Abstract
Rhodococcus sp. strain RHA1, a soil bacterium related to Mycobacterium tuberculosis, degrades an exceptionally broad range of organic compounds. Transcriptomic analysis of cholesterol-grown RHA1 revealed a catabolic pathway predicted to proceed via 4-androstene-3,17-dione and 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3,4-DHSA). Inactivation of each of the hsaC, supAB, and mce4 genes in RHA1 substantiated their roles in cholesterol catabolism. Moreover, the hsaC− mutant accumulated 3,4-DHSA, indicating that HsaCRHA1, formerly annotated as a biphenyl-degrading dioxygenase, catalyzes the oxygenolytic cleavage of steroid ring A. Bioinformatic analyses revealed that 51 rhodococcal genes specifically expressed during growth on cholesterol, including all predicted to specify the catabolism of rings A and B, are conserved within an 82-gene cluster in M. tuberculosis H37Rv and Mycobacterium bovis bacillus Calmette–Guérin. M. bovis bacillus Calmette–Guérin grew on cholesterol, and hsaC and kshA were up-regulated under these conditions. Heterologously produced HsaCH37Rv and HsaDH37Rv transformed 3,4-DHSA and its ring-cleaved product, respectively, with apparent specificities ≈40-fold higher than for the corresponding biphenyl metabolites. Overall, we annotated 28 RHA1 genes and proposed physiological roles for a similar number of mycobacterial genes. During survival of M. tuberculosis in the macrophage, these genes are specifically expressed, and many appear to be essential. We have delineated a complete suite of genes necessary for microbial steroid degradation, and pathogenic mycobacteria have been shown to catabolize cholesterol. The results suggest that cholesterol metabolism is central to M. tuberculosis's unusual ability to survive in macrophages and provide insights into potential targets for novel therapeutics.
Keywords: catabolic pathway, oxygenase, Rhodococcus, steroid degradation
Rhodococci are a genus of GC-rich, mycolic acid-producing bacteria within the order Actinomycetales that includes Mycobacterium (1). Rhodococci degrade a broad range of organic compounds, particularly hydrophobic ones, thereby playing a key role in the global carbon cycle. Analysis of the 9.7-Mb genome of RHA1 (www.rhodococcus.ca) reveals that this organism harbors a diverse armamentarium of enzymes (2), consistent with the catabolic versatility of the genus. These catabolic activities, together with robust and rapid rhodococcal growth, are of great interest to pharmaceutical, environmental, chemical, and energy industries (3).
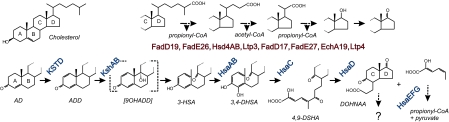
The bacterial catabolism of steroids has attracted considerable attention (3) in part as a potential means of producing bioactive steroids from natural, low-cost sterols such as β-sitosterol and cholesterol. A pathway responsible for the aerobic degradation of the latter via 4-androstene-3,17-dione (AD) and 3-hydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3-HSA) may be pieced together from biochemical and genetic studies in diverse bacteria (Fig. 1). In some Mycobacterium (4) and Rhodococcus (5, 6) species, the aliphatic side chain at C17 is removed via a process similar to β-oxidation involving progressively shorter carboxylic acids. In these strains, 3-ketosteroid Δ1-dehydrogenase (KSTD) and 3-ketosteroid 9α-hydroxylase catalyze the opening of ring B and aromatization of ring A to yield 3-HSA (3, 7–9). The subsequent degradation of 3-HSA to 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid (DOHNAA) via oxygenolytic cleavage of ring A is specified by the tes genes in the testosterone-degrading strain Comamonas testosteroni TA441 (10, 11). In Rhodococcus equi, the propionate moiety of DOHNAA is removed via β-oxidation (12). Many of the genes involved in steroid catabolism have yet to be identified, and many of the pathway enzymes are poorly characterized, particularly those involved in degrading the bicycloalkanone originating from rings C and D. Detailed knowledge of steroid catabolism is essential to engineering strains for the biotransformation of sterols.
Fig. 1.
The deduced cholesterol catabolic pathway of Rhodococcus sp. RHA1, M. tuberculosis H37Rv, and M. bovis bacillus Calmette–Guérin. The enzymatic steps of side-chain degradation and ring opening are depicted. The latter are important for H37Rv survival in the macrophage (Fig. 2). Dashed arrows indicate multiple enzymatic steps. The compound in brackets undergoes nonenzymatic hydrolysis. Genes responsible for the degradation of rings C and D in RHA1 are not conserved in H37Rv or bacillus Calmette–Guérin. ADD, 1,4-androstadiene-3,17-dione; 9OHADD, 9α-hydroxy-1,4-androstadiene-3,17-dione; KshAB, 3-ketosteroid 9α-hydroxylase.
Recent genomic analyses revealed that rhodococci may be useful models for many mycobacterial processes: ≈60% of the 3,999 genes of Mycobacterium tuberculosis H37Rv are conserved in RHA1, including many of unknown function (2). M. tuberculosis is the leading cause of mortality from bacterial infection, killing 2 million to 3 million people worldwide each year, and extensive drug resistant strains such as XDR-TB are now emerging (ref. 13 and www.who.int/mediacentre/factsheets/fs104/en/index.html). One poorly characterized aspect of mycobacterial physiology that contributes to the prevalence of tuberculosis is the bacterium's unusual ability to survive for long periods of time, and even to replicate, in the normally hostile environment of the macrophage (14, 15). The mechanisms enabling this persistence are poorly understood, but are logical targets for novel therapeutic agents. Transposon site hybridization (TraSH), a genomewide microarray-based technique, identified 126 genes that appear to be necessary for survival of H37Rv in macrophages under conditions that model the immune response (16) and many others that are critical for in vivo survival in mice (17). Further, transcriptomic studies have identified suites of genes that are specifically up-regulated during survival in the macrophage (18). Despite the importance of these genes, their physiological roles are largely unknown.
