Abstract
Fetal progenitor cells enter the maternal circulation during pregnancy and can persist for decades. We aimed to determine the role of these cells in tissue inflammation during pregnancy. WT female mice were mated to males transgenic for the EGFP (ubiquitous) or the luciferase gene controlled by the VEGF receptor 2 (VEGFR2; V-Luc) promoter. A contact hypersensitivity reaction was triggered during such pregnancies. Fetal cells were tracked by using real-time quantitative amplification of the transgene (real-time PCR), Y chromosome in situ hybridization (FISH), immunofluorescence or in vivo bioluminescence imaging. Real-time PCR disclosed fetal cells in the inflamed areas in all tested mice (17/17) with higher frequency and numbers in the inflamed compared with the control areas (P = 0.01). Double labeling demonstrated CD31+ EGFP+ fetal cells organized as blood vessels. In WT pregnant mice bearing V-Luc fetuses, a specific luciferase activity signal could be detected at the hypersensitivity site only, demonstrating the elective presence of VEGFR2-expressing fetal cells. In conclusion, using various techniques, we found the presence of fetal endothelial cells lining blood vessels in maternal sites of inflammation. These results imply that fetal endothelial progenitor cells are acquired by the mother and participate in maternal angiogenesis during pregnancy.
Keywords: angiogenesis, chimerism, gestation, fetal stem cells
Fetal cells enter the maternal circulation during all human pregnancies (1, 2). Trophoblasts (3), nucleated erythroblasts, and leukocytes (4), but also hematopoietic progenitor cells (5, 6), or in rare cases mesenchymal stem cells (7) of fetal origin, have been identified in maternal blood during pregnancy. More interestingly, persisting fetal cells, including hematopoietic progenitor cells, can be detected in a large proportion of women decades after delivery (8–10). These microchimeric cells can be found in the circulation and in a variety of tissues in women as well as in animal models of fetal cell microchimerism (11–13).
The lack of full histocompatibility between the mother and the fetus has led to the hypothesis that the persisting fetal cells could trigger a graft vs. host-type immune reaction in the mother (14). In accordance with these findings, epidemiological studies have shown an association between the number of circulating fetal cells and systemic sclerosis (SSc), an autoimmune disease with clinical features of chronic graft vs. host reaction (15, 16). However, these results remain controversial (17), and therefore the immune role of chimeric cells in SSc remains putative (18). Besides, various studies demonstrated that in maternal tissues affected with nonimmune diseases, the detected fetal cells disclosed the phenotype of the host organ (13, 19–21). The variety of observed phenotypes suggested the transfer of a multipotent population of cells. Therefore, microchimeric cells could be recruited to sites of maternal injury to help in the repair process (22).
In previous studies on the immune consequences of microchimeric fetal cells during pregnancy, our group had demonstrated the presence of fetal cells in 60% of lesional skin from women affected with an unexplained inflammatory skin disease of pregnancy called the polymorphic eruptions of pregnancy (23). Based on these results, our present objective was to assess in a mouse model the possibility for skin inflammation to recruit fetal cells during pregnancy. We report here the increased presence of fetal cells in areas of maternal contact dermatitis. We also present evidence that these fetal cells organize as blood vessels and express markers of endothelial cells proving their participation in maternal new vessel formation.
Results
Fetal Microchimeric Cells Are More Frequent in the Inflamed Skin.
To assess the ability of maternal inflammation to recruit fetal cells, we induced contact-hypersensitivity skin reactions using oxazolone in virgin WT pregnant mice bearing transgenic fetuses. In all mice, the right ear challenged with oxazolone was inflamed with redness and swelling, although the left ear was normal. The inflammation was also verified by histological analysis that disclosed features of contact hypersensitivity (CHS) in similar levels in all mice [supporting information (SI) Table 2 and SI Fig. 6].
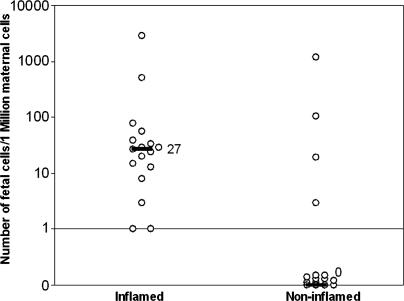
Virgin C57BL/6 (B6) female mice, named G1–G15 and U1–U2, were mated with EGFP+ or U-GFP+ males, respectively, and underwent CHS on their right ears (Table 1). All mice had at least one transgenic fetus at the day of death. We used a real-time quantitative PCR assay to detect and quantify the number of transgenic fetal cells present in the right and the left ears of these 17 mice. In all right inflamed ears, microchimeric cells were detected, and the median frequency of fetal cells was 27 per million maternal genome equivalent (GE) (range, 1–2,857 GE). Fetal cells could also be detected in the left control ears but at a significantly lower median frequency (0 per million maternal GE, P < 0.001; Table 1 and Fig. 1). In addition, the level of fetal cell microchimerism was higher in the inflamed ears when compared with the ears of female mice bearing transgenic fetuses that were exposed to the vehicle alone (n = 7, median = 0, P < 0.001; SI Table 3).
