Abstract
The mammalian nuclear pore complex (NPC) is an ≈120-MDa proteinaceous assembly consisting of ≈30 proteins and is the sole gate in the nuclear envelope. The human protooncogene Nup214 was first identified as a target for chromosomal translocation involved in leukemogenesis. Nup214 is located on the cytoplasmic face of the NPC and is implicated in anchoring the cytoplasmic filaments of the NPC and recruiting the RNA helicase Ddx19. Here, we present the crystal structure of the human Nup214 N-terminal domain at 1.65-Å resolution. The structure reveals a seven-bladed β-propeller followed by a 30-residue C-terminal extended peptide segment, which folds back onto the β-propeller and binds to its bottom face. The β-propeller repeats lack any recognizable sequence motif and are distinguished by extensive insertions between the canonical β-strands. We propose a mechanism by which the C-terminal peptide extension is involved in NPC assembly.
Keywords: β-propeller, mRNA export, leukemia, Dbp5/Ddx19, nuclear pore complex
The selective transport of proteins or of protein–nucleic acid complexes across the double membrane of the nuclear envelope is achieved solely through the nuclear pore complex (NPC) (1, 2). Recent proteomic studies in yeast and mammalian cells have revealed that the NPC consists of ≈30 nucleoporins (nups) that form an ≈120-MDa assembly in mammalian cells (3, 4). Electron microscopic reconstructions have uncovered that the NPC consists of a central core with an 8-fold nucleocytoplasmic axis of symmetry and a two-fold axis of symmetry in the plane of the nuclear envelope (3, 5). A nuclear basket structure and pseudosymmetric cytoplasmic filaments flank this symmetric core. Homology studies have suggested that the nups are constructed from only a small number of major structural domains: coiled-coils, FG repeats, α-helical solenoids, β-propellers, and zinc fingers (6). The vertebrate NPC is significantly larger than its yeast counterpart, but its most distinct feature is the cell cycle-dependent reversible disassembly. In cells that undergo an open mitosis, the NPCs disassemble into distinct subcomplexes from which they reassemble at the completion of the cell division, a process that is poorly understood (7).
The human Nup214/CAN gene was first identified as a target for chromosomal translocations involved in leukemogenesis (8, 9). Fusions of Nup214 to the tyrosine kinase ABL retain the N-terminal portion of Nup214 [supporting information (SI) Fig. 6] and result in a deregulated kinase fusion protein that has been implicated in T cell acute lymphoblastic leukemia (10). Fusion proteins of the C-terminal portion of Nup214 to DEK (11) and SET (12) (SI Fig. 6) give rise to acute myeloid leukemia (9).
Nup214 is positioned at the cytoplasmic side of the NPC (8), interacts with proteins of the mRNA export machinery (13, 14), and, in a subcomplex with Nup88, anchors the cytoplasmic fibrils to the NPC (15). An N-terminal region of Nup214 has been found to interact with the DEAD box helicase Ddx19, and mutations in Ddx19 that disrupt binding to Nup214 inhibit mRNA export (16).
Because high-resolution structural information of the NPC is unavailable, the principles that govern the assembly of the nups into the NPC remain poorly understood. To deepen our understanding of the architecture of the NPC, the regulation of its assembly, and the involvement of nups in leukemogenesis, we have determined the crystal structure of the N-terminal domain (NTD) of the human protooncogene Nup214 at a resolution of 1.65 Å. The structure reveals that the NTD consists of two distinct structural domains, an N-terminal seven-bladed β-propeller that is followed by a 30-residue C-terminal extended peptide segment (CTE) that binds to the bottom face of the β-propeller. We find that the β-propeller contains numerous insertions that form long loops as well as additional strands that pack against the β-propeller. In contrast to many previously determined β-propeller structures, the NTD β-propeller lacks any repeated sequence motifs within the primary sequence. Analysis of the surface conservation of the NTD within higher eukaryotes identifies several highly conserved putative protein–protein interaction sites. The CTE binds with low affinity to the β-propeller, thereby partially covering the highly conserved bottom face. We speculate that the binding of this extension may be dynamic in nature and that the CTE and these conserved regions play a regulatory role in NPC assembly.
Results
Structure Determination.
