Abstract
The plastid envelope of higher plant chloroplasts is a focal point of plant metabolism. It is involved in numerous pathways, including tetrapyrrole biosynthesis and protein translocation. Chloroplasts need to import a large number of proteins from the cytosol because most are encoded in the nucleus. Here we report that a loss-of-function mutation in the outer plastid envelope 16-kDa protein (oep16) gene causes a conditional seedling lethal phenotype related to defects in import and assembly of NADPH:protochlorophyllide (Pchlide) oxidoreductase A. In the isolated knockout mutant of Arabidopsis thaliana, excess Pchlide accumulated in the dark operated as photosensitizer and provoked cell death during greening. Our results highlight the essential role of the substrate-dependent plastid import pathway of precursor Pchlide oxidoreductase A for seedling survival and the avoidance of developmentally programmed porphyria in higher plants.
Keywords: cell death, greening, plastid biogenesis, protein translocation
In plants, light provides an important environmental signal and trigger for the production of photosynthetically active chloroplasts (1, 2). In dark-grown angiosperms, plastid development is arrested at a state that leads only to the formation of so-called etioplasts. These organelles are devoid of chlorophyll and are incapable of photosynthetic function. Once dark-grown plants break through the soil after germination and reach the sunlight, they begin to synthesize chlorophyll and assemble the photosynthetic apparatus. The synthesis of chlorophyll from protochlorophyllide (Pchlide) is, in angiosperms, a light-dependent reaction and an essential step for the establishment of photosynthetically active chloroplasts. In barley, it is catalyzed by two closely related light-activated enzymes, NADPH:Pchlide oxidoreductases A and B (PORA and PORB) (3), which form supramolecular light-harvesting structures designated LHPP in the prolamellar body of etioplasts (4, 5). Dark-stable PORA:PORB-Pchlide-NADPH supracomplexes are poised such that absorption of a photon by Pchlide b bound to PORA leads to energy transfer onto PORB-bound Pchlide a and its subsequent reduction, resulting in the formation of Chlide a (4, 5). Like other free tetrapyrroles, Pchlide not bound to POR could operate as a photosensitizer (6–8). Angiosperm plants, therefore, have evolved efficient mechanisms to keep the level of these potentially phototoxic compounds low and thereby avoid porphyrias (9–12). One such mechanism is feedback control of Pchlide synthesis by heme and Pchlide (10, 11). Another factor is the fluorescent (FLU) protein, which depresses Pchlide synthesis in darkness (12). After LHPP's dissociation, which is induced by light and correlates with the dispersal of the prolamellar body (13), PORA gains activity as a Pchlide b-reducing enzyme such that another part of the light energy is quenched in a nonhazardous way (4, 5).
Being encoded in the nucleus, PORA and PORB are synthesized as larger precursors (pPORA and pPORB) and are imported into the plastids through specific protein translocon complexes in the outer and inner envelope membranes (14–18). Whereas pPORB enters the plastids via the general protein import apparatus comprising the presequence receptor TOC159 and translocation channel protein TOC75 (17, 18), pPORA uses a Pchlide-dependent translocon named the PTC complex, which is distinctive from the general protein import site (14, 17). Among the identified PTC proteins was a 16-kDa protein related to a group of amino acid and preprotein transporters found in free-living bacteria and endosymbiotic mitochondria and chloroplasts (14, 17). Several lines of evidence verified that the identified barley PTC16 gene is an ortholog of the previously characterized pea and Arabidopsis OEP16 (At2g28900) gene (17).
In the present work a reverse genetic approach was taken to dissect the function of the Arabidopsis OEP16 (At2g28900) gene in planta. Using a knockout line of Arabidopsis thaliana that is deficient in the OEP16 gene we demonstrate that PTC16/OEP16 is involved in pPORA import. Interestingly, dark-grown Atoep16 plants resembled etiolated flu plants and accumulated free Pchlide. After a dark-to-light shift, this pigment operated as photosensitizer and caused rapid bleaching and cell death. Our results underscore the essential role of the substrate-dependent import pathway of pPORA, which couples protein translocation to pigment biosynthesis in the plastid envelope, for Pchlide homoeostasis and cell viability during seedling de-etiolation.