We investigated the cholesterol catabolic pathway in RHA1 by comparing the transcriptomes of cholesterol- and pyruvate-grown cells. Targeted gene deletion was used to substantiate key catabolic steps. Bioinformatic analyses enabled annotation of many of the cholesterol catabolic genes and also revealed their presence in M. tuberculosis and Mycobacterium bovis. Conditions to grow M. bovis on cholesterol were developed, and the expression of two pathway genes was shown by quantitative RT-PCR. Two of the M. tuberculosis pathway enzymes were heterologously produced and shown to efficiently catalyze the predicted transformations of steroid ring A. The results are discussed with respect to the survival of M. tuberculosis in the macrophage.
Results
The Cholesterol Transcriptome of RHA1.
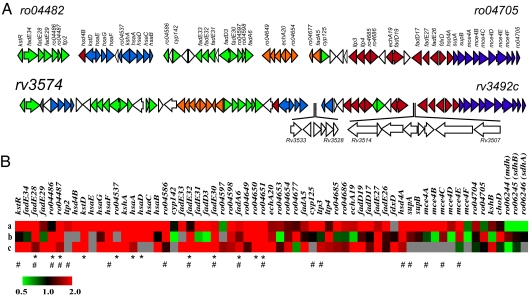
In liquid medium containing 2 mM cholesterol as the sole organic substrate, RHA1 grew to a density of 2 × 108 cells per ml. Microarray analysis revealed 572 genes that were up-regulated at least 2-fold during growth on cholesterol compared with on pyruvate. Many of the up-regulated genes are scattered throughout the 9.7-Mb genome (www.rhodococcus.ca) and likely reflect a general physiological adaptation of the bacterium to growth on a highly hydrophobic, polycyclic compound. However, six clusters of up-regulated genes were clearly discerned [supporting information (SI) Table 3]. The most striking of these was a cluster of 51 genes that occur within a 235-kb stretch of RHA1's 7.9-Mb chromosome (ro04482–ro04705; Fig. 2 A). As discussed below, these genes encode proteins with significant sequence identity with enzymes involved in the catabolism of steroid rings A and B by C. testosteroni TA441 (10, 11) and Rhodococcus erythropolis SQ1 (7, 8). A second cluster of chromosomal genes (ro06687–ro06698) also appear to be involved in cholesterol catabolism. The four other gene clusters (ro00440–ro00453, ro03461–ro03464, ro08053–ro08060, and ro10126–ro10162) do not appear to be directly involved in steroid catabolism, and some are described elsewhere (19).
Fig. 2.
The cholesterol catabolic genes of Rhodococcus sp. RHA1 and M. tuberculosis H37Rv: comparison of their organization and their activities in different studies. (A) Genes in the physical map are color-coded according to assigned function: purple, uptake; red, side-chain degradation; blue, cleavage of rings A and B; orange, degradation of the DOHNAA propionate moiety; green, degradation of rings C and D. White arrows represent genes for which no reciprocal homologue is present. The nucleotide sequences of the M. tuberculosis H37Rv and M. bovis bacillus Calmette–Guérin clusters share 96% identity. (B) Heat map indicating correlation between gene expression (fold difference) during growth of RHA1 on cholesterol versus pyruvate (a), effect of gene disruption on H37Rv survival in IFN-γ-activated macrophages according to TraSH analysis (reciprocal of ratio) (16) (b), and gene expression in H37Rv after 48 h of growth in IFN-γ-activated macrophages (18) (c). M. tuberculosis genes predicted as essential for survival in the macrophage (16, 32) and in vivo in mice (17) are indicated by * and #, respectively.
Annotation of Cholesterol Catabolic Genes.
Among the genes that were up-regulated during growth on cholesterol, the annotation of those predicted to specify cholesterol catabolism is summarized in SI Table 4 Most of these comprise the 51 genes of the ro04482–ro04705 cluster (Fig. 2 A), and most of the encoded proteins have such sufficient sequence similarity to well characterized enzymes that their function can be confidently predicted. Thus, sequences of KshA, KshB, and KSTD (Fig. 1) are 40–69% identical to those of orthologs in R. erythropolis SQ1 (SI Table 4) that act sequentially to transform AD to 3-HSA (3, 7, 8). Further degradation of 3-HSA was predicted to be specified by seven genes, annotated here as hsa, that are clustered with kstD and kshA (Figs. 1 and 2 A). The encoded proteins share significant amino acid sequence similarity (30–60%;SI Table 4) with the tes-encoded enzymes of C. testosteroni TA441 that transform 3-HSA during growth on testosterone (10, 11). HsaC and HsaD were previously annotated as BphC5 and BphD2 in RHA1, respectively, because of the former's ability to catalyze the extradiol cleavage of 2,3-dihydroxybiphenyl (DHB) and their sequence similarity to the corresponding biphenyl catabolic enzymes (20). However, HsaC shares greater sequence identity with TesB of C. testosteroni TA441 (11) than with extradiol dioxygenases that preferentially cleave DHB. Moreover, quantitative RT-PCR analyses confirmed that hsaC was up-regulated 15.4-fold during growth of RHA1 on cholesterol as compared with either biphenyl or pyruvate.