Table 1.
Fetal microchimeric cells in the inflamed and noninflamed tissues
| Mice | Fetal per million maternal GE |
|
|---|---|---|
| Inflamed | Control | |
| G1 | 1 | 0 |
| G2 | 1 | 1,166 |
| G3 | 56 | 0 |
| G4 | 20 | 3 |
| G5 | 29 | 0 |
| G6 | 13 | 103 |
| G7 | 15 | 19 |
| G8 | 2,857 | 0 |
| G9 | 29 | 0 |
| G10 | 39 | 0 |
| G11 | 24 | 0 |
| G12 | 8 | 0 |
| G13 | 27 | 0 |
| G14 | 34 | 0 |
| G15 | 3 | 0 |
| U1 | 511 | 0 |
| U2 | 78 | 0 |
| Median | 27 | 0 |
G1–G15, C57BL/6J virgin females were mated with EGFP+ transgenic male mice. U1 and U2, C57BL/6J virgin females were mated with U-GFP+ transgenic males. On day 10 of gestation, females were sensitized with oxazolone solution. Six days after sensitization, the right ear was challenged by oxazolone and the left ear with vehicle alone. Numbers represent the quantity of fetal per million maternal genome equivalents (GE) amplified after 3 days of challenge with oxazolone.
Fig. 1.
Frequency of fetal cells in maternal inflamed and noninflamed tissues. C57BL/6J virgin females were mated with males transgenic for the EGFP. During pregnancy, a CHS reaction was triggered by cutaneous application of oxazolone on previously sensitized mice. From each mouse, inflamed and noninflamed (control) tissues were recovered and assessed for the presence of fetal cells using real-time quantitative PCR amplification of the egfp transgene. Each dot represents an individual mouse. The median number of fetal cells per million maternal cells (black lines) was significantly higher in inflamed compared with the control tissue (27 vs. 0 per million maternal cells; P < 0.01).
Localization of Fetal Cells in Maternal CHS.
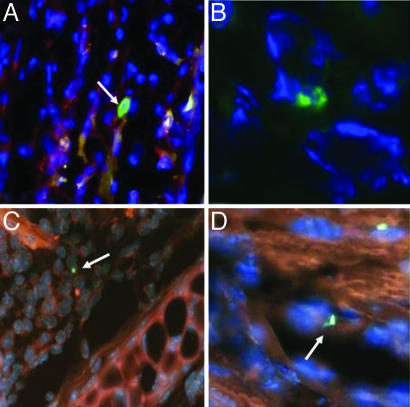
We then used complementary techniques to confirm the presence of fetal cells. We identified fetal cells using anti-EGFP immunofluorescence in the maternal inflamed tissue sections (Fig. 2 A and B). In contrast, EGFP-positive cells were never found in WT tissue or in sections from ears where the egfp transgene could not be detected by PCR. In addition, assuming that 50% of the fetuses were male, we examined paraffin-embedded sections of the maternal inflamed skin for the presence of Y chromosome-positive fetal cells using FISH. No Y chromosome signal could be detected on tissue sections from virgin female mice. The inflamed ear of three tested mice displayed cells with a Y chromosome signal within intact nuclear borders (Fig. 2 C and D). We also performed FISH using X and Y chromosome probes simultaneously. Each microchimeric Y-positive cell had only one X chromosome (SI Fig. 7), ruling out the possibility of fused nuclei between maternal and fetal cells. Using these two in situ techniques, we observed that all fetal cells were exclusively in the dermis. The fetal cells were consistently found in areas of inflammation (Fig. 2 A and C) or in the walls of blood vessels (Fig. 2 B and D).
Fig. 2.
Fetal microchimeric cells locate in the inflammatory infiltrate and in maternal vessel walls during pregnancy. Inflamed ears were obtained from female pregnant mice bearing EGFP transgenic or male fetuses. (A and B) Photomicrographs representing immunofluorescence staining with anti-EGFP antibody (×400 and ×1,000 magnification, respectively). EGFP+ cells (white arrows) are labeled with fluorescein (green); nuclei are labeled with DAPI (blue). (C and D) Photomicrographs representing Y chromosome FISH. Y chromosomes labeled with fluorescein (green) were detected in intact nuclei (DAPI) of male fetal cells in the general background fluorescence (orange-red). Fetal cells (arrows) were localized in the inflammatory infiltrate (A and C) or the walls of blood vessels (B and D).
Phenotype of Fetal Cells in Maternal CHS During Pregnancy.
