Abstract
Electron transfer dissociation (ETD) is a recently introduced mass spectrometric technique that provides a more comprehensive coverage of peptide sequences and posttranslational modifications. Here, we evaluated the use of ETD for a global phosphoproteome analysis. In all, we identified a total of 1,435 phosphorylation sites from human embryonic kidney 293T cells, of which 1,141 (≈80%) were not previously described. A detailed comparison of ETD and collision-induced dissociation (CID) modes showed that ETD identified 60% more phosphopeptides than CID, with an average of 40% more fragment ions that facilitated localization of phosphorylation sites. Although our data indicate that ETD is superior to CID for phosphorylation analysis, the two methods can be effectively combined in alternating ETD and CID modes for a more comprehensive analysis. Combining ETD and CID, from this single study, we were able to identify 80% of the known phosphorylation sites in >1,000 phosphorylated peptides analyzed. A hierarchical clustering of the identified phosphorylation sites allowed us to discover 15 phosphorylation motifs that have not been reported previously. Overall, ETD is an excellent method for localization of phosphorylation sites and should be an integral component of any strategy for comprehensive phosphorylation analysis.
Keywords: bioinformatics, motifs, phosphorylation, signal transduction, systems biology
Most cellular processes are regulated by posttranslational modifications of proteins. For some posttranslational modifications (e.g., acetylation and tyrosine phosphorylation), identifying the modified amino acid is relatively straightforward because they are quite stable in the presence of the energy required for collision-induced dissociation (CID) experiