps per final particle (Table 1). Here, there is a clear advantage in adopting a series of intermediary distributions between μ1 and the target distribution. Finally, as an indication of the maximum possible efficiency of ABC samplers for this example, performing rejection sampling with sampling distribution equal to the posterior distribution requires >21 data-generation steps per final particle. Note that each particle must still satisfy steps REJ2 and REJ3, so we do not automatically accept every particle proposed. This gives algorithm ABC-PRC 28% of maximum possible efficiency in this case, compared with only 5% for rejection sampling.
Analysis of Tuberculosis Transmission Rates.
We now reimplement an analysis of tuberculosis transmission rates originally investigated using algorithm ABC-MCMC (7). The aim of this study was to estimate three compound parameters of biological interest, namely, the reproductive value, the doubling time, and the net transmission rate. The data used come from a study of tuberculosis isolates collected in San Francisco during the early 1990s by Small et al. (25). These consist of 473 isolates genotypes using the IS6110 marker; the resulting DNA fingerprints can be grouped into 326 distinct genotypes as follows
where nk indicates there were k clusters of size n. The ABC-MCMC method was used in conjunction with a stochastic model of transmission, which is an extension of a birth and death process to include mutation of the marker. Simulating samples from this model allows comparisons with the actual data through two summary statistics: g, the number of distinct genotypes in the sample, and H, the gene diversity. An informative prior was used for the mutation rate taken from published estimates of the rate for IS6110. Further details can be found in ref. 7.
Tanaka et al. (7) previously implemented the ABC-MCMC algorithm with tolerance ϵ = 0.0025. Three Markov chains with an average acceptance rate of ≈0.3% were thinned and combined to form the final sample, utilizing >2.5 million data-generation steps. (Actually, more were used, as one chain became “stuck” in a distributional tail for most of its length, as illustrated in Fig. 1, and had to be rerun.)
We illustrate algorithm ABC-PRC with a sequence of 10 distributions, defined by ϵ1 = 1 and for t = 2,…, 9, ϵt = ½(3ϵt−1 − ϵ10) is taken to be halfway between the previous tolerance and the target of ϵ10 = 0.0025. Ten distributions were selected so that successive distributions were reasonably similar. We adopt Kt(θt|θt−1) = Lt−1(θt−1|θt) as the ABC-MCMC Gaussian random walk proposal of Tanaka et al. (7) with a slightly larger step for the mutation parameter.
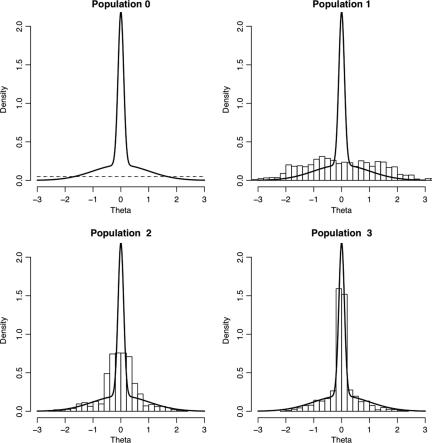
Based on a population size of N = 1,000, Fig. 3 illustrates smoothed posterior distributions of the quantities of interest: (Figure 3 Left) the net transmission rate (α − δ) is the rate of increase of the number of infectious cases in the population; the doubling time [log(2)/(α − δ)] is the required duration for the number of infectious cases in the population to double; (Figure 3 Right) the reproductive value (α/δ) is the expected number of new infectious cases produced by a single infectious case while the primary case is still infectious. As is evident, the results of the ABC-MCMC and ABC-PRC algorithms are essentially indistinguishable.
Fig. 3.
(Left to Right) Posterior distribution of f(α − δ|x0) (net transmission rate) f(log(2)/(α − δ)|x0) (doubling time) and f(α/δ|x0) (reproductive value) for both ABC-MCMC (solid) and ABC-PRC (dash) samplers.