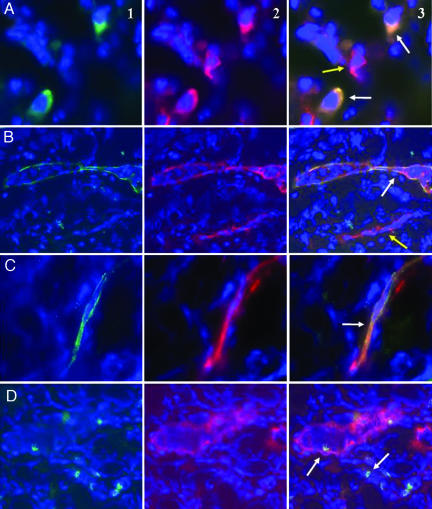
We used double labeling to identify the phenotype of EGFP-positive fetal cells on three inflamed maternal ear specimen (G8, U1, and U2). Some fetal cells inside the inflammatory infiltrate expressed CD45, the common leukocyte antigen, and could be found as leukocytes inside the maternal blood vessels (Fig. 3 A and D). Colocalization with CD31 antibody staining also showed that fetal EGFP expressing cells could be organized as vascular endothelial cells usually isolated in a vessel wall and surrounded with maternal endothelial cells (EGFP negative) (Fig. 3C and SI Fig. 8). Interestingly, some blood vessels in inflamed ears were completely formed by the green fetal cells expressing CD31 (Fig. 3B). Some of these vessels of fetal origin were filled with EGFP-negative cells (presumably maternal), suggesting they were connected to maternal circulation. These fetal leukocytes and endothelial cells were identical in morphology to observations made in EGFP transgenic animals after staining with CD45 or CD31 (SI Fig. 9). Only few fetal cells were seen in noninflamed ears; they did not stain with CD45 or CD31 (SI Fig. 10).
Fig. 3.
Fetal cells express the endothelial marker CD31 and organize as blood vessel endothelium in maternal inflamed tissues during pregnancy. Inflamed ears were obtained from female pregnant mice bearing EGFP transgenic and presumably male fetuses. Sections were counterstained with DAPI (blue) to identify nuclei and labeled by anti-EGFP (green) and anti-CD45 or -CD31 (red) antibodies. (A) Photomicrograph showing two EGFP+ fetal cells in the maternal inflammatory infiltrate (A1, ×1,000 magnification). These two fetal cells were CD45+ (white arrows). Adjacent to these, a maternal CD45+ cell can be visualized as well (yellow arrow; A2 and A3, ×1,000 magnification). (B) Photomicrograph showing an EGFP+ blood vessel containing EGFP− maternal cells (B1, ×200 magnification). B2 and B3, CD31 staining colocalizes with the EGFP staining (white arrow). Yellow arrow shows a maternal CD31+ blood vessel that does not stain with anti-EGFP. (C) Photomicrograph showing a single endothelial cell of fetal origin stained with anti-EGFP (C1) and anti-CD31 (C2). Both stainings colocalize in the cytoplasm of the fetal cell (C3, ×400 magnification). (D) Photomicrographs showing EGFP+ cells (white arrows, D1) in the inflammation around a blood vessel stained with CD31 (D2). The signals do not colocalize. Of note, an EGFP+ cell can be detected inside the maternal blood vessel (D3).
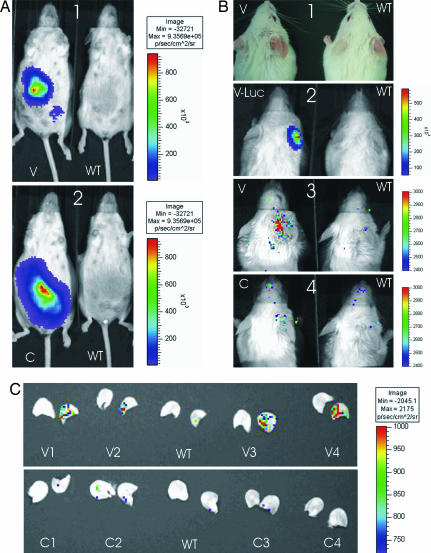
Given the results of this double labeling, we used another model to assess the vascular phenotype of the fetal cells during maternal inflammation. The V-Luc transgenic mouse expresses the luciferase gene under the control of the VEGF receptor 2 (VEGFR2) promoter, mainly in endothelial cells. We therefore assumed that fetal transgenic cells with a vascular phenotype during maternal inflammation should express luciferase and would be detectable using in vivo imaging. We mated eight FVB/N WT female mice (named here V1–V8) with V-Luc males. We also used four WT FVB/N female mice (named C1–C4) mated with C-Luc males, three FVB/N female mice mated with syngenic males (WT1–WT3), one FVB/N (WT4), and two V-Luc transgenic males (V-Luc1 and -2; SI Table 2). Because the luciferase transgenic males are hemizygous, we examined each pregnant mouse for the presence of luciferase transgenic fetuses by ventral imaging at 14 and 18 days after mating. All mice mated with luciferase transgenic males had transgenic fetuses and were eligible for further evaluation (Fig. 4 A1 and A2). No signal was detected on the abdomen of mice mated with WT males.
Fig. 4.