Results and Discussion
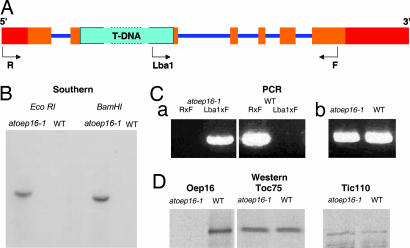
At2g28900 (AtOEP16) contains six exons and five introns (Fig. 1A), and we used reverse genetics to determine its role in Arabidopsis. A respective Arabidopsis mutant was obtained from the Salk Institute Genomic Analysis Laboratory collection (19) carrying a T-DNA insertion SALK_024018 that disrupts the AtOEP16 locus (Fig. 1 A and C). Sequencing of the T-DNA/AtOEP16 junction established that the insertion disrupted the gene 6 bp upstream of the 3′ end of exon 2. We refer to this line as Atoep16-1 throughout the rest of the article. DNA gel blot analyses yielded a single T-DNA hybridizing band in genomic DNA isolated from light-grown homozygous Atoep16-1 plants that had been digested with two different restriction enzymes (Fig. 1B), demonstrating that Atoep16-1 contained a single T-DNA insertion in the genome. Expression studies showed that neither OEP16 transcript nor OEP16 protein was detectable in Atoep16-1 plants, and thus the mutant was null with respect to the Atoep16-1:1 gene (Fig. 1D).
Fig. 1.
Description of the Atoep16-1 T-DNA insertion line (SALK_024018.50.90.X). (A) Diagram of the Atoep16-1:1 gene and respective T-DNA insertion. 5′ and 3′ untranslated regions are in red, exons are in ochre, and introns are in violet. The 4.5-kb T-DNA insertion (not drawn to scale) is shown in turquoise. R, F, and Lba1 mark primers used for PCR analyses. (B) DNA gel blot analysis of the Atoep16-1 line. Genomic DNA (10 μg) from homozygous Atoep16-1 or wild-type plants was digested with EcoRI and BamHI, respectively, and the filter-bound DNA fragments were hybridized to a DNA probe corresponding to the kanamycin-resistance gene of the T-DNA using standard procedures. (C) Confirmation of the T-DNA insertion by PCR using the following primers. (a) Lba1, 5′-ATGGTTCACGTAGTGGGCCATCG-3′; R (reverse primer), 5′-ATCCACCGTTAAAAGCCCCTT-3′; F, (forward primer), 5′-AACGAACTGAGAAGCGGTTGC-3′. (b) Primers specific for the adenine phosphoryl transferase gene of wild-type and Atoep16-1 plants. (D) Western blot analysis of OEP16 protein expression in chloroplasts of Atoep16-1 and wild-type plants. Protein was prepared from isolated chloroplasts and subjected to Western blotting using antisera against OEP16 and the 75- and 110-kDa proteins of the outer and inner envelope membrane translocases of chloroplasts TOC75 and TIC110, respectively.