It was more difficult to assign specific roles to the numerous β-oxidation genes of the ro04482–ro04705 cluster. Most of these gene products share greatest sequence identity with homologs that occur in M. tuberculosis H37Rv and were annotated accordingly (SI Table 4).One set of these genes (hsd4A, hsd4B, fadD19, fadE26, and ltp3) is highly up-regulated and encodes all of the enzymes necessary to perform one full cycle of β-oxidation. Hsd4A and Hsd4B share intriguing sequence similarity with the eukaryotic multifunctional 17β-hydroxysteroid dehydrogenase IV (17βHSD4) involved in peroxisome-related disorders (21). Hsd4A is homologous to the N-terminal domain of 17βHSD4, which acts as a 17β-hydroxysteroid dehydrogenase and, with branched fatty acids and bile acids, as a d-3-hydroxyacyl-CoA dehydrogenase. Hsd4B is homologous to the central domain of 17βHSD4, which is a 2-enoyl acyl-CoA hydratase proposed to be involved in cholesterol side-chain shortening. Accordingly, we predict that these RHA1 genes specify at least one cycle of β-oxidative transformation of the C17 side chain to propionyl-CoA and acetyl-CoA. A second near-complete set of β-oxidation genes (echA19, fadD17, fadE27, and ltp4) are up-regulated to a lesser extent, but are likely also involved in side-chain degradation. The bifunctional Hsd4A likely transforms the 17β-hydroxysteroid resulting from cleavage of the cholesterol side chain.
A third cluster of up-regulated genes related to β-oxidation, including fadE28, is similar to those involved in testosterone catabolism by C. testosteroni TA441 (10, 11). These genes may be involved in the degradation of the DOHNAA originating from steroid rings C and D (Fig. 1), as this part of the molecule is common to testosterone and cholesterol, whereas the C17 side chain is not. This set of genes is preceded by a gene encoding a TetR-type transcriptional regulator similar to the KstR (32% identity) repressor of kstD (7), suggesting that the RHA1 genes are also regulated by steroids. The propionate moiety of DOHNAA is likely degraded by β-oxidation encoded by the gene cluster that includes echA20. This cluster includes genes encoding a two-subunit, ATP-dependent CoA transferase of the type thought to initiate β-oxidation (22).
The ro04482–ro04705 cluster also includes eight genes that appear to encode a multicomponent cholesterol uptake system: supAB and mce4ABCDEF. Conserved domain data revealed the presence of a domain related to an ABC-transport system involved in resistance to organic solvents in both SupA and SupB (23). The mce cluster is one of two such clusters in RHA1 that are highly similar to the four sets of “mammalian cell entry” (mce) genes of M. tuberculosis H37Rv (24). Mce proteins are critical virulence factors in M. tuberculosis (16), although the exact role of these genes is unknown. Heterologously expressed mce1A enhanced the entry of Escherichia coli into nonphagocytic HeLa cells (25), whereas mce1− and mce4− strains of M. tuberculosis H37Rv showed attenuated survival in mice (26). It has been proposed that Mce proteins are components of transport systems that translocate lipids between the bacterial cell and its host (26). Consistent with this proposed role, Mce1A is expressed at the cell surface of M. tuberculosis H37Rv (27). Indeed, signal sequences are predicted for all of the Mce4 proteins of RHA1 except Mce4C [SignalP (28)], indicating that these proteins are secreted or surface-exposed proteins. In summary, the 51 up-regulated genes of the ro04482–ro04705 cluster appear to include all of those necessary to specify the catabolism of cholesterol to DOHNAA.
The separate ro06687–ro06698 gene cluster, induced on cholesterol, includes genes typical of those encoding cycloalkanone catabolism (SI Table 4). These include ro06698 and ro06693, which encode a probable monooxygenase and lactone hydrolase, respectively. We predict that these genes are involved in degrading the steroid ring D of DOHNAA.
Annotation of the cholesterol catabolic genes further revealed that these genes are but one of four sets in RHA1 that appear to specify the catabolism steroid-like compounds. Each of these sets encodes homologs of all ring-degrading enzymes: 3-ketosteroid 9α-hydroxylase, KstD, HsaAB, HsaC, HsaD, and at least one cyclohexanone monooxygenase. Sequence analyses revealed that all of the KshA homologs (ro02490, ro04538, ro05811, and ro09003) share at least 52% amino acid sequence identity with KshA of R. erythropolis SQ1 (8). Phylogenetic analyses (SI Fig. 3A) revealed that these enzymes define a subclass of Rieske nonheme oxygenases. Similarly, all of the HsaC homologs (ro02488, ro04541, ro05803, and ro09005) share at least 37% amino acid sequence identity and key active-site residues with TesB of C. testosteroni TA441. These enzymes constitute a subclass of type I extradiol dioxygenases (SI Fig. 3B) distinct from those involved in biphenyl and naphthalene catabolism. Similar analyses of HsaA and HsaD revealed comparable relationships (data not shown): for each type of enzyme, the known steroid-degrading homologs constitute a distinct subclass. None of the additional three sets of genes were up-regulated in RHA1 during growth on cholesterol and so appear to encode degradation of other steroids.
Mutational Analysis of Cholesterol Catabolic Genes.
The critical role of Mce4A–Mce4F and SupAB proteins in cholesterol catabolism was confirmed by unmarked in-frame gene deletion of the entire mce4ABCDEF gene cluster and the supAB genes, respectively, in RHA1. Both the mce4 and sup mutants were severely impaired in the ability to grow on cholesterol in liquid mineral medium (Table 1). By contrast, growth on AD was not affected, supporting our hypothesis that Mce4 and SupAB are specifically involved in the uptake of cholesterol in RHA1. The doubling times of RHA1 and the mutants on AD (≈12 h) were approximately three times longer than on pyruvate or benzoate.
Table 1.