Fetal cells express VEGFR2 in maternal inflamed ear during pregnancy. (A) WT female mice bearing WT fetuses (A1 and A2 Right), V-Luc (A1 Left), or C-Luc transgenic fetuses (A2 Left) were imaged ventrally during the third week of gestation. Transgenic fetuses were visualized in all cases, although no signal could be detected from WT fetuses. (B) After triggering a CHS reaction (B1), animals were imaged dorsally with WT females bearing WT fetuses (B1–B4 Right). Bioluminescence reflecting luciferase activity can be detected on the inflamed ear of a V-Luc transgenic male (B2 Left) tested as a positive control and a WT female bearing V-Luc fetuses (B3 Left). The signal level on mice bearing WT or C-Luc fetuses did not reach significance (B2–B4 Right; B4 Left). (C) To further prove the origin of the bioluminescent signal, ears were sectioned and imaged. The right and left ears of each mouse were imaged in that same order. The signal on the inflamed ears recovered from mice bearing V-Luc fetuses displayed a specific signal (V1–V4, Upper). No specific signal could be detected on ears from mice bearing WT or C-Luc fetuses.
On the day of sensitization with oxazolone, no signal was visualized on the right or left ears. After triggering the CHS reaction, the inflamed ears of transgenic V-Luc males displayed high levels of luciferase signal (mean signal intensity, 40,606 photons per sec per cm2; Fig. 4B2; SI Table 4). On the ears without inflammation, no specific signal was detected. Similarly, WT mice bearing WT fetuses had no specific signal on any ear (Fig. 4B; SI Table 4).
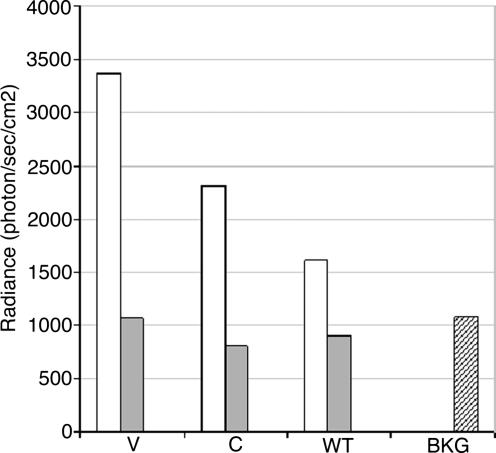
In contrast to mice bearing WT fetuses, V1–V8 had a specific signal on the inflamed ear. The signal intensity was significantly higher on the inflamed compared with the left ears (3,362 vs. 1,059 photons per sec per cm2; P = 0.004). It was also significantly higher than the signal level on the inflamed ears of WT1–WT4 mice (3,362 vs. 1,507 photons per sec per cm2, P = 0.02) (Fig. 4B3; Fig. 5; SI Table 4). C1–C4 mice had a milder signal on the right ear that could hardly be considered as specific (Fig. 4B4; Fig. 5; SI Table 4) and was not significantly different from the WT controls (2,306 vs. 1,507 photons per s per cm2; P = 0.19). Finally, to confirm that the observed signal originated from ears, these were cut and exposed under the camera (Fig. 4C). The specific luciferase signal was identified on the right ears from mice V1–V8, whereas there was no specific signal on the left ears or on ears from mice WT1–WT4 or C1–C4.
Fig. 5.
Quantification of the bioluminescent signal in females bearing luciferase transgenic fetuses. WT female mice bearing WT fetuses (WT), V-Luc fetuses (V), or C-Luc transgenic fetuses (C) were imaged after triggering a contact dermatitis on the right ear. The signal was quantified for each individual mouse, and the average is represented for the right (white bars) and left (gray bars) ears after deduction of the background level (BKG) and compared among different groups. In mice bearing V-Luc fetuses, the signal on the inflamed ear was significantly higher compared with the left or right ear of mice bearing WT fetuses. V, the average radiance on the right ears of the mice V1–V8; C, the average radiance on the right ears of the mice C1–C4; WT, the average radiance on the right ears of the mice WT1–WT4. BKG, the average of signal on the back of WT mice.
Discussion
Using various techniques, our results show that during pregnancy, maternal inflammation recruits fetal cells that can form blood vessel endothelium. With a sensitive real-time quantitative PCR assay, we found that fetal cells were significantly more frequent in maternal inflamed tissues, although they may on occasion be found in the noninflamed ear, as reported (19). The preferential homing or amplification of the chimeric cells in the inflamed ears was also demonstrated by in vivo imaging. The localization and morphology of the fetal cells were determined by in situ techniques such as anti-EGFP immunofluorescence and Y chromosome FISH. Both consistently showed that the chimeric cells could either be in inflammatory infiltrates or in vessel walls. Finally, double labeling and promoter-specific reporter expression analysis of luciferase using in vivo imaging again were consistent, showing that fetal cells had mainly an endothelial phenotype, because they expressed CD31 and VEGFR2. Some other cells located in the inflammatory infiltrates were leukocytes and expressed CD45.
Although the correlation between these different techniques has not always been reported (24), we consistently found microchimeric cells in specimens from inflamed areas. In particular, we could not reliably detect fetal cell bioluminescent signal in female mice bearing C-Luc fetuses. This may be due to the basal level of expression of luciferase in the fetal-derived chimeric cells. Of note, the bioluminenscence signal in females with C-luc fetuses contrasts with the strong specific signal observed in female mice bearing V-Luc fetuses, demonstrating the relative importance of VEGFR2 expression among fetal microchimeric cells. During murine pregnancy, fetal cell-free DNA can also be detected in maternal plasma (25). The detection of intact nucleated fetal cells by immunofluorescence and FISH rules out the possibility that the amplified egfp sequences were only due to cell-free fetal DNA.