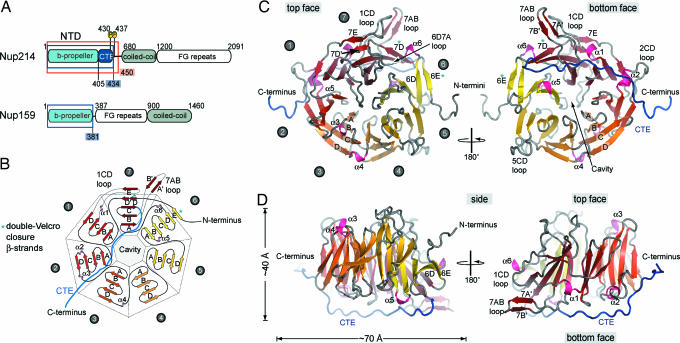
To investigate and clarify the domain organization of Nup214, we performed primary structure and sequence conservation analysis as well as secondary structure predictions for the 2,090-residue human Nup214. We identified the approximate domain boundaries of three structurally distinct domains (Fig. 1A) and designed a series of expression constructs for the N-terminal all-β-sheet region. We identified a stable fragment composed of residues 1–450 that exists as a monomer in solution, as determined by gel-filtration analysis (data not shown), which we termed the Nup214 NTD. Crystals of the ≈50-kDa NTD appeared in the orthorhombic space group P212121. The structure was solved by multiple isomorphous replacement anomalous scattering, using x-ray diffraction data from two heavy-metal derivatives. The final model contains residues 1–434 and was refined to a 1.65-Å resolution with an Rwork and an Rfree of 19.7% and 23.7%, respectively.
Fig. 1.
The structure of the NTD of human Nup214. (A) Domain structure of Nup214 and Nup159. The construct used for crystallization is boxed red, and two phosphorylation sites of the NTD are indicated. Residues observed in the crystal structures are boxed in blue. (B) Schematic representation of the NTD structure. The blades of the β-propeller are labeled from 1 to 7. The CTE is shown in blue, and β-strands forming the double-Velcro closure are indicated with an asterisk. (C) Ribbon representation of the NTD structure. A 180°-rotated view is shown on the right. As a reference, the strands of blade 3 are labeled A–D. The blades of the β-propeller and the CTE are labeled as in B. The helical insertions are shown in pink. (D) Ribbon representation of side views of the structure of the NTD. The view on the right is rotated by 180°.
Architectural Overview of the NTD.
The structure of the NTD can be divided into two parts: an N-terminal canonical seven-bladed β-propeller domain followed by a CTE (Fig. 1 B–D). The NTD creates an elliptical, disk-shaped molecule with an overall diameter of ≈70 Å and a thickness of ≈40 Å (Fig. 1 C and D). The canonical core of the β-propeller is generated by four antiparallel β-strands in each of the seven blades, that by convention are termed A to D from the inside to the outside of the β-propeller (Fig. 1 B and C). The loops are named according to the β-strands that they connect, and the face that contains the N-terminal ends of the innermost β-strands is defined as the top face (Fig. 1 C and D). The blades are pseudosymmetrically arranged around a seven-fold axis and create a central cavity at the bottom face of the NTD (Fig. 1 B and C). Typically, the β-propellers of many proteins are generated by conserved sequence motifs, such as the ≈40-residue WD repeat, in which very short loops connect the β-strands. However, in the NTD β-propeller long insertions between the β-strands form extensive loops protruding on both faces of the molecule (Fig. 1 C and D). The lack of such short connecting loops significantly loosens the restraints on the primary sequence, and consequently, no sequence repeat motif can be identified in the NTD β-propeller. The 30-residue CTE (residues 405–434) folds back onto the β-propeller, adopts an extended conformation, and is bound to the bottom face (Fig. 1 C and D).
Unique Structural Features of the NTD.
The canonical features of the Nup214 β-propeller, as first seen in the structure of methylamine dehydrogenase (17), are complemented by special features, such as the central cavity, structural additions to the β-propeller fold, and a double-Velcro closure (Fig. 1 B–D). The circular packing of the blades forms a central, water-filled tunnel connecting the top and the bottom in the vast majority of β- propellers, as seen in the Gtβ subunit of the heterotrimeric G protein complex transducin (18, 19). However, in the Nup214 β-propeller, the channel is closed at the top face, generating a ≈30-Å deep, conical cavity with a diameter of ≈15 Å at its opening that is filled with ≈50 well ordered water molecules.