Relative algorithm efficiencies can again be measured by the mean number of data-generation steps per final particle. Table 2 lists the number of data-generation instances in ABC-PRC and ABC-REJ algorithms. For algorithm ABC-REJ, this amounts to a mean of 7,206.3 data-generation steps per particle. In contrast, algorithm ABC-PRC yields a mean of 1,421.3 data-generation steps per particle, >5 times more efficient.
Table 2.
Mean number of data-generation steps per final particle for each population, based on 1,000 particles, under algorithms ABC-PRC and ABC-REJ
| t | ϵ | ABC-PRC | ABC-REJ | |
|---|---|---|---|---|
| 1 | 1.000 | 2.595 | ||
| 2 | 0.5013 | 8.284 | ||
| 3 | 0.2519 | 8.341 | ||
| 4 | 0.1272 | 7.432 | ||
| 5 | 0.0648 | 10.031 | ||
| 6 | 0.0337 | 17.056 | ||
| 7 | 0.0181 | 34.178 | ||
| 8 | 0.0102 | 72.704 | ||
| 9 | 0.0064 | 171.656 | ||
| 10 | 0.0025 | 1,089.006 | 7,206.333 | |
| Total | 1,421.283 | 7,206.333 |
Comparisons to the original ABC-MCMC analysis of Tanaka et al. (7) can also be made in terms of the number of data- generation steps required to generate one uncorrelated particle. Here, the Markov nature of the sampler and the very low acceptance rates induce a strongly dependent sequence. Thinning this sequence so that there were no significant (marginal) autocorrelations above lag 10 resulted in using 8,834 data- generations steps per realization. Repeating this so that there were no significant autocorrelations at any lag yielded 67 uncorrelated particles, corresponding to ≈27,313 data-generation steps per final realization. By this measure, algorithm ABC-PRC is ≈20 times more efficient than the MCMC implementation.
Discussion
Likelihood-free samplers for Bayesian computation are growing in importance, particularly in population genetics and epidemiology, so it is crucial that efficient and accessible algorithms are made available to the practitioner. Existing MCMC algorithms exhibit an inherent inefficiency in their construction, whereas rejection methods are wasteful when sampling and target distributions are mismatched. Through an SMC approach, we may circumvent these problems and generate improved sampling, particularly in distributional tails, while achieving large computational savings.
Evaluations of certain user-specified aspects of algorithm ABC-PRC have not been presented, although these have been studied for SMC algorithms in general, and the necessity of their specification could be considered a disadvantage of the SMC approach. The incorporation of measures other than effective sample size to determine the optimal resampling time is given by Chen et al. (26) and forward and backward kernel choice by Del Moral et al. (17). Jasra et al. (27) give a study of various tolerance schedules and the number of distributions, f1,…, fT. It seems credible that the tolerance schedule and distribution number could be determined dynamically based on one-step-ahead estimates of distributional change, ft−1 → ft, and the required computation (number of data-generation steps). This could be considered one method of selecting the final tolerance ϵT.
Acknowledgments
We thank two anonymous referees whose suggestions have strongly improved the final form of this article. This work was supported by the Australian Research Council through the Discovery Grant scheme (DP0664970 and DP0556732) and by the Faculty of Science, University of New South Wales.
Abbreviations
- ABC
approximate Bayesian computation
- MCMC
Markov chain Monte Carlo
- PRC
partial rejection control
- SMC
sequential Monte Carlo
- ESS
effective sample size.
Appendix
We briefly justify the use of partial rejection control in deriving algorithm ABC-PRC. Following ref. 17, a generic sequential Monte Carlo algorithm is implemented as follows
SMC Algorithm
-
SMC1. Identify the sequence of distributions f1,…, fT, where fT corresponds to the target distribution, and initial sampling distribution μ1.
Set population indicator t = 1.
SMC2. Set particle indicator i = 1.
-
SMC2.1. If t = 1, sample θt(i) ∼ μ1(θ) independently from μ1.
If t > 1, perturb each particle to θt(i) ∼ Kt(θ|θt−1(i)) according to a Markov transition kernel Kt.