Previous studies also demonstrated the specific homing of fetal microchimeric cells to maternal sites of injury. Fetal cells have been detected in the lesional epidermis of women affected with polymorphic eruptions of pregnancy but not in normal skin from control pregnant women (23). This benign skin inflammation occurs exclusively in the third trimester during pregnancy. Similarly, in murine models of microchimerism, Imaizumi et al. (26) showed that during and after pregnancy, mice with thyroiditis had a higher frequency of detectable microchimerism than controls in the thyroid. The phenotype of fetal cells was not determined in these studies. In mice with brain-induced damage after pregnancy, cerebral lesions also significantly increased the number of fetal microchimeric cells in the brain (27).
Recently, many studies have demonstrated the capacity of fetal cells acquired during pregnancy to differentiate and participate in maternal tissue repair processes. In studies of women with various diseases and a prior pregnancy, fetal cells with various phenotypes have been found. The microchimeric cells express markers of epithelia, hepatocytes, leukocytes, or cardiomyocytes (13, 21, 28). Similar results have been found in mouse models of fetal cell microchimerism where fetal-derived cells can express markers of neurons, glial cells (27), hepatocytes, or renal tubular epithelium (29). In our study, we demonstrate that, during pregnancy, fetal cells can give rise to endothelial cells and form blood vessels in maternal inflamed tissues. In accordance with previous studies showing a variety of different phenotypes in fetal microchimeric cells, our results point again toward the transfer to the mother of a population of pregnancy-associated progenitor cells (22). It seems highly unlikely that the observed fetal-derived blood vessels resulted from the apposition of individual rare microchimeric fetal cells. A more logical explanation resides in the proliferation of an endothelial progenitor cell (EPCs) giving rise to daughter cells that form a blood vessel. The present study implies that fetal EPCs are acquired by pregnant mice and home specifically to maternal inflammatory tissues.
The capacity of fetal cells to express vascular markers such as CD31 or VEGFR2 and to organize as blood vessels connected to the maternal circulation strongly suggests the presence of an EPC. The expression of VEGFR2 by EPCs has been reported. This receptor is particularly involved in the homing of the EPCs to sites of ischemia and tumors (30). The homing of VEGFR2-expressing cells in sites of inflammation has also been recently shown (31). After CHS, VEGFR2-positive cells were found in the inflamed skin with a peak at day 2. However, these cells did not seem to persist in the skin, because their signal vanished along with the inflammation. In our study, we did not assess the long-term presence of fetal cells long after the inflammation.
Unlike hematopoietic progenitor (5, 6) or mesenchymal stem cells (7), fetal EPCs have not yet been described in maternal blood during pregnancy. However, trophoblastic cells acquire endothelial properties during pregnancy (32). During pregnancy, in the spiral artery invasion process, fetal cytotrophoblastic cells line along the maternal endothelium. They express markers of cell adhesion specific to endothelial cells (33), produce and respond to VEGF-A through VEGFR1 (34), and finally synthesize important molecules of hemostasis (35). Although the expression of VEGFR2 is lost by invasive cytotrophoblastic cells, this population could give rise to the phenomenon described in our study, because they can be found in maternal circulation during pregnancy (3). Trophoblast cells with endothelial properties could therefore participate in the angiogenesis triggered by maternal inflammation. However, this hypothesis is not supported by the endothelial morphology of the fetal cells detected in our study. Overall, it remains unclear whether the pregnancy-associated progenitor cells transferred to the mother during pregnancy consist of the addition of multiple progenitors with different progeny, or whether the observed plasticity results from the differentiation of a single multipotent cell population.
The capacity of fetal cells to participate in maternal angiogenesis, as demonstrated here, most probably with an EPC capacity, opens new perspectives. First, fetal endothelial cells could also participate in maternal angiogenesis occurring in other circumstances than inflammation during pregnancy, such as wound healing or tumor development. Second, the description and possible further isolation of a placental EPC could be the basis for cell therapy in different ischemic vascular diseases, as has been proposed (36). In conclusion, we describe the capacity of fetal cells to participate in maternal new vessel formation during pregnancy. We believe this phenomenon relates to the presence of a fetal EPC.
Materials and Methods
Mice.
WT virgin C57BL/6 or FVB/N female mice (6–12 weeks old) were bred to syngenic male controls or to various transgenic males for reporter genes. In the resulting gestations, 50% of the fetuses are transgenic. Fetal cells harboring the reporter transgene would be tracked in the maternal WT tissues.
We used four different strains of transgenic male mice (1). The C57BL/6-Tg(ACTbEGFP)1Osb/J mouse (named EGFP+), transgenic for the enhanced GFP under the control of the chicken β-actin promoter, was kindly provided by M. Okabe (Osaka University, Osaka, Japan) (2, 37). The C57BL/6-Tg(UBC-GFP)30Scha/J (named U-GFP+) mouse expresses the GFP in all tissues under the control of the human ubiquitin C promoter. The GFP transgene is therefore ubiquitously expressed but more specifically in the hematopoietic system (3). The transgenic mouse FVB/NTg (Vegfr2-Luc)Xen (named V-Luc) expresses the luciferase reporter with the promoter of the second receptor of the murine VEGFR2 (4). Finally, the FVB/NTg (CMV-Luc)Xen transgenic mouse (named C-Luc) expresses luciferase under the control of a CMV ubiquitous promoter. The luciferase transgenic mice were purchased from Xenogen (Alameda, CA).