Depending on the growth conditions, Atoep16-1 seedlings exhibited different phenotypes (Fig. 2). If grown in darkness and exposed to white light, the mutant rapidly bleached and finally died (Fig. 2B). In plants kept under continuous white light right from the beginning of germination, no phenotype was detectable, and the plants looked like the wild type (Fig. 2, compare C and F). Etiolated Atoep16-1 plants examined under blue light with a Leica MZ12 fluorescence microscope showed a strong red fluorescence indicative of the presence of free porphyrin pigments in darkness (Fig. 2A, compare with the wild type shown in D). These results were reminiscent of findings reported for the flu mutant of Arabidopsis that contains elevated levels of red-fluorescing Pchlide in darkness (12). Pchlide is present in a free form in etiolated flu plants, which triggers singlet oxygen formation and cell death upon illumination (12). When dark-grown Atoep16-1 seedlings were exposed to white light, very similar defects were observed, including bleaching and cell death (Fig. 2 B versus E). However, the time courses shown in Fig. 2G revealed some differences in cell death progression (Fig. 2G). We assumed that this difference may be reflective of the actual level and/or composition of free porphyrin pigment(s) in Atoep16-1 versus flu plants. Pigment analyses (20) indeed showed that whereas flu plants accumulate ≈8.5-fold-higher levels of total Pchlide than wild-type plants, Atoep16-1 plants contained only ≈4.5-fold-higher pigment levels. Interestingly, the composition of pigments was also altered in Atoep16-1 versus flu and wild-type plants (Fig. 2H). Whereas Atoep16-1 plants accumulated Pchlide a, flu plants contained elevated levels of Pchlide b [Fig. 2H; for details of pigment identification, see supporting information (SI) Fig. 8].
Fig. 2.
Growth-dependent cell death phenotype of the Atoep16-1 mutant. (A–F) Atoep16-1 (A–C) and wild-type (D–F) plants were grown in the dark (A and D) under a nonpermissive dark-to-light shift (B and E) or under permissive continuous white light (C and F) and inspected under a microscope. Accumulation of free, red-fluorescing Pchlide was monitored under blue light (400–500 nm) (A and D). Free, excited Pchlide molecules present in etiolated Atoep16-1 (A) but not in wild-type (D) plants caused photooxidative damages, as a result of which the seedlings died (B versus E, respectively). (G) Seedling survival rates of wild-type (open circles), Atoep16-1 (triangles), and flu (filled circles) plants during a nonpermissive dark-to-light shift. Of a number of 300 seedlings in three independent experiments, the indicated percentages survived, whereas the remainder died as a result of pigment-sensitized photooxidation. (H) Pigment accumulation in etiolated wild-type (gray line), Atoep16-1 (blue line), and flu (red line) plants. The different peaks were identified as Pchlide b (peak eluting at 12.5 min) and Pchlide a (peak eluting at 15 min) by using synthetic standards and absorbance measurements as well as mass spectrometry (SI Fig. 8).
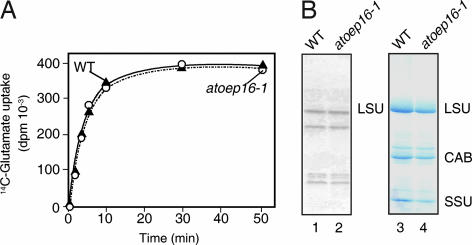
OEP16 is a transmembrane channel protein of the outer plastid envelope membrane implicated in amino acid (21) and/or polypeptide transport (17). When we performed radioisotope- labeling studies with 14C-glutamate, 14C-glutamine, and 14C-glycine, and isolated chloroplasts and etioplasts (22), no difference in amino acid uptake was found for Atoep16-1 versus wild-type plants (Fig. 3A and data not shown). Also, no large differences were apparent in the patterns of plastid-encoded, [35S]methionine-labeled (23) proteins (Fig. 3B). Furthermore, the red fluorescence phenotype in darkness implies that Atoep16 plants were not impaired in glutamate- and 5-aminolevulinic acid (5-ALA)-dependent Pchlide synthesis. If OEP16 would be involved in amino acid uptake, an inhibition, but not an elevation, of glutamate-dependent synthesis of 5-ALA and Pchlide should have occurred in etiolated Atoep16-1 plants, which was obviously not observed (see Fig. 2). We thus concluded that Atoep16-1 was not affected in bulk amino acid uptake into isolated plastids.
Fig. 3.