Growth yields of RHA1 and mutants on different organic substrates
| Protein | Cholesterol, 1 mM | AD, 1 mM | Pyruvate, 20 mM | Benzoate, 20 mM |
|---|---|---|---|---|
| WT | 73 (5) | 77 (12) | 170 (20) | 470 (60) |
| ΔsupAB | 1 (1) | 75 (11) | 170 (20) | 520 (50) |
| Δmce4 | 3 (1) | 78 (14) | 150 (40) | 440 (90) |
Growth yields are expressed as micrograms of protein per milliliter of culture medium and are averages of triplicate cultures.Values in parentheses are standard errors.
To substantiate the predicted role of HsaC in catalyzing the extradiol cleavage of 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3,4-DHSA), a catechol, hsaC was deleted. In liquid media, the hsaC− mutant grew on cholesterol at a rate that was 60% that of the WT strain and developed a pink color. By contrast, growth on pyruvate was not affected. The slower growth on cholesterol may be caused by either degradation of the C17 side chain or complementary activity of one of the HsaC homologs in RHA1 (SI Fig. 3B). The pink color is consistent with the accumulation and nonenzymatic oxidation of a catechol. To identify the latter, metabolites were extracted from the supernatant of hsaC− cells incubated in the presence of cholesterol. HPLC analysis revealed a major metabolite, which, when derivatized with tri-methyl-silane (TMS), yielded a compound with a molecular ion m/z = 460 (SI Fig. 4). The molecular ion and its fragmentation pattern correspond to those predicted for TMS-derivatized 3,4-DHSA. Finally, transformation of the metabolite with HsaCH37Rv as described below yielded a product with a pH-dependent spectrum essentially identical to that reported for 4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid (4,9-DSHA) (9) (ε392 = 7.64 mM−1·cm−1 at pH 8.0), confirming the metabolite's identity as 3,4-DHSA (Fig. 1).
Conservation of the Cholesterol Catabolic Pathway in Mycobacteria.
Further bioinformatic analyses revealed that 58 genes of the ro04482–ro04705 cluster in RHA1, including the 51 that were up-regulated during growth on cholesterol, are conserved together with much of their putative operonic structure within an 82-gene cluster in the genomes of M. tuberculosis H37Rv (Rv3492c–Rv3574; Fig. 2 A) and M. bovis bacillus Calmette–Guérin (Bcg3556c–Bcg3639; www.sanger.ac.uk/Projects/M_bovis) as well as within an 80-gene cluster in Mycobacterium avium (subsp. paratuberculosis) (Map0571–Map0491; ref. 29). As noted above, these genes appear to be sufficient to specify the uptake of cholesterol, the β-oxidation of the branched side chain at C17, and the catabolism of rings A and B to central metabolites via 3-HSA to yield DOHNAA (Fig. 1). The sequence identities of the RHA1, H37Rv, and bacillus Calmette–Guérin homologs are summarized in SI Table 4 Phylogenetic analyses revealed that among the four sets of steroid-degrading enzymes in RHA1 the mycobacterial enzymes are most similar to those involved in cholesterol catabolism (SI Fig. 3).
Cholesterol Catabolism in M. bovis Bacillus Calmette–Guérin.
Initial attempts to grow bacillus Calmette–Guérin on cholesterol as the sole energy source met with limited success, as for other pathogenic mycobacteria (30). However, with a liquid minimal medium containing asparagine, citrate, and Triton (18), the final growth yield of bacillus Calmette–Guérin was proportional to the initial concentration of cholesterol in the medium. Thus, in medium supplemented with 0, 0.25, and 0.5 mM cholesterol, respectively, the overall protein yields were 22 ± 7, 46 ± 9, and 70 ± 4 μg/ml. Further modification of the medium to reflect host factors or improve the availability of the cholesterol to the bacterium may improve growth.
To investigate whether the predicted cholesterol catabolic pathway is involved in this growth of bacillus Calmette–Guérin, quantitative RT-PCR analyses were performed on kshA and hsaC with sigA as a control. Normalized transcript levels were significantly higher in cultures growing on cholesterol (n = 4) than on glucose (n = 5) for both kshA (P < 0.005) and hsaC (P < 0.05), with relative fold differences of 3.7 and 2.4, respectively. Similar results were observed when comparing cholesterol- to pyruvate-grown cells. The relative fold differences for kshA and hsaC were very similar to the expression ratios determined for these genes (4.6 and 2.1, respectively) using the microarray to compare RHA1 growing on cholesterol versus on pyruvate (SI Table 3); although, a slightly higher fold difference was determined for hsaC in RHA1 with quantitative RT-PCR. The relative fold differences for kshA and hsaC were also very similar to those determined for these genes using a microarray to compare H37Rv growing in macrophages versus in vitro (18).
The Catalytic Activities of HsaCH37Rv and HsaDH37Rv.
To substantiate the predicted cholesterol catabolic pathway in M. tuberculosis H37Rv, the activities of two central enzymes, HsaCH37Rv and HsaDH37Rv, were investigated. These enzymes were targeted in part because they were previously annotated as putative biphenyl-degrading enzymes (20). Accordingly, HsaCH37Rv and HsaDH37Rv were heterologously expressed in E. coli, and their steady-state kinetic parameters were evaluated with cell extracts. As summarized in Table 2, the enzymes preferentially transformed cholesterol metabolites as compared with biphenyl metabolites. Specifically, cell extracts containing HsaCH37Rv catalyzed the extradiol cleavage of 3,4-DHSA with an apparent specificity 44-fold higher than for DHB. Similarly, extracts containing HsaDH37Rv catalyzed the hydrolysis of 4,9-DSHA with an apparent specificity 34-fold higher than for 2-hydroxy-6-oxo-6-phenylpentadienoate (HOPDA). Equivalent extracts prepared from cells that contained the empty vector did not detectably transform either the steroid or biphenyl metabolites. These results strongly support the predicted roles of the mycobacterial enzymes in steroid metabolism and also indicate that the aliphatic side chain of cholesterol is removed before ring degradation.