-
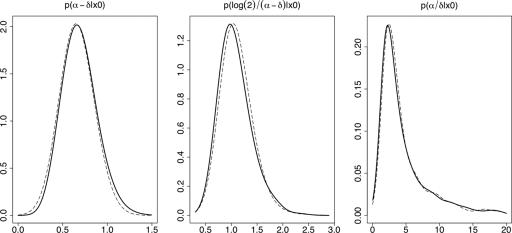
SMC2.2. Evaluate weights Wt(i) for each particle according to
where ft denotes the intermediate distribution at step t and Lt−1 is a backward transition kernel.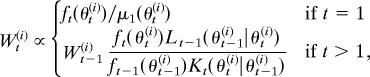
If i < N, increment i = i + 1 and go to SMC2.1.
-
SMC3. Normalize the weights so that Σi=1N Wt(i) = 1.
If ESS = |Σi=1N (Wt(i))2]−1 < E, then resample with replacement, the particles {θt(i)} with weights {Wt(i)} to obtain a new population {θt(i)}, and set weights {Wt(i) = 1/N}.
SMC4. If t < T, increment t = t + 1 and go to SMC2.
Algorithm SMC can be justified intuitively by considering the final weight of particle θT(i), assuming no weight normalization for clarity
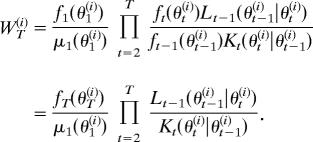 |
The ratio fT(θT(i))/μ1(θ1(i) is immediately identifiable as the standard importance sampling weight with μ1 as the sampling distribution. The product of kernel ratios term evaluates the ratio of probabilities of moving from θT(i) → θ1(i) (numerator) and from θ1(i) → θT(i) (denominator).
Recognizing that many particles of small weight will have minimal impact on the final population, fT, (e.g., they may be lost in the resampling step SMC3), partial rejection control aims to remove them at an earlier stage according to the following scheme (see chapters 2 and 3 of ref. 16).
Given a particle population {θt(i)} with weights {Wt(i)}, a small threshold, c, is specified such that all particles with weights greater than c remain unchanged. For those particles with weights smaller than c, there is a chance [with probability min (1, Wt(i)/c)] that these particles also remain unchanged, otherwise they are replaced by a particle from the previous population {θt−1(i)} chosen according to weights {Wt−1(i)}. This particle is then propagated from distribution ft−1 to ft (via Kt) as before, where its weight is then compared to the threshold, c, once more. This process is repeated until all particles have passed the threshold. PRC is performed within SMC algorithms before the population resampling step (SMC3). See ref. 16 for a justification of this approach.
For a particle population {θt(i)} with weights {Wt(i)} and t > 1, this process is given in algorithmic form by
PRC Algorithm
A1. Set threshold value c > 0 and particle indicator i = 1.
-
A2. With probability min{1, Wt(i)/c}, set weight Wt(i) = max{Wt(i), c} and go to A4.
Otherwise, go to A3.
-
A3. Sample a new particle, θ*, from {θt−1(i)} with probability proportional to {Wt−1(i)}.
Perturb the particle to θt(i) ∼ Kt(θ|θ*) according to a Markov transition kernel Kt.
Go to A2.
A4. If i < N, increment i = i + 1 and go to A2.
A5. Normalize weights according to Σi–1N Wt(i) = 1.
PRC may benefit the SMC algorithm in the ABC setting as follows: The minimum computational specification for the sequence {Bt} in the posterior (Eq. 1) is Bt = 1 for all t. In this setting, large numbers of particles will have identically zero weight, as ρ(S(x), S(x0)) > ϵt occurs with high probability. Suppose we then implement the PRC algorithm for some c > 0 such that only identically zero weights are smaller than c. This will remove those particles for which ρ(S(x), S(x0)) > ϵt and replace them with particles for which ρ(S(x), S(x0)) ≤ ϵt, which then belong to ft.