Amino acid uptake into Atoep16-1 and wild-type chloroplasts. (A) Isolated chloroplasts were incubated with 14C-glutamate for various time intervals, and uptake of radioactivity was determined. (B) After labeling isolated chloroplasts with [35S]methionine for 5 min (lanes 1 and 2) or 20 min (data not shown), protein was extracted and precipitated with trichloroacetic acid, separated by SDS/PAGE, and detected by either autoradiography (lanes 1 and 2) or Coomassie staining (lanes 3 and 4). LSU and SSU, large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase; CAB, chlorophyll a/b-binding protein of photosystem II.
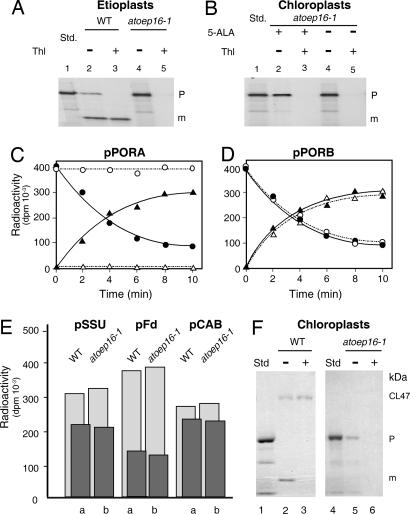
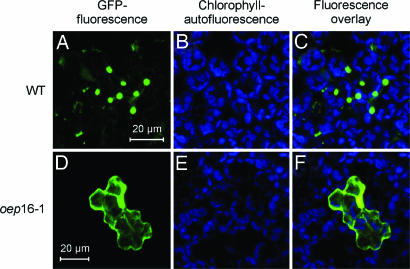
Alternatively, OEP16 could be involved in polypeptide transport (17). This idea was tested by performing in vitro import experiments (24). Fig. 4 shows that no differences in import of the precursors to the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (pSSU), ferredoxin (pFd), the light-harvesting chlorophyll a/b binding protein LHCII (pCAB), as well as pPORB were detectable for isolated etioplasts and chloroplasts of Atoep16-1 and wild-type plants. Similar amounts of the different tested precursors were taken up and processed to mature size (Fig. 4 D and E). Only import of pPORA was blocked in vitro, as evidenced by the unaltered precursor levels and the concomitant lack of the mature PORA in etioplasts from Atoep16-1 versus wild-type plants (Fig. 4A). Likewise, no import was detectable with 5-ALA-treated, Pchlide-containing Atoep16-1 as compared with wild-type chloroplasts (Fig. 4 B and C). Chemical cross-linking (17) using a chimeric protein that consisted of the transit peptide of pPORA (transA) and a cytosolic dihydrofolate reductase (DHFR) reporter protein of mouse revealed that the precursor was able to bind Atoep16-1 plastids; nevertheless, the precursor could not enter a productive import pathway (Fig. 4F). Transient assays in leaf epidermis cells expressing transit peptide fusions of pPORA (transA) with GFP (transA-GFP) indeed confirmed the import defect of Atoep16 plants (Fig. 5).
Fig. 4.