The structural additions to the canonical β-propeller fold of the NTD include extensive loops containing helical turns and noncanonical β-strands. The remarkably extensive loops, which are up to 23 residues long, are the interblade connector 6D7A as well as loop 7AB and the loops that connect strands C and D of blades 1, 2, and 5 (Figs. 1C and 2B). Two α-helices, α1 and α2, and four 310-helices, α3–α6, can be found in these loops. Helix α1 is inserted between strands C and D of blade 1 and is located on the side of the β-propeller. Helix α6 is also located on the side and inserted in the 6D7A loop. The helices α3 and α4 are located on the top face within loops 2D3A and 3D4A, respectively, whereas helices α2 and α5 are located on the bottom face of the β-propeller and are part of the 2CD and 6AB loop (Fig. 1 B–D).
Fig. 2.
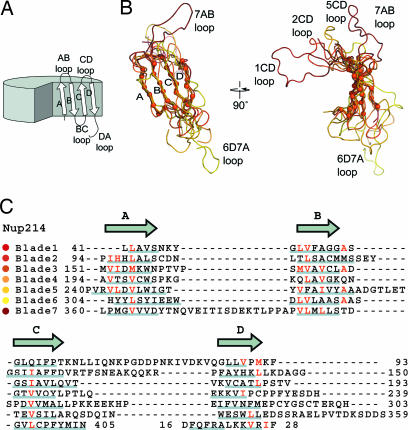
Superposition of the NTD β-propeller blades. (A) Schematic drawing of a β-propeller fold indicating the β-strands and loops of one β-propeller blade. (B) Coil representation of the structural alignment of the seven blades of the β-propeller. Blades are colored as in Fig. 1. As a reference, the Cα atoms of blade 2 are shown as orange spheres. A 90°-rotated view is shown on the right. (C) Structure-based sequence alignment of the blades. The β-strands are indicated above the sequence. Similar residues are shown in red, and the residues of each blade that participate in β-sheet hydrogen bonds are underlined in gray.
In the NTD β-propeller, two five-stranded blades are created by noncanonical β-strands (Fig. 1 B–D). Binding of the very N-terminal β-strand 6E to 6D in a parallel fashion forms the five-stranded blade 6. This interaction creates the noncanonical double-Velcro closure (Fig. 1 B and C). Blade 7 is the second five-stranded blade. The β-strand 7E, which is located in the 23-residue 1CD loop, binds in a parallel fashion to strand 7D′ (Fig. 1C Left). Instead of forming part of the bottom face of the β-propeller, this interaction causes the 1CD loop to be pushed toward the side. Similar, the 19-residue 7AB loop, which is stabilized by two short accessory β-strands (7A′ and 7B′), flips to the side of the β-propeller. Almost parallel, both loops protrude ≈25 Å from the side of the β-propeller and form hydrogen bonds to each other (Fig. 1 C and D, Right).
The Sequence Repeat of the NTD β-Propeller.
In the vast majority of cases, the pseudo-seven-fold symmetry of β-propeller fold-containing proteins is reflected in the primary sequence (20). For example, the primary sequence of the β-propeller of the heterotrimeric G protein subunit Gtβ contains seven WD repeats (21). Each of the WD repeats contains a conserved GH dipeptide ≈20 residues from its N terminus and a conserved WD dipeptide at its C terminus. The conserved tryptophan of the WD dipeptide is buried in a hydrophobic environment between β-sheets and usually interacts with conserved aspartate, histidine, and serine or threonine residues within the same repeat. The hydrogen bond arrangement of these residues stabilizes the interactions between the β-strands in each β-sheet and creates a rigid molecule (20). In addition to the WD repeat, other clearly defined sequence motifs, such as the kelch motif, the AxSPD, or the YWTD repeats, can be found in other β-propeller proteins (18).
To identify structurally equivalent positions within the blades and to determine whether the Nup214 β-propeller contains a sequence repeat, we superimposed all seven blades of the β-propeller (Fig. 2 A and B). We found that whereas the canonical β-strands of the seven blades align very well structurally with an rmsd of ≈1.2 Å, the loops connecting the β-strands are structurally diverse (Fig. 2B). Extensive insertions between the β-strands cause the length of the sequence repeat to vary between 43 and 64 residues. Apart from hydrophobic side chains in the central residues of β-strands A, B, and C, no recognizable sequence motif can be identified by this structural alignment (Fig. 2C).