Table 2.
Steady-state kinetic parameters of HsaCH37Rv and HsaDH37Rv for steroid and biphenyl metabolites
| Enzyme | Substrate | Km, μM | Vmax, μM·s−1 | Vmax/ Km, s−1 |
|---|---|---|---|---|
| HsaCH37Rv | 3,4-DHSA | 0.9 (0.5) | 12 (4) | 790 (370) |
| DHB | 8.5 (0.8) | 2.5 (0.4) | 18 (3) | |
| HsaDH37Rv | 4,9-DSHA | 4 (1) | 0.06 (0.02) | 1.0 (0.2) |
| HOPDA | 19 (6) | 0.009 (0.003) | 0.028 (0.007) |
Parameters were normalized to the amount of cellular extract (milligrams of protein content) used in the assays. Values in parentheses represent standard errors.
Discussion
The current study identified clusters of genes that encode the catabolism of cholesterol in RHA1. These were initially identified through bioinformatic analyses of genes that were up-regulated during growth on cholesterol. Moreover, the involvement of Mce4 and SupAB proteins in cholesterol catabolism and the role of HsaCRHA1, an extradiol dioxygenase, were substantiated by using gene deletion and characterization of the resultant mutants. Steroids such as cholesterol are ubiquitous in plants, animals, and some microbes and likely comprise an important energy source for saprophytic bacteria, particularly actinomycetes that efficiently use hydrophobic substrates. Although various aspects of steroid catabolism have been described in different bacteria (4 –12,22), in this study the genes of an entire catabolic pathway are delineated in a single organism. The identified genes include several involved in sterol uptake and side-chain degradation that are particularly good targets for cell and enzyme engineering studies. Thus, sterol uptake is believed to be rate-limiting, yet its mechanism is poorly understood. Similarly, efficient sterol side-chain degradation is critical for high yield production of steroid intermediates, particularly as most sterols used in microbial transformations consist of mixtures of compounds with slightly different side chains that are transformed with different efficiencies. Overall, this study facilitates the development of whole-cell biotransformation processes for the synthesis of industrially relevant steroid compounds.
A second important contribution of the current study is the discovery that the cholesterol catabolic pathway is conserved in related pathogenic actinomycetes, including M. tuberculosis, M. bovis, and M. avium. Thus, the latter appear to have retained the capacity for cholesterol metabolism and exploited it to survive in their hosts. Consistent with our bioinformatic predictions, M. bovis bacillus Calmette–Guérin used cholesterol as a carbon and energy source, and genes encoding the ring-degrading enzymes KshA and HsaC were up-regulated during this utilization. The substrate of the pathway in M. tuberculosis was verified by demonstrating the apparent specificity of HsaCH37Rv, an extradiol dioxygenase, and HsaDH37Rv, a C C bond hydrolase, for the steroid metabolites. These enzymes had been annotated as a probable DHB dioxygenase (NP_218085) and a HOPDA (CAB07143), respectively. The current study further suggests that these enzymes do not play a direct role in mycobacterial cell wall synthesis as recently suggested (31). Of the pathway proteins conserved in RHA1 and M. tuberculosis those with the lowest amino acid sequence identities are the Mce4 proteins. It is possible that the latter have different functions in the two organisms. However, our findings in RHA1 are consistent with the recent proposal that the supAB and mce4 genes encode a lipid-transport system (26). Moreover, some of the genes that were functionally linked to this system in that study include several cholesterol catabolic genes. Thus, the low sequence identities of the RHA1 and mycobacterial Mce4 homologs may instead reflect the different environments from which these two strains must scavenge cholesterol.
C bond hydrolase, for the steroid metabolites. These enzymes had been annotated as a probable DHB dioxygenase (NP_218085) and a HOPDA (CAB07143), respectively. The current study further suggests that these enzymes do not play a direct role in mycobacterial cell wall synthesis as recently suggested (31). Of the pathway proteins conserved in RHA1 and M. tuberculosis those with the lowest amino acid sequence identities are the Mce4 proteins. It is possible that the latter have different functions in the two organisms. However, our findings in RHA1 are consistent with the recent proposal that the supAB and mce4 genes encode a lipid-transport system (26). Moreover, some of the genes that were functionally linked to this system in that study include several cholesterol catabolic genes. Thus, the low sequence identities of the RHA1 and mycobacterial Mce4 homologs may instead reflect the different environments from which these two strains must scavenge cholesterol.
Several lines of evidence indicate that the identified steroid catabolic pathway is essential for the survival of M. tuberculosis in the macrophage. First, 41 of the pathway genes, including those specifying catabolism of rings A and B, are among those specifically up-regulated during survival in the macrophage (Fig. 2 B and ref. 18). Second, TraSH analyses predict that at least 11 of the pathway genes are essential for M. tuberculosis H37Rv to survive in the macrophage under conditions that model the immune response (Fig. 2B) (16). Most of the 11 encode enzymes such as KSTD, HsaA, and HsaD, which are involved in the degradation of steroid rings A and B (Fig. 1). Intriguingly, cholesterol catabolic genes that were not identified in TraSH studies have functions that may be complemented by other similar genes in M. tuberculosis H37Rv. These include KshB and HsaB, the respective reductase components of the AD(D) and the 3-HSA hydroxylases. Some of the TraSH mutants, such as mce4, displayed a progressive in vivo growth defect 2–4 weeks after infection in mice (17). Moreover, the essentiality of some of these genes has been substantiated. Thus, a ΔyrbE4A (i.e., supA) and Δmce4 mutants show attenuated survival of M. tuberculosis H37Rv in mice (17, 26). Similarly, inactivation mt3626 of M. tuberculosis CDC1551 (rv3527 in H37Rv; Fig. 2 B), a gene of unknown function adjacent to kshA and clustered with the hsa genes, had an impaired ability to arrest phagosome acidification and resulted in attenuated survival (32). Clearly, the essential nature of the cholesterol catabolic genes needs to be further substantiated. However, the available evidence suggests that cholesterol uptake and metabolism are important for M. tuberculosis to be able to persist in the macrophage for longer periods of time.