In vitro import of 35S precursors into plastids isolated from Atoep16-1 and wild-type seedlings. (A) Import of 35S-pPORA into etioplasts of Atoep16-1 and wild-type plants. The incubations were performed in the presence of 2 mM Mg-ATP. The autoradiograms show precursor (P) and mature (m) protein levels in plastids treated with (+) or without (−) thermolysin (Thl). Std defines input translation standards. (B) As in A, but showing import data of 35S-pPORA for 5-ALA-pretreated, Pchlide-containing, and mock-incubated Atoep16-1 chloroplasts. (C and D) Time courses of import of 35S-pPORA (C) and 35S-pPORB (D) into 5-ALA-treated, Pchlide-containing wild-type (filled circles and filled triangles) and Atoep16-1 (open circles and open triangles) chloroplasts. Precursor and mature proteins are marked by open and filled circles as well as open and filled triangles, respectively. Quantification of radioactivity in protein was done with a PhosphorImager. (E) Quantitative import data for the small subunit precursor of ribulose-1,5-bisphosphate carboxylase/oxygenase (pSSU), precursor ferredoxin (pFd), and the chlorophyll a/b-binding protein precursor of photosystem II (pCAB). Light and dark gray columns refer to precursor and mature protein levels after 15-min import reactions into 5-ALA-treated, Pchlide-containing chloroplasts of wild-type (a) and Atoep16 (b) plants. (F) Cross-linking of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB)-activated 35S-transA-DHFR, consisting of the Pchlide-responsive transit peptide of pPORA (transA) and a DHFR reporter protein of mouse, to isolated 5-ALA-pretreated, Pchlide-containing wild-type (lanes 1–3) and Atoep16-1 (lanes 4–6) chloroplasts, obtained at 2 mM Mg-ATP and 0.1 mM Mg-GTP. CL47 defines 47-kDa products established between Cys-80 in the DHFR domain of the 31-kDa precursor (P) and Cys-71 in OEP16, and m stands for the imported, mature, 25-kDa DHFR (lane 2). Lane 3 shows a respective immunoprecipitation using OEP16 antiserum. Lanes 5 and 6 depict cross-link products and immunoprecipitates corresponding to those in lanes 2 and 3, respectively, for Atoep16 chloroplasts. In lanes 1 and 4 are shown input translation standards (Std). Note the lack of CL47 and imported, mature DHFR in Atoep16 chloroplasts.
Fig. 5.
Transient expression and plastid import of TransA-GFP in Arabidopsis leaf epidermis cells of Atoep16-1 and wild-type plants. DNA for transA-GFP, encoding transA and the GFP, was transformed into leaf epidermis cells of Arabidopsis wild-type and atoep16 plants. After 24 h in darkness, GFP (A and D) and chlorophyll (B and E) fluorescences were monitored between 505–530 nm and 575–650 nm, respectively, by using an excitation wavelength of 488 nm. (C) Merge of A and B. (F) Merge of D and E. As subcellular localization controls, the naked GFP moiety lacking a transit peptide attached to it (not imported into either atoep16 or wild-type plastids) and transB-GFP (imported into both plastid types) were used (data not shown).
That knockout in the Atoep16-1:1 gene did not pleiotropically affect all of the different cytosolic precursors that need to be taken up by the plastid compartment is further supported by the growth rescue of Atoep16-1 plants in continuous white light (compare Fig. 2C). Like pSSU, pFd, pCAB, and pPORB, the enzymes that are involved in 5-ALA synthesis and subsequent steps of Pchlide production were also obviously not affected by the lack of the OEP16 protein. Otherwise, no excess pigment would have accumulated in dark-grown Atoep16-1 plants. All enzymes of the C5 pathway leading to chlorophyll are nucleus-encoded and imported posttranslationally from the cytoplasm (1).
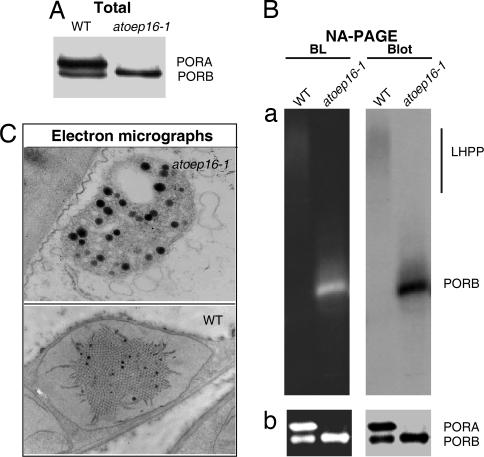
Protein gel blot analyses showed a lack of PORA and PORA-containing larger LHPP complexes in Atoep16-1 versus wild-type plants (Fig. 6A and B). Electron microscopy highlighted a complete lack of the prolamellar body in Atoep16-1 as compared with wild-type etioplasts (Fig. 6C). Together, these findings conclusively showed that PORA, as part of larger complexes in the prolamellar body, played an essential role for plant survival during seedling de-etiolation and greening. Free Pchlide not bound to the PORA, by contrast, operated as photosensitizer and, by triplet–triplet interchange, provoked singlet oxygen production and cell death. Similar observations have been made for photobleached flu plants (12). In a sense, these findings are reminiscent of genetically inherited porphyrias in humans where perturbations in heme metabolism trigger cell death, including nuclear condensation, chromatin separation, DNA breakdown, and release of cytochrome c from mitochondria into the cytosol (25). However, porphyrin-dependent, plastid-dependent cell death in plants also exhibits unique properties, as illustrated by recent work of Wagner et al. (26) on the executer mutant of Arabidopsis.