CHS Induction.
Oxazolone (Sigma-Aldrich, St. Louis, MO) was used to induce CHS reactions as described (31). Briefly, on day 10 after mating, pregnant females were shaved and sensitized by a topical application on the abdomen of 50 μl of 2% oxazolone solution in acetone/olive oil, 4:1 vol/vol. Six days after sensitization, the right ear was challenged with 10 μl of 2% oxazolone solution in the same vehicle, whereas the left ear was treated with vehicle alone. The ear challenge was repeated once per day during 3 days. For each mouse, the level of inflammation was evaluated histologically after blinding to the exposure type (vehicle alone or oxazolone). The intensity of the edema, the infiltrate, and the epidermal lesions were graded from absent to strong. We also analyzed a control group of mice that were not sensitized and that were exposed to vehicle alone on the left ear.
In Vivo Imaging of Luciferase Activity.
In vivo imaging was performed as described (38) by using an IVIS100 imaging system (Xenogen, Alameda, CA). Bioluminescent images were collected 1 day after the last oxazolone application. Female mice bearing transgenic Vegfr2-luciferase, CMV-Luciferase, or WT FVB/N fetuses were injected i.p. with 120–150 mg/kg luciferin (Xenogen). Ten minutes after injection, mice were imaged for 1–5 min under anesthesia by isoflurane inhalation. Photons emitted from specific regions were quantified by using the LivingImage software (Xenogen). For each image, the background radiance was estimated on the back of a WT mouse and deducted from all measured signal on the image. In vivo luciferase activity is measured in photons per second per cm2.
Tissue Collection.
Mice were killed at day 18 of gestation. Fetuses were assessed under UV light (365-nm wavelength) or CCD camera for the expression of EGFP or luciferase, respectively. This procedure allowed us to ensure that each female mouse had at least one transgenic fetus. On each pregnant mouse, the right and left ears were collected and cut into three parts. One was immediately frozen at −80°C for further DNA extraction. The two others were fixed in 4% formaldehyde overnight, embedded in either paraffin or optimum cutting temperature compound, and further snap-frozen in liquid nitrogen.
DNA Extraction and Real-Time PCR Amplification.
Genomic DNA extraction was performed on all samples by using an isopropanol precipitation technique. In brief, samples were incubated overnight at 56°C in a solution containing 100 mM Tris 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl, and 100 μg/ml proteinase K. After centrifugation, DNA was precipitated with isopropanol (vol:vol) washed in 70°C ethanol, air-dried, and resuspended in Tris-EDTA buffer.
The real-time quantitative amplification of the egfp transgene was performed as described by using a FAM-TAMRA reporter quencher Taqman system (39). A standard curve was established by dilution of known quantities of genomic DNA of splenocytes from an EGFP transgenic animal. On each sample, we first amplified the genomic sequence of the apolipoprotein b (apob) gene as an internal control for the quantification of total amplifiable DNA. The egfp amplification assay was then applied to 100,000 pg of DNA from each sample per well. Each sample was analyzed in five wells. All samples were run on an ABI 7300 Sequence Detection System with the SDS Ver. 1.9 software (Applied Biosystems, Foster City, CA).
Each hemizygous fetal EGFP+ mouse cell contains only one copy of the egfp gene (40). We were able to amplify the egfp transgene in as low as 6.4 pg of EGFP+ mouse genomic DNA, which is the equivalent of one cell. The estimation of the number of fetal EGFP+ cells in the tissue maternal was based on the quantity of egfp transgene reported to the quantity of apob gene detected in the maternal tissues. A tissue was considered positive for fetal-cell microchimerism if the amount of EGFP transgene detected was equivalent to at least one genome.
Immunofluorescence Staining.
Seven-micrometer sections were obtained from the frozen samples with or without formaldehyde fixation. Briefly, after rehydration and permeabilization (Triton X-100), sections were blocked by using 20% normal goat serum (DakoCytomation, Carpenteria, CA). Primary antibodies were incubated for 3 h at room temperature. After washes, secondary antibodies were incubated for 40 min. Slides were then washed, counterstained with 0.3 μg/ml DAPI, and observed under a fluorescence microscope (Leica, Deerfield, IL) with a QImaging digital camera (Media Cybernetics, Silver Spring, MD). Primary antibodies used were a rabbit anti-GFP polyclonal antibody (1:40; Chemicon International, Temecula, CA), purified rat anti-mouse CD45 (1:10; BD PharMingen, San Diego, CA), and purified rat anti-mouse CD31 (PECAM-1) monoclonal antibody (1/10 BD PharMingen). The secondary antibodies used were goat anti-rabbit IgG labeled with FITC and goat anti-rat IgG labeled with Texas red (1:100; Jackson ImmunoResearch, West Grove, PA).