The deduced cholesterol catabolic pathway is consistent with at least two features of M. tuberculosis pathogenicity. First, cholesterol is essential for the phagocytosis of the bacterium by the macrophage and the inhibition of phagosome maturation (33–35). For example, depletion of cholesterol from macrophages abrogates the receptor-specific uptake of mycobacteria. Moreover, cholesterol depletion overcomes the block in phagosome maturation of M. avium-infected macrophages (35), further suggesting that cholesterol might play a similar role in other mycobacterial pathogens. Second, the large number of oxygenases in the pathway is consistent with the observation that tuberculosis infections are associated with the most O2-rich sites within the body (36). More specifically, the cholesterol catabolic genes encode six oxygenases, including two associated cytochromes P450 of unknown function. Reactivation of the disease occurs most frequently in the upper pulmonary lobes, likely the most oxygenated regions of the body (36).
At least two differences between the deduced cholesterol catabolic pathways in RHA1 and the pathogenic mycobacteria suggest distinct metabolism of cholesterol rings C and D. First, the Baeyer-Villiger monooxygenase and hydrolase typically associated with the ring fission of cycloalkanones, and whose genes are up-regulated in the RHA1 cholesterol transcriptome, are not conserved in the mycobacteria. Second, the mycobacterial hsa operon includes an N-acetyl transferase gene (24, 31). Thus, it is possible that pathogenic mycobacteria transform this portion of the cholesterol molecule for an alternate function such as signaling or cell wall integrity. Moreover, the cholesterol metabolic enzymes reported herein may also transform other host steroids or their derivatives, such as vitamin D, recently shown to mediate an innate immune response to mycobacteria (37). Nevertheless, the identified mycobacterial pathway transforms most of the cholesterol molecule to central metabolites, consistent with growth of bacillus Calmette–Guérin on cholesterol in vitro and suggesting that the sterol is an important source of energy for M. tuberculosis during its survival in the macrophage. The essential nature of the cholesterol catabolic enzymes in vivo makes them promising targets for the development of therapeutic agents to combat XDR-TB and other strains, particularly as many of these enzymes have no human homolog.
Materials and Methods
Bacterial Growth.
RHA1 was grown at 30°C on a shaker in one of two minimal media: W minimal salt medium (38) plus 20 mM pyruvate or 2 mM cholesterol or a similar medium supplemented with a different mineral solution (39) plus cholesterol, AD, pyruvate, or benzoate as indicated. RHA1 cells were harvested at midlog phase (OD600 of 1.0 for pyruvate and 2.0 for cholesterol). Bacillus Calmette–Guérin was grown at 37°C on a tube roller (10 rpm) in screw-capped 15-ml vials filled with 10 ml of liquid medium containing 0.5 g/ml asparagine, 1 g/ml KH2PO4, 2.5 g/ml Na 2PO4, 10 mg/l MgSO4·7H2O, 0.5 mg/liter CaCl2, 0.1 mg/liter ZnSO4, 50 mg/liter ferric ammonium citrate, and 0.5 ml/liter Triton wR1339 (Tyloxapol) (18) plus the indicated amount of cholesterol, 10 mM pyruvate, or 10 mM glucose. Total protein content of cultures was determined in cells disrupted by sonication (10 cycles of 30 s at 6 μm) by using the Bradford protein assay (BioRad, Hercules, CA) and BSA as standard.
RNA Extraction and Microarray.
RNA was isolated from RHA1 as described (19). RNA was similarly isolated from bacillus Calmette–Guérin except that both the RNeasy Plus and RNeasy Mini Kits (Qiagen, Valencia, CA) were used, and the sample was treated with 2 units of TURBO DNase (Ambion, Austin, TX). The RHA1 transcriptome was analyzed by using indirectly labeled cDNA and a microarray containing 70-mer probes for 8,313 genes as described (19). Data were analyzed by using GeneSpring (Agilent Technologies, Santa Clara, CA) and MeV 3.1 (The Institute for Genomic Research, Rockville, MD). For each condition, RNA was extracted from each of three independently grown cultures. Data were averaged and normalized by using Locally Weighted Linear Regression (Lowess). Details of the microarray design, transcriptomic experimental design and transcriptomic data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo).
Quantitative RT-PCR.
RT-PCR was performed as described (19) with TaqMan probes, and cDNA was synthesized by using the ThermoScript RT-PCR System (Invitrogen, Carlsbad, CA) and random hexamers. All oligonucleotide and probe sequences are provided in SI Table 3. The gene-encoding DNA polymerase IV and σ A were used as internal standards in the multiplex reactions performed by using RHA1 and bacillus Calmette–Guérin cDNA, respectively (19). The Ct values were normalized (ΔCt) by subtracting those of the internal standard. Significant differences in ΔCt values were tested by using a two-sample t test assuming unequal variances. Relative fold differences were calculated as 2−ΔΔCt, where ΔΔCt = ΔCt treatment − ΔCt control.