This process is equivalent to deriving the entire population {θt(i)} one particle at a time, by taking random draws from the previous population, perturbing the particle, and accepting the particle if ρ(S(x), S(x0)) ≤ ϵt. That is, incorporating the data-generation process, we can replace step SMC2 in algorithm SMC above with step PRC2 in algorithm ABC-PRC. Accordingly, we are able to maximize algorithm efficiency in that we obtain a new particle on each occasion for which ρ(S(x), S(x0)) ≤ ϵt, rather than wasting extra data-generation steps in overevaluating likelihood values.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bortot P, Coles SG, Sisson SA. J Am Stat Assoc. 2007 in press. [Google Scholar]
- 2.Marjoram P, Molitor J, Plagnol V, Tavaré S. Proc Natl Acad Sci USA. 2003;100:15324–15328. doi: 10.1073/pnas.0306899100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaumont MA, Zhang W, Balding DJ. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- 5.Marjoram P, Tavaré S. Nat Rev Genet. 2006;7:759–770. doi: 10.1038/nrg1961. [DOI] [PubMed] [Google Scholar]
- 6.Butler A, Glasbey C, Allcroft A, Wanless S. A Latent Gaussian Model for Compositional Data with Structural Zeroes. Edinburgh: Biomathematics and Statistics Scotland; 2006. technical report. [Google Scholar]
- 7.Tanaka MM, Francis AR, Luciani F, Sisson SA. Genetics. 2006;173:1511–1520. doi: 10.1534/genetics.106.055574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leman SC, Chen Y, Stajich JE, Noor MAF, Uyenoyama MK. Genetics. 2006;171:1419–1436. doi: 10.1534/genetics.104.040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornton K, Andolfatto P. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert C, Casella G. Monte Carlo Statistical Methods. 2nd Ed. New York: Springer; 2004. [Google Scholar]
- 11.Gilks WR, Richardson S, Spiegelhalter DJ, editors. Markov Chain Monte Carlo in Practice. London: Chapman and Hall; 1995. [Google Scholar]
- 12.Tavaré S, Balding DJ, Griffiths RC, Donnelly P. Genetics. 1997;145:505–518. doi: 10.1093/genetics/145.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowles MK, Carlin BP. J Am Stat Assoc. 1996;91:883–904. [Google Scholar]
- 14.Gelman A, Meng XL. Stat Sci. 1998;13:163–185. [Google Scholar]
- 15.Neal R. Stat Comput. 2001;11:125–139. [Google Scholar]
- 16.Liu JS. Monte Carlo Strategies in Scientific Computing. New York: Springer; 2001. [Google Scholar]
- 17.Del Moral P, Doucet A, Jasra A. J R Stat Soc B. 2006;68:411–436. [Google Scholar]
- 18.Liu J, Chen R. J Am Stat Assoc. 1998;93:1032–1044. [Google Scholar]
- 19.Givens GH, Raftery AE. J Am Stat Assoc. 1996;91:132–141. [Google Scholar]
- 20.Doucet A, de Freitas N, Gordon N, editors. Sequential Monte Carlo Methods in Practice. New York: Springer; 2001. [Google Scholar]
- 21.Del Moral P. Feynman-Kac Formulae: Genealogical and Interacting Particle Systems with Applications. New York: Springer; 2004. [Google Scholar]
- 22.Kunsch HR. Ann Stat. 2005;33:1983–2021. [Google Scholar]
- 23.Chopin N. Ann Stat. 2004;32:2385–2411. [Google Scholar]
- 24.Peters GW. Cambridge, UK: Univ of Cambridge; 2005. MA thesis. [Google Scholar]
- 25.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Xie J, Liu JS. J R Stat Soc B. 2005;67:199–217. [Google Scholar]
- 27.Jasra A, Stephens DA, Holmes CC. On Population-Based Simulation for Static Inference. Cambridge, UK: Univ of Cambridge; 2006. technical report. [Google Scholar]