CTE.
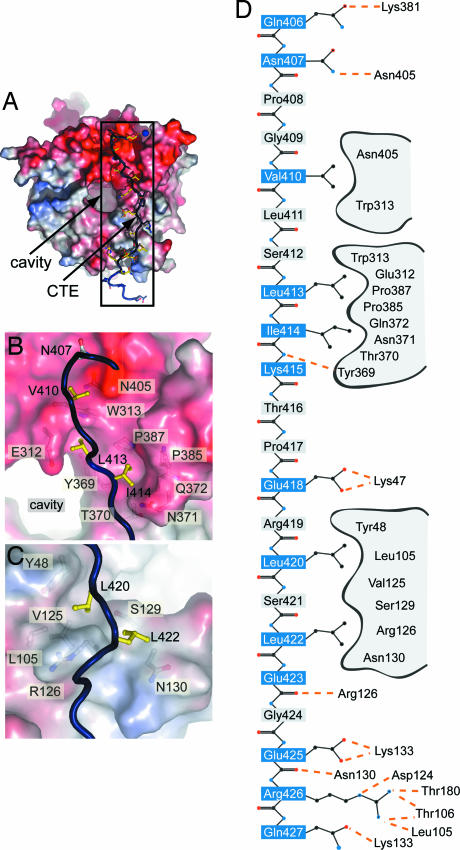
The 30-residue CTE binds in an extended conformation to the bottom face of the β-propeller and buries ≈15% (≈2,500 Å2) of the surface area of the β-propeller (Fig. 3A). The majority of the interactions between the β-propeller and the CTE are ionic in nature. Eight of the CTE residues form hydrogen bonds with residues of the β-propeller, whereas only five are involved in hydrophobic van der Waals contacts. Hydrogen bonds with the β-propeller are formed by the CTE residues Gln-406, Asn-407, Lys-415, Glu-418, Glu-423, Glu-425, Arg-426, and Gln-427, whereas the CTE residues Val-410, Leu-413, Ile 414, Leu-420, and Leu-422 form hydrophobic interactions. The side chains of the remaining residues of the CTE face away from the interface with the β-propeller (Fig. 3 B–D and SI Fig. 7 A and B).
Fig. 3.
CTE binding to the bottom face of the β-propeller. (A) The surface of the Nup214 β-propeller is colored according to the electrostatic potential from −10 kBT (red) to + 10 kBT (blue). The CTE is shown in blue coil representation with the side chains in ball-and-stick representation. The black box indicates the region magnified in D. (B) Hydrophobic interactions of CTE residues Val-410, Leu-413, and Leu-414 (yellow). (C) Interactions of Leu-420 and Leu-422 (yellow) with residues of the β-propeller. Hydrophobic pocket-forming residues are shown in gray. The surface of the β-propeller is colored as in A. (D) Schematic representation of the contacts between the β-propeller and the CTE. Hydrogen and ionic bonds are indicated by orange dashed lines and van der Waals contacts with gray grooves.
The carbonyl group of Gln-406 forms a water-mediated hydrogen bond to the ε-amino group of Lys-381, and the amide nitrogen atom of Asn-407 forms a hydrogen bond with the side-chain carbonyl group of Asn-405. Val-410 forms close van der Waals contacts with a groove in the β-propeller surface formed by Asn-405 and Trp-313 (Fig. 3B). Leu-413 and Ile-414 bind to the hydrophobic pockets formed by Glu-312, Trp-313, Tyr-369, and Pro-387, and Tyr-369, Thr-370, Asn-371, Gln-372, Pro-385, and Pro-387, respectively (Fig. 3B). The main-chain amide of Lys-415 interacts with the main-chain carbonyl of Tyr-369, and the carboxylate group of Glu-418 forms a salt bridge with the ε-amino group of Lys-47. The hydrophobic residues Leu-420 and Leu-422 are both lodged into a hydrophobic pocket formed by Tyr-48, Leu-105, Val-125, Arg-126, Ser-129, and Asn-130 (Fig. 3C). The main-chain carbonyl moiety of Glu-423 forms a hydrogen bond with Arg-126, whereas the side chain faces away from the surface. The carboxylate group of Glu-425 interacts with the side-chain amide moiety of Asn-130 and forms a salt bridge with the ε-amino group of Lys-133. Arg-426 forms the most intimate interactions with the β-propeller surface. The guanidinium group of Arg-426 interacts with the main-chain carbonyl moiety of Thr-180, the hydroxyl group of Thr-106, and the main-chain carbonyl of Leu-105 as well as with Asp-124 by means of a salt bridge. The side-chain carbonyl group of Gln-427 forms a hydrogen bond to the ε-amino group of Lys-133 (Fig. 3D).