Gene Replacement and Deletion.
The hsaC gene was replaced in RHA1 with an apramycin resistance marker, apraR, using a procedure in which the gene was first replaced in a fosmid by using λ-RED-based methodology and then in RHA1 by using the modified fosmid and allelic exchange (39). The parent fosmid, RF00128O15, contained 38.3 kb of RHA1 DNA including the hsaADCB cluster. The oligonucleotides used to generate the resistance cassette used to replace hsaC were hsaC-for1 and hsaC-rev1 (SI Table 5). The six mce4 genes and the supAB genes were deleted separately in RHA1 by using the sacB counterselection system essentially as described (7). Oligonucleotides used to amplify the upstream and downstream region of the six mce4 genes were ro04698-F and ro04698-R(SpeI), and ro04703-F(SpeI) and ro04703-R(HindII), respectively. The upstream and downstream region of the supAB genes were amplified by using oligonucleotides SupA-F and SupA-R(SpeI), and Sup4B-F(SpeI) and SupB-R (SI Table 5). Gene replacements and deletions were verified by using a series of PCRs using: (i) primers matching sequences within the target gene(s), (ii) primers matching sequences flanking the target gene, and, when appropriate, (iii) primers matching a region within apraR.
Cloning and Expression of Mtb Genes.
The hsaCH37Rv and hsaDH37Rvgenes were amplified by PCR using Expand High Fidelity DNA polymerase (Roche Diagnostics, Indianapolis, IN) and cloned essentially as described for dbfB (40). The genes were amplified by using M. tuberculosis H37Rv genomic DNA and either Hcmt-F and Hcmt-R or Hdmt-F and Hdmt-R (SI Table 5). The amplicons were digested with NdeI and BamHI and cloned into similarly digested pT7–7, and their respective nucleotide sequences were confirmed to yield pT7HC1 and pT7HD1. HsaCH37Rv and HsaD H37Rvwere produced by using E. coli GJ1158 transformed with pT7HC1 and pT7HD1, respectively, as described for DbfB (40).
Enzyme Assays.
HsaC H37Rv and HsaD H37Rv activities in cellular extracts were measured by following the formation (HsaC) or consumption (HsaD) of the ring-cleaved product on a Cary 5000 spectrophotometer (Varian, Walnut Creek, CA) equipped with a thermojacketed cuvette holder, essentially as described for biphenyl catabolic enzymes (40). Experiments were performed by using 20 mM 3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid, 80 mM sodium chloride, pH 8.0 at 25.0 ± 0.1°C. Concentrations of 4,9-DSHA (ε 392 = 7.64 mM−1·cm−1) and HOPDA (ε 434 =32.5 mM−1·cm−1) were monitored at 392 and 434 nm, respectively. Initial velocities were determined from a least-squares analysis of the linear portion of the progress curves by using the kinetics module of Cary software. Steady-state rate equations were fit to data as described (40).
Metabolite Preparation and Characterization.
Culture supernatant was acidified by using 0.5% orthophosphoric acid then extracted twice with 0.5 volume of ethyl acetate. The ethyl acetate extracts were pooled, dried with anhydrous magnesium sulfate, and evaporated to dryness with a rotary evaporator. The residue was dissolved in a 7:3 mixture of methanol/water containing 0.5% phosphoric acid and purified by HPLC with a 2695 separation module (Waters, Milford, MA) and a Prodigy 10-μm ODS-Prep column (21.2 × 250 mm; Phenomenex, Torrance, CA). Metabolites were eluted by using the same methanol/water solvent at a flow rate of 5 ml/min. The eluate was monitored at 280 nm. The retention time of the major metabolite was ≈21 min. The fractions containing this metabolite were pooled, added to 10 volumes of water, and extracted as described above. The metabolite was derivatized by using Sylon BFT (Supelco, Bellefonte, PA) and analyzed by using a 6890 gas chromatograph (Agilent Technologies) and 5973N mass-selective detector (Agilent Technologies) in electron ionization mode. The extinction coefficient of 4,9-DHSA was determined with an oxygraph assay (40).
Supplementary Material
Acknowledgments
We thank Pascal D. Fortin for help in cloning hsaC and hsaD; Yossef Av-Gay (University of British Columbia) for the gift of H37Rv genomic DNA; Christine Florizone, Gordon R. Stewart, Matthew J. Myhre, and Jie Liu for skilled technical assistance; and Charles Thompson for critically reading the manuscript. This work was supported by grants from Genome Canada (to L.D.E. and W.W.M.), Genome BC (to L.D.E. and W.W.M.), the Natural Sciences and Engineering Research Council of Canada (to L.D.E. and W.W.M.), the Integration of Biosynthesis and Organic Synthesis Program of Advanced Chemical Technologies for Sustainability (to M.H.W.), and the Wellcome Trust (to E.S. and M.C.A.). K.Y. received a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada. M.H.W. received a traineeship from the European Graduate College program of the Groningen Biomolecular Sciences and Biotechnology Institute.
Abbreviations
- 3-HSA
3-hydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione
- 3,4-DHSA
3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione
- 4,9-DSHA
4,5–9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid
- AD
4-androstene-3,17-dione
- DHB
2,3-dihydroxybiphenyl
- DOHNAA
9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid
- KSTD
3-ketosteroid Δ1-dehydrogenase
- HOPDA
2-hydroxy-6-oxo-6-phenylpentadienoate
- TraSH
transposon site hybridization.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605728104/DC1.
This article is a PNAS direct submission. W.R.J. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE6709).