FISH for the Y and X Chromosomes.
FISH was performed on paraffin-embedded tissue sections as described (41). We used FITC-labeled Y chromosome and rhodamine-labeled X chromosome whole-paint Starfish probes (Cambio, Cambridge, U.K.). Cells containing one Y chromosome inside the nucleus with intact borders were considered as male.
Statistical Analyses.
For each mouse, the quantitative estimation of the frequency of fetal cells (real-time quantitative PCR amplification of the egfp transgene) or the fetal luciferase bioluminescent signal was considered as a continuous variable. The values obtained for the right and left ears were compared within each group or among different groups of mice using the nonparametric Mann–Whitney–Wilcoxon test or Student's t test. A P value <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Marie-Josée Espié for technical help. We thank Dr. Claude Carnaud for helpful discussion and Dr. Marco Giovannini for authorizing access to in vivo imaging. This work was supported by the Assistance Publique-Hôpitaux de Paris (Grant CRC04026), the Société Française de Dermatologie, and the Fondation pour la Recherche Médicale (Grant INE20050303543), Paris, France.
Abbreviations
- CHS
contact hypersensitivity
- VEGFR2
VEGF receptor 2
- EPC
endothelial progenitor cell.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606490104/DC1.
This article is a PNAS direct submission.
References
- 1.Krabchi K, Gros-Louis F, Yan J, Bronsard M, Masse J, Forest JC, Drouin R. Clin Genet. 2001;60:145–150. doi: 10.1034/j.1399-0004.2001.600209.x. [DOI] [PubMed] [Google Scholar]
- 2.Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, Lee TH. Transfusion. 2001;41:1524–1530. doi: 10.1046/j.1537-2995.2001.41121524.x. [DOI] [PubMed] [Google Scholar]
- 3.van Wijk IJ, van Vugt JM, Mulders MA, Konst AA, Weima SM, Oudejans CB. Am J Obstet Gynecol. 1996;174:871–878. doi: 10.1016/s0002-9378(96)70315-0. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi DW. Br J Haematol. 1999;105:574–583. doi: 10.1046/j.1365-2141.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 5.Guetta E, Gordon D, Simchen MJ, Goldman B, Barkai G. Blood Cells Mol Dis. 2003;30:13–21. doi: 10.1016/s1079-9796(03)00008-1. [DOI] [PubMed] [Google Scholar]
- 6.Osada H, Doi S, Fukushima T, Nakauchi H, Seki K, Sekiya S. Transfusion. 2001;41:499–503. doi: 10.1046/j.1537-2995.2001.41040499.x. [DOI] [PubMed] [Google Scholar]
- 7.O'Donoghue K, Choolani M, Chan J, De La FJ, Kumar S, Campagnoli C, Bennett PR, Roberts IA, Fisk NM. Mol Hum Reprod. 2003;9:497–502. doi: 10.1093/molehr/gag063. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Proc Natl Acad Sci USA. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert NC, Lo YM, Erickson TD, Tylee TS, Guthrie KA, Furst DE, Nelson JL. Blood. 2002;100:2845–2851. doi: 10.1182/blood-2002-01-0295. [DOI] [PubMed] [Google Scholar]
- 10.Artlett CM, Cox LA, Ramos RC, Dennis TN, Fortunato RA, Hummers LK, Jimenez SA, Smith JB. Clin Immunol. 2002;103:303–308. doi: 10.1006/clim.2002.5222. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KL, Nelson JL, Furst DE, McSweeney PA, Roberts DJ, Zhen DK, Bianchi DW. Arthritis Rheum. 2001;44:1848–1854. doi: 10.1002/1529-0131(200108)44:8<1848::AID-ART323>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez DF, Leapley AC, Lee CI, Ultsch MN, Tarantal AF. Transplantation. 2005;79:142–146. doi: 10.1097/01.tp.0000144468.71962.aa. [DOI] [PubMed] [Google Scholar]
- 13.Khosrotehrani K, Johnson KL, Cha DH, Salomon RN, Bianchi DW. J Am Med Assoc. 2004;292:75–80. doi: 10.1001/jama.292.1.75. [DOI] [PubMed] [Google Scholar]
- 14.Nelson JL. Arthritis Rheum. 1996;39:191–194. doi: 10.1002/art.1780390203. [DOI] [PubMed] [Google Scholar]
- 15.Nelson JL, Furst DE, Maloney S, Gooley T, Evans PC, Smith A, Bean MA, Ober C, Bianchi DW. Lancet. 1998;351:559–562. doi: 10.1016/S0140-6736(97)08357-8. [DOI] [PubMed] [Google Scholar]
- 16.Artlett CM, Smith JB, Jimenez SA. N Engl J Med. 1998;338:1186–1191. doi: 10.1056/NEJM199804233381704. [DOI] [PubMed] [Google Scholar]