CTEs have been found in other β-propeller-containing proteins. For example, in Coronin-1, a regulator of actin cytoskeletal dynamics in lymphocytes, the binding of the Coronin-1 CTE to the bottom face of its β-propeller is primarily achieved by intimate hydrophobic packing that involves a tryptophan and a tyrosine residue of the CTE (22). This interaction has been shown to be crucial for the stability of the protein (23). To determine whether the Nup214 CTE is required for the stability of the NTD, we expressed a C-terminal truncation mutant that contains the β-propeller (residues 1–405) but lacks the CTE (NTD1–405). NTD1–405 was indistinguishable from the crystallized NTD in its expression level, stability, and behavior on a gel-filtration column. This observation strongly suggests that the CTE is not required for the architectural integrity of the NTD. The highly charged nature of the CTE-β-propeller interface does not appear to lock the CTE in a bound conformation. Therefore, we hypothesized that the interaction between the CTE and the β-propeller is dynamic in nature, and we tested whether the CTE is able to dissociate from the β-propeller. We tested the binding of a synthetic 28-residue CTE peptide (residues 405–432) to the NTD1–405 β-propeller on a gel-filtration column, but we were not able to see any interaction (data not shown). Because isothermal calorimetry (ITC) has generally a much higher sensitivity, we then attempted to measure the binding affinity between the CTE peptide and the NTD1–405 with ITC. However, the two domains did not interact with measurable affinity under the condition tested, suggesting that the affinity was below the ITC detection limit (data not shown). The finding that the CTE is bound to the β-propeller in our crystal structure, together with our inability to measure the intermolecular binding affinity suggests that the interaction between the β-propeller and the CTE is entropically favored through intramolecular interactions. In addition, the CTE contains several putative phosphorylation sites, and phosphorylation of Ser-430 and Thr-437 of the Nup214 CTE was found in a large-scale characterization of HeLa nuclear phosphoproteins (24). Ser-430 is located at the end of the CTE, faces away from the β-propeller, and is accessible for phosphorylation (Fig. 3D). Thus, the CTE–β-propeller interaction may be regulated and dynamic in nature.
Comparison with the Yeast Nup159.
Although the ≈120-MDa vertebrate and the ≈60-MDa yeast NPC share a conserved architecture and use a similar number of nups (4), several vertebrate nups lack a homolog in yeast, undergo vertebrate specific modifications, or have several homologous nups in yeast (2). When we compare the sequence of the human Nup214 with its yeast homolog Nup159, it is clear that the various domains are rearranged (Fig. 1A).
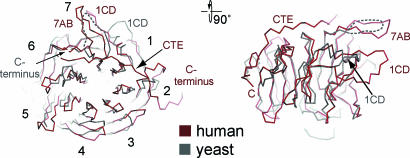
Recently, the crystal structure of the β-propeller of the yeast Nup159 was determined (25). However, although the expression construct of the yeast Nup159 contained additional residues at the C-terminal end of the core β-propeller, those residues were disordered in the crystal structure, suggesting that this region does not interact with the β-propeller. Because of the low sequence conservation between the human and yeast homologs, the prediction of the existence of the CTE in lower eukaryotes such as yeast is difficult (SI Fig. 8). The comparison between the structures reveals that, apart from the canonical β-propeller folds, both proteins are quite different. The yeast structure lacks the CTE as well as the significantly more extensive and complex decoration of the human NTD. When we compare the β-propeller core of the two homologs, we find that both align reasonably well with an rmsd of ≈1.5 Å (Fig. 4). A structural alignment of the Nup214 β-propeller with the only other available nup β-propeller structure, Nup133, shows that they align equally well and also with an rmsd of ≈1.5 Å. We furthermore tested how well other β-propeller proteins that have completely different functions, such as Gβ (21), the murine Coronin-1 (22), and the yeast Sif2 WD propeller (26), align and revealed that their β-propeller cores align with comparable rmsd values of ≈1.7Å. This analysis demonstrates that the human Nup214 β-propeller is not significantly more closely related to the β-propeller core of Nup159 than to the β-propeller of Nup133, Gβ, Coronin-1, or Sif2. Although only ≈280 residues are required to construct a minimal seven-bladed β-propeller fold, the nup β-propellers have been predicted to contain 400–500 residues (27). Thus, we speculate that the specific functions of the nup β-propeller are not conferred by their β-propeller core but rather by the unique structural features, such as long loop insertions and additional structural elements that decorate the core of the β-propeller fold and dominate the surface properties of the domain.