References
- 1.Gurtler V, Mayall BC, Seviour R. FEMS Microbiol Rev. 2004;28:377–403. doi: 10.1016/j.femsre.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 2.McLeod M, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, et al. Proc Natl Acad Sci USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van der Geize R, Dijkhuizen L. Curr Opin Microbiol. 2004;7:255–261. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Wovcha MG, Antosz FJ, Knight JC, Kominek LA, Pyke TR. Biochim Biophys Acta. 1978;531:308–321. doi: 10.1016/0005-2760(78)90213-8. [DOI] [PubMed] [Google Scholar]
- 5.Murohisa T, Iida M. J Ferment Bioeng. 1993;75:13–17. [Google Scholar]
- 6.Murohisa T, Iida M. J Ferment Bioeng. 1993;76:174–177. [Google Scholar]
- 7.Van der Geize R, Hessels GI, Van Gerwen R, Van der Meijden P, Dijkhuizen L. FEMS Microbiol Lett. 2001;205:197–202. doi: 10.1016/s0378-1097(01)00464-5. [DOI] [PubMed] [Google Scholar]
- 8.Van der Geize R, Hessels GI, Van Gerwen R, Van der Meijden P, Dijkhuizen L. Mol Microbiol. 2002;45:1007–1018. doi: 10.1046/j.1365-2958.2002.03069.x. [DOI] [PubMed] [Google Scholar]
- 9.Gibson DT, Wang KC, Sih CJ, Whitlock H., Jr J Biol Chem. 1966;241:551–559. [PubMed] [Google Scholar]
- 10.Horinouchi M, Hayashi T, Yamamoto T, Kudo T. Appl Environ Microbiol. 2003;69:4421–4430. doi: 10.1128/AEM.69.8.4421-4430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horinouchi M, Kurita T, Yamamoto T, Hatori E, Hayashi T, Kudo T. Biochem Biophys Res Commun. 2004;324:597–604. doi: 10.1016/j.bbrc.2004.09.096. [DOI] [PubMed] [Google Scholar]
- 12.Miclo A, Germain P. Appl Microbiol Biotechnol. 1992;36:456–460. doi: 10.1007/BF00172190. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization. Global TB Fact Sheet. Geneva: WHO; 2005. [Google Scholar]
- 14.Zhang Y. Annu Rev Pharmacol Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 15.Clark-Curtiss JE, Haydel SE. Annu Rev Microbiol. 2003;57:517–549. doi: 10.1146/annurev.micro.57.030502.090903. [DOI] [PubMed] [Google Scholar]
- 16.Rengarajan J, Bloom BR, Rubin EJ. Proc Natl Acad Sci USA. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sassetti CM, Rubin EJ. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, et al. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonçalves ER, Hara H, Miyazawa D, Davies J, Eltis LD, Mohn WW. Appl Environ Microbiol. 2006;72:6183–6193. doi: 10.1128/AEM.00947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai M, Masai E, Asami H, Sugiyama K, Kimbara K, Fukuda M. J Biosci Bioeng. 2002;93:421–427. doi: 10.1016/s1389-1723(02)80078-0. [DOI] [PubMed] [Google Scholar]
- 21.Mindnich R, Moller G, Adamski J. Mol Cell Endocrinol. 2004;218:7–20. doi: 10.1016/j.mce.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Miclo A, Germain P. Appl Microbiol Biotechnol. 1990;32:594–599. [Google Scholar]
- 23.Marchler-Bauer A, Anderson J.B., DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, et al. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 25.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 26.Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. Proc Natl Acad Sci USA. 2006;103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitale S, Ehrt S, Kawamura I, Fujimura T, Shimono N, Anand N, Lu S, Cohen-Gould L, Riley LW. Cell Microbiol. 2001;3:247–254. doi: 10.1046/j.1462-5822.2001.00110.x. [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. Proc Natl Acad Sci USA. 2005;102:12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Av-Gay Y, Sobouti R. Can J Microbiol. 2000;46:826–831. [PubMed] [Google Scholar]
- 31.Anderton MC, Bhakta S, Besra GS, Jeavons P, Eltis LD, Sim E. Mol Microbiol. 2006;59:181–192. doi: 10.1111/j.1365-2958.2005.04945.x. [DOI] [PubMed] [Google Scholar]
- 32.Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, Russell DG. Proc Natl Acad Sci USA. 2004;101:13642–13647. doi: 10.1073/pnas.0401657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatfield J, Pieters J. Science. 2000;288:1647–1650. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- 34.Peyron P, Bordier C, N′Diaye EN, Maridonneau-Parini I. J Immunol. 2000;165:5186–5191. doi: 10.4049/jimmunol.165.9.5186. [DOI] [PubMed] [Google Scholar]
- 35.De Chastellier C, Thilo L. Cell Microbiol. 2006;8:242–256. doi: 10.1111/j.1462-5822.2005.00617.x. [DOI] [PubMed] [Google Scholar]
- 36.Adler JJ, Rose DN. In: Tuberculosis. Rom WN, Garay SM, editors. Boston: Little, Brown, and Co; 1996. pp. 129–140. [Google Scholar]
- 37.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, et al. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 38.Seto M, Kimbara K, Shimura M, Hatta T, Fukuda M, Yano K. Appl Environ Microbiol. 1995;61:3353–3358. doi: 10.1128/aem.61.9.3353-3358.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patrauchan MA, Florizone C, Dosanjh M, Mohn WW, Davies J, Eltis LD. J Bacteriol. 2005;187:4050–4063. doi: 10.1128/JB.187.12.4050-4063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortin PD, Lo ATF, Haro MA, Kaschabek SR, Reineke W, Eltis LD. J Bacteriol. 2005;187:415–421. doi: 10.1128/JB.187.2.415-421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.