- 17.Gannage M, Amoura Z, Lantz O, Piette JC, Caillat-Zucman S. Eur J Immunol. 2002;32:3405–3413. doi: 10.1002/1521-4141(200212)32:12<3405::AID-IMMU3405>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 18.Khosrotehrani K, Bianchi DW. Curr Opin Obstet Gynecol. 2003;15:195–199. doi: 10.1097/00001703-200304000-00014. [DOI] [PubMed] [Google Scholar]
- 19.Srivatsa B, Srivatsa S, Johnson KL, Samura O, Lee SL, Bianchi DW. Lancet. 2001;358:2034–2038. doi: 10.1016/S0140-6736(01)07099-4. [DOI] [PubMed] [Google Scholar]
- 20.Johnson KL, Samura O, Nelson JL, McDonnell M, Bianchi DW. Hepatology. 2002;36:1295–1297. doi: 10.1053/jhep.2002.35622. [DOI] [PubMed] [Google Scholar]
- 21.Bayes-Genis A, Bellosillo B, de la Calle, Salido M, Roura S, Ristol FS, Soler C, Martinez M, Espinet B, Serrano S, et al. J Heart Lung Transplant. 2005;24:2179–2183. doi: 10.1016/j.healun.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Khosrotehrani K, Bianchi DW. J Cell Sci. 2005;118:1559–1563. doi: 10.1242/jcs.02332. [DOI] [PubMed] [Google Scholar]
- 23.Aractingi S, Berkane N, Bertheau P, Le Goué C, Dausset J, Uzan S, Carosella ED. Lancet. 1998;352:1898–1901. doi: 10.1016/S0140-6736(98)05121-6. [DOI] [PubMed] [Google Scholar]
- 24.Mezey E, Nagy A, Szalayova I, Key S, Bratincsak A, Baffi J, Shahar T. Science. 2003;299:1184. doi: 10.1126/science.1079318. [DOI] [PubMed] [Google Scholar]
- 25.Khosrotehrani K, Wataganara T, Bianchi DW, Johnson KL. Hum Reprod. 2004;19:2460–2464. doi: 10.1093/humrep/deh445. [DOI] [PubMed] [Google Scholar]
- 26.Imaizumi M, Pritsker A, Unger P, Davies TF. Endocrinology. 2002;143:247–253. doi: 10.1210/endo.143.1.8563. [DOI] [PubMed] [Google Scholar]
- 27.Tan XW, Liao H, Sun L, Okabe M, Xiao ZC, Dawe GS. Stem Cells. 2005;23:1443–1452. doi: 10.1634/stemcells.2004-0169. [DOI] [PubMed] [Google Scholar]
- 28.Guettier C, Sebagh M, Buard J, Feneux D, Ortin-Serrano M, Gigou M, Tricottet V, Reynes M, Samuel D, Feray C. Hepatology. 2005;42:35–43. doi: 10.1002/hep.20761. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Iwatani H, Ito T, Horimoto N, Yamato M, Matsui I, Imai E, Hori M. Biochem Biophys Res Commun. 2004;325:961–967. doi: 10.1016/j.bbrc.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Sharpe EE, Maupin AB, Teleron AA, Pyle AL, Carmeliet P, Young PP. FASEB J. 2006;20:1495–1497. doi: 10.1096/fj.05-5137fje. [DOI] [PubMed] [Google Scholar]
- 31.Zhang N, Fang Z, Contag PR, Purchio AF, West DB. Blood. 2004;103:617–626. doi: 10.1182/blood-2003-06-1820. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Genbacev O, Fisher SJ. Ann NY Acad Sci. 2003;995:73–83. doi: 10.1111/j.1749-6632.2003.tb03211.x. :73–83. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y, Fisher SJ, Janatpour M, Genbacev O, Dejana E, Wheelock M, Damsky CH. J Clin Invest. 1997;99:2139–2151. doi: 10.1172/JCI119387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, McMaster M, Woo K, Janatpour M, Perry J, Karpanen T, Alitalo K, Damsky C, Fisher SJ. Am J Pathol. 2002;160:1405–1423. doi: 10.1016/S0002-9440(10)62567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sood R, Kalloway S, Mast AE, Hillard CJ, Weiler H. Blood. 2006;107:3173–3180. doi: 10.1182/blood-2005-10-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishida A, Ohya Y, Sakuda H, Ohshiro K, Higashiuesato Y, Nakaema M, Matsubara S, Yakabi S, Kakihana A, Ueda M, et al. Circ J. 2005;69:1260–1265. doi: 10.1253/circj.69.1260. [DOI] [PubMed] [Google Scholar]
- 37.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 38.Contag PR, Olomu IN, Stevenson DK, Contag CH. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 39.Khosrotehrani K, Johnson KL, Guegan S, Stroh H, Bianchi DW. J Reprod Immunol. 2005;66:1–12. doi: 10.1016/j.jri.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Nakanishi T, Kuroiwa A, Yamada S, Isotani A, Yamashita A, Tairaka A, Hayashi T, Takagi T, Ikawa M, et al. Genomics. 2002;80:564–574. doi: 10.1006/geno.2002.7008. [DOI] [PubMed] [Google Scholar]
- 41.Johnson KL, Zhen DK, Bianchi DW. BioTechniques. 2000;29:1220–1224. doi: 10.2144/00296st01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.