Fig. 6.
POR expression in etiolated Atoep16-1 and wild-type seedlings. (A) PORA and PORB protein levels in Atoep16-1 and wild-type etioplasts determined by SDS/PAGE and Western blotting. (B) Detection of light-dissociable PORA:PORB-Pchlide-NADPH supracomplexes indicative of LHPP in solubilized membrane fractions of Atoep16-1 and wild-type etioplasts before (a) and after (b) a 1-msec flash of white light. Protein detection was made by nondenaturing, analytical PAGE (NA-PAGE) and blue light (BL)-induced pigment autofluorescence (Left) and protein gel blot (Blot) analysis (Right) using a POR antiserum. (C) Transmission electron micrographs of Atoep16-1 and wild-type etioplasts. Black dots represent plastoglobules formed in excess in Atoep16-1 versus wild-type etioplasts.
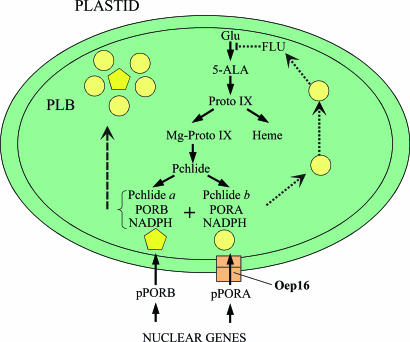
Our results suggest the working model described in Fig. 7, explaining the possible interaction of OEP16, PORA, and FLU during greening. Accordingly, PORA-Pchlide b-NADPH ternary complexes not assembled into PORA:PORB suprastructures after import through OEP16 could provide the link to the FLU protein and help fine-tune the size of the prolamellar body. In wild-type plants, PORA would sense the amount of Pchlide, and once enough LHPP is made, excess PORA-Pchlide b-NADPH ternary complexes could bind to and block the activity of the FLU protein, thereby inhibiting glutamyl-tRNA reductase. By contrast, this negative feedback would no longer operate in the OEP16-deficient Atoep16-1 mutant line because of the lack of imported PORA. It is clear that the intimate interaction of all players is required for successful greening. Any change in Pchlide homoeostasis has deleterious effects on plant growth.
Fig. 7.
Working model of pPORA import, LHPP assembly, and FLU-mediated regulation of Chl biosynthesis. Yellow circles and pentagons illustrate PORA-Pchlide b-NADPH and PORB-Pchlide a-NADPH ternary complexes, respectively. The ochre box highlights Oep16 as part of the Ptc complex. Proto IX, protoporphyrin IX.
Materials and Methods
Plant Growth.
Seeds of the Atoep16 line (SALK_024018) were obtained from the Salk Institute Genomic Analysis Laboratory collection (19). Seeds were germinated on half-concentrated Murashige–Skoog–agar medium on Petri dishes for 4.5 days. To trigger photooxidative damages, dark-grown seedlings were exposed to white light at 125 μE·m−2. Alternatively, seeds were germinated on soil and grown to maturity in continuous white light.
Pigment Measurements.
High-performance liquid chromatography of acetone-extracted pigments was carried out by using a C18 reverse-phase silica gel column (Hypersil ODS, 5 μm; Shandon HPLC, Chreshire, U.K.) and synthetic Pchlides a and b as standards (5, 20). Pigments were detected and quantified at 455 nm, the Soret band of Pchlide b (5, 20).