Fig. 4.
Structural comparison of the NTD of the human Nup214 and its yeast homolog Nup159. Cα trace of a structural superposition of the Nup214 NTD (ruby) and the Nup159 β-propeller (gray).
Conserved Features of the NTD.
The vast majority of β-propeller fold-containing proteins are known to mediate protein–protein interactions (18). Several surface areas have been found to serve as docking platforms. For example, the interblade grooves on the side of the β-propeller are often used in peptide interactions, whereas the central channel often binds ligands or prosthetic groups (18). The β-propeller fold imposes few restraints on its primary sequence and is able to tolerate insertions between the canonical β-strands. These extended loops protrude from the top and bottom face of the β-propeller and can create a variety of binding and regulation sites.
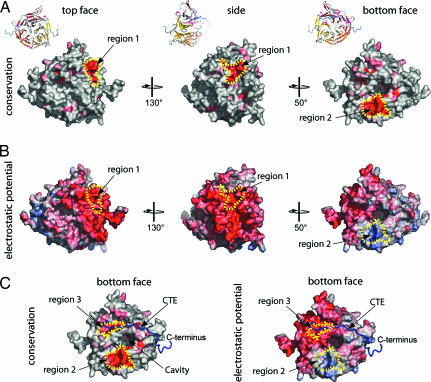
Several proteins have been found to interact directly with Nup214. The Nup88≈Nup214 NPC subcomplex is involved in the anchoring of the cytoplasmic filaments (28), whereas the recruitment of Ddx19 by Nup214 is a crucial step of the mRNA export pathway (16). To identify possible protein interaction sites on the surface of the NTD, we analyzed its conservation and electrostatic potential. Although the sequences of Nup214 homologs are not very conserved within eukaryotes, sequences can be aligned reliably within higher eukaryotes, and the majority of the conserved residues were found to be part of the core β-strands (SI Fig. 9). Nevertheless, three regions on the NTD surface were pinpointed. Region 1 covers approximately one-third of the side surface of the NTD (Fig. 5A and B) and has an invariant core formed by the 5D6A loop (Tyr-295), the 6BC loop (Glu-325), and the 6D7A loop (Asp-345, Arg-348, and Glu-350). The 6D7A loop of the Nup159 β-propeller has been shown to interact directly with the DEAD box helicase Dbp5, and a mutation of two residues within this loop (V323E/I326E) has been shown to disrupt the formation of a complex between the two proteins (25). Although the yeast and human homologs display considerable sequence conservation within this loop, residues that have been identified to be crucial for Nup159≈Dbp5 complex formation are not conserved in Nup214. In fact, one of the two Nup159 residues (V323E) that disrupt the Nup159≈Dbp5 complex when mutated is already a glutamate in the human Nup212/CAN β-propeller (Glu-350) (SI Fig. 8). The primary sequence of yeast Dbp5 and its human homolog Ddx19 are ≈45% identical, and although it seems plausible that the same region of the surface of the Nup214 β-propeller binds to Ddx19, the requirement of different residues for complex formation appears necessary.
Fig. 5.
Conserved features of the NTD. (A) Surface representation showing conservation of residues within higher eukaryotes. The conserved surface is shaded from gray (<70% identity) to red (100% identity) according to the alignment in SI Fig. 8. (B) Electrostatic potential of the NTD surface (colored as in Fig. 3A). (C) Surface representation of the bottom face of the Nup2141–405 showing the surface conservation within higher eukaryotes (Left) and electrostatic potential (Right). The Cα trace of the CTE is shown in a blue coil representation.