Analysis of Amino Acid Uptake into Isolated Plastids.
Uptake of 14C amino acids into isolated chloroplasts was monitored by using the filter paper disk method of Mans and Novelli (22). Pulse-labeling of chloroplast protein with [35S]methionine (1.87 MBq per 50-μl assay, 37 TBq/mmol; Amersham Pharmacia, Uppsala, Sweden) was carried out according to Mullet et al. (23). Protein was extracted with trichloroacetic acid, depleted of chlorophyll, and washed and run on 10–20% SDS/polyacrylamide gradients (27). Protein detection was made by Coomassie staining and autoradiography or by Western blotting (28).
Protein Import Assay in Vitro and in Planta.
Protein import into isolated Arabidopsis chloroplasts and etioplasts was studied as described by using cDNA-encoded, wheat germ-translated 35S precursors (24). Briefly, 35S precursors were synthesized from corresponding cDNA clones by coupled transcription/translation and incubated with isolated, energy-depleted Arabidopsis plastids from wild-type and Atoep16-1 plants. Plastids were treated with thermolysin (29) after import to degrade unimported precursors. Chemical cross-linking of 5,5′-dithiobis(2-nitrobenzoic acid)-derivatized 35S-transA-DHFR, consisting of the transit peptide of pPORA (transA) and a cytosolic dihydrofolate reductase (DHFR) reporter protein of mouse, was carried out as described (17).
TransA-GFP derivatives were produced as described in SI Methods. Transient expression of TransA-GFP derivatives in Arabidopsis leaf epidermis cells was performed after ballistic bombardment by using a pneumatic particle inflow gun according to Finer et al. (30). The conditions of bombardment were adjusted to helium pressure of 6.5 bar, at a 12-cm target distance, with a disperse grid at 7 cm, using 1 μm of gold microcarriers (Bio-Rad, Hercules, CA). After bombardment, the plantlets were kept under sterile conditions and incubated for 24 h in darkness. Confocal laser scanning microscopy was carried out by using a LSM 510 Meta microscope (Zeiss, Jena, Germany) with krypton/argon laser excitation at 488 nm and an emission wavelength window from 505 to 530 nm. LSM 510 Meta software release 3.2 (Zeiss, Oberkochen, Germany) and Photoshop 7 (Adobe Systems, San Jose, CA) were used for image acquisition and processing. Nondenaturing, analytical PAGE was carried out as described (31).
Miscellaneous.
Electron microscopy was carried out by using ultrathin sections of leaf tissues prepared from etiolated plants using a Zeiss 109 electron microscope.
Supplementary Material
Acknowledgments
We are indebted to L. Reinbothe for editorial work. This work was supported by a Chaire d'Excellence research project grant from the French Ministry of Research (to C.R.). This is scientific paper no. 1201-06 from the College of Agricultural, Human, and Natural Resource Sciences of Washington State University.
Abbreviations
- Pchlide
protochlorophyllide
- pPORA/B
precursor Pchlide oxidoreductases A and B
- DHFR
dihydrofolate reductase
- 5-ALA
5-aminolevulinic acid
- trans A
transit peptide of pPORA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610934104/DC1.
References
- 1.von Wettstein D, Gough S, Kannangara CG. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirk JTO, Tilney-Basset RAE. The Plastids: Their Chemistry, Structure, Growth, and Inheritance. Amsterdam: Elsevier North-Holland Biomedical Press; 1978. [Google Scholar]
- 3.Holtorf H, Reinbothe S, Reinbothe C, Bereza B, Apel K. Proc Natl Acad Sci USA. 1995;92:3254–3258. doi: 10.1073/pnas.92.8.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinbothe C, Lebedev N, Reinbothe S. Nature. 1999;397:80–84. [Google Scholar]
- 5.Reinbothe S, Pollmann S, Reinbothe C. J Biol Chem. 2003;278:807–815. doi: 10.1074/jbc.M209738200. [DOI] [PubMed] [Google Scholar]
- 6.Rebeiz CA, Nandihalli UB, Velu J. Photochem Photobiol. 1990;52:1099–1117. [Google Scholar]
- 7.Matringe M, Camadro JM, Labbe P, Scalla R. Biochem J. 1989;260:231–235. doi: 10.1042/bj2600231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mock H-P, Keetman U, Kruse E, Rank B, Grimm B. Plant Physiol. 1998;116:107–116. [Google Scholar]
- 9.Beale SI, Weinstein JD. In: Biosynthesis of Heme and Chorophyll. Dailey HA, editor. New York: McGraw–Hill; 1990. pp. 287–391. [Google Scholar]
- 10.Pontoppidan B, Kannangara CG. Eur J Biochem. 1994;225:529–537. doi: 10.1111/j.1432-1033.1994.00529.x. [DOI] [PubMed] [Google Scholar]
- 11.Vothknecht UC, Kannangara CG, von Wettstein D. Phytochemistry. 1998;47:513–519. doi: 10.1016/s0031-9422(97)00538-4. [DOI] [PubMed] [Google Scholar]
- 12.Meskauskiene R, Nater M, Gosling D, Kessler F, op den Camp R, Apel K. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahn A. Plant Physiol. 1968;43:1781–1785. doi: 10.1104/pp.43.11.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinbothe S, Quigley F, Gray J, Schemenewitz A, Reinbothe C. Proc Natl Acad Sci USA. 2004;101:2197–2202. doi: 10.1073/pnas.0307284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinbothe S, Runge S, Reinbothe C, van Cleve B, Apel K. Plant Cell. 1995;7:161–172. doi: 10.1105/tpc.7.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinbothe S, Reinbothe C, Holtorf H, Apel K. Plant Cell. 1995;7:1933–1940. doi: 10.1105/tpc.7.11.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinbothe S, Quigley F, Springer A, Schemenewitz A, Reinbothe C. Proc Natl Acad Sci USA. 2004;101:2203–2208. doi: 10.1073/pnas.0301962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronsson H, Sohrt K, Soll J. Biol Chem. 2000;381:1263–1267. doi: 10.1515/BC.2000.155. [DOI] [PubMed] [Google Scholar]
- 19.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 20.Reinbothe S, Pollmann S, Reinbothe C. J Biol Chem. 2003;278:800–806. doi: 10.1074/jbc.M209737200. [DOI] [PubMed] [Google Scholar]
- 21.Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R. Proc Natl Acad Sci USA. 1997;94:9504–9509. doi: 10.1073/pnas.94.17.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mans RJ, Novelli GD. Biochem Biophys Res Commun. 1960;3:540–543. doi: 10.1016/0006-291x(60)90171-6. [DOI] [PubMed] [Google Scholar]
- 23.Mullet JE, Klein RR, Grossman AR. Eur J Biochem. 1986;155:331–338. doi: 10.1111/j.1432-1033.1986.tb09495.x. [DOI] [PubMed] [Google Scholar]
- 24.Reinbothe S, Pollmann S, Springer A, James RJ, Tichtinsky G, Reinbothe C. Plant J. 2005;42:1–12. doi: 10.1111/j.1365-313X.2005.02353.x. [DOI] [PubMed] [Google Scholar]
- 25.Moore MR, McColl KEL, Rimington C, Goldberg SA. Disease of Porphyrin Metabolism. New York: Plenum; 1990. [Google Scholar]
- 26.Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Wursch M, Laloi C, Nater M, Apel K. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Towbin M, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cline K, Werner-Washburne M, Andrews J, Keegstra K. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finer JJ, Vain P, Jones MW, McMullen MD. Plant Cell Rep. 1992;11:323–328. doi: 10.1007/BF00233358. [DOI] [PubMed] [Google Scholar]
- 31.Reinbothe S, Krauspe R, Parthier B. J Plant Physiol. 1990;137:81–87. doi: 10.1007/BF02411535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.