The other conserved regions, regions 2 and 3, are located at the bottom face of the β-propeller (Fig. 5C). Region 2 is formed by the 4AB loop, has a positive electrostatic potential, and is the most conserved region within higher eukaryotes; region 3 is formed by Phe-17, Trp-313, Trp-341, and Pro-387 and has a negative electrostatic potential (Fig. 5C). Interestingly, the 4AB loop within region 2 is lysine-rich and invariant within higher eukaryotes, but it shares no sequence conservation with any of its yeast homologs (SI Figs. 8 and 9). Thus, we speculate that a vertebrate-specific protein might bind to this positively charged, highly conserved 4AB loop. In addition, Nup214 has been found to be highly posttranslationally modified and, in particular, to be phosphorylated during interphase and hyperphosphorylated during mitosis (29). The sequence of the 4AB loop (WSPKGKQL) resembles the consensus site for a number of Ser/Thr protein kinases, and we identify Ser-201 within this loop as a putative serine phosphorylation site by using a biocomputational analysis with NetPhos (30) (SI Fig. 9).
Implications of the CTE in the Dynamic Reassembly of the NPC.
In the cell, many examples have been identified in which the interaction of two proteins is facilitated by an unstructured peptide segment (31). An intriguing possibility for the function of the CTE can be derived by comparing Nup214 with karyopherins (kaps), the mobile transport factors that facilitate nucleocytoplasmic transport. The binding of the NLS-containing cargo protein to kap-α is regulated by the heterodimerization of kap-α and kap-β. In the absence of kap-β, the N-terminal peptide segment of kap-α (auto-NLS) binds intramolecularly to its NLS-binding site, thereby preventing the NLS of cargo protein from binding. In the presence of kap-β, the heterodimerization between kap-α and kap-β, which is facilitated by the auto-NLS of kap-α, frees the binding site for the NLS of the cargo protein. After transport, the binding of Ran≈GTP to kap-β triggers the dissociation of the heterotrimeric complex by displacing the auto-NLS of kap-α from kap-β (SI Fig. 10A) (31).
Based on this system of regulation, we propose a model for the involvement of flexible peptide segments, such as the Nup214 CTE, in the assembly of the NPC (SI Fig. 10B). Because the CTE binds with low affinity to the essentially invariant bottom face of the β-propeller, we speculate that the NTD exists in two states: a closed state that we observe in our crystal structure with the CTE bound to the β-propeller, and a presumed open state, in which the CTE is released from the β-propeller. The binding of another protein to the CTE would release the CTE from the β-propeller and expose the highly conserved surface of the bottom face. In this open state, the conserved region 2 and 3 would then be available for the binding of yet another protein, resulting in the formation of a heterotrimeric complex. In contrast to the dissociation of the nuclear import complex through Ran·GTP binding, the complex could be dissociated by posttranslational modifications. Support for this idea comes from the observation that the CTE of Nup214 has been found to be phosphorylated at two sites (Ser-430 and Thr-437) in vivo (29). Furthermore, the bottom face of the β-propeller contains a putative phosphorylation site within the 4AB loop, suggesting that the heterotrimeric complex could be dissociated by Ser/Thr phosphorylation.
Conclusions
The NPC is one of the largest known proteinaceous assemblies in the cell. At present, we have a very limited knowledge of the architectural principles that govern the assembly of the NPC from the structural domains of the nups. Approximately two-thirds of the ≈30 nups contain predicted β-propeller or α-solenoid domains or a combination of both, and almost all of these nups contain predicted, unstructured regions directly adjacent to the structured domains (6). The results presented in this work strongly suggest that although the nup β-propeller domains share a conserved structural design, each one is uniquely decorated with structural elements that dictate their surface properties. Furthermore, the demonstrated lack of conserved sequence motifs within the nup β-propeller repeats makes the discrimination between the core elements and the unique features in structure predictions difficult. Ultimately, only high-resolution structural analysis can provide this information.
We suggest that unique structural features, such as the CTE, exist in a number of nups. Thus, it appears plausible that the fully assembled NPC is strapped together by numerous phosphorylation-dependent auto-NLS-like interactions. Such a model could help explain the close correlation of the reversible disassembly of the NPC to the cell cycle, which itself is tightly controlled by phosphorylation through an interplay of several Ser/Thr kinases. However, further structures of nups and studies of the interactions of Nup214 as well as cell cycle-dependent posttranslational modifications of Nup214 are required. The structure of the NTD presented here provides the framework for the design of in vivo studies that are aimed at revealing deeper insights into the involvement of Nup214 in the assembly of the NPC, the nuclear export of mRNAs, as well as leukemogenesis.
Methods
The details of molecular cloning, expression, purification, crystallization, x-ray diffraction data collection, and structure determination are described in the SI Methods. In short, the Nup214 NTD has been expressed by using a pET28a vector (GE Healthcare, Piscataway, NJ) modified to contain a PreScission protease-cleavable N-terminal hexahistidine tag (GE Healthcare) (32). Native and anomalous diffraction data of crystals of the NTD were collected at the National Synchrotron Light Source, Brookhaven National Laboratory (Upton, NY), beamline X9a, and the Advanced Light Source, Lawrence Berkeley National Laboratory (Berkeley, CA), beamline 8.2.1. Phases were determined in SHARP (33), followed by density modification in DM (34). A model including residues 1–434 was built with the program O (35) and refined with CNS (36). Data collection and refinement statistics are summarized in SI Table 1.
Supplementary Material
Acknowledgments
We dedicate this paper to Henning Friedrich. We thank the Kazusa DNA Research Institute for reagents, B. Manjasetty (National Synchrotron Light Source) and C. Ralston (Advanced Light Source) for excellent scientific support and help with x-ray measurements; J. Janz for help with ITC measurements; S. Etherton and S. Lawrie for help with editing the manuscript; and K. Hsia, M. Kampmann, I. Melcak, V. Nagy, A. Patke, and P. Stavropoulos for comments on the manuscript. Peptide synthesis was performed by The Rockefeller University Protein Center. This work was supported by a Predoctoral Fellowship from the Women in Science Foundation (to J.N.) and a Grant from the Leukemia and Lymphoma Society (to A.H.). G.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- CTE
C-terminal extended peptide segment
- ITC
isothermal calorimetry
- kap
karyopherin
- NPC
nuclear pore complex
- NTD
N-terminal domain
- nup
nucleoporin.
Footnotes
The authors declare no conflict of interest.
Data deposition: The structure factors and atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2OIT).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610828104/DC1.
References
- 1.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichelt R, Holzenburg A, Buhle EL, Jr, Jarnik M, Engel A, Aebi U. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rout MP, Blobel G. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. J Mol Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 6.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hetzer MW, Walther TC, Mattaj IW. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 8.Kraemer D, Wozniak RW, Blobel G, Radu A. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, Stul M, Dutta B, Boeckx N, et al. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 11.Fu GK, Grosveld G, Markovitz DM. Proc Natl Acad Sci USA. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suntharalingam M, Wente SR. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vellieux FM, Huitema F, Groendijk H, Kalk KH, Jzn JF, Jongejan JA, Duine JA, Petratos K, Drenth J, Hol WG. EMBO J. 1989;8:2171–2178. doi: 10.1002/j.1460-2075.1989.tb08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paoli M. Prog Biophys Mol Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 19.Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 20.Fulop V, Jones DT. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 21.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 22.Appleton BA, Wu P, Wiesmann C. Structure (London) 2006;14:87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Gatfield J, Albrecht I, Zanolari B, Steinmetz MO, Pieters J. Mol Biol Cell. 2005;16:2786–2798. doi: 10.1091/mbc.E05-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weirich CS, Erzberger JP, Berger JM, Weis K. Mol Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Cerna D, Wilson DK. J Mol Biol. 2005;351:923–935. doi: 10.1016/j.jmb.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 27.Berke IC, Boehmer T, Blobel G, Schwartz TU. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin JC. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 30.Blom N, Gammeltoft S, Brunak S. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 31.Hoelz A, Blobel G. Nature. 2004;432:815–816. doi: 10.1038/432815a. [DOI] [PubMed] [Google Scholar]
- 32.Hoelz A, Nairn AC, Kuriyan J. Mol Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 33.de la Fortelle ED, Bricogne G. Methods Enzymol. 1997;276:476–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project No 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore G. M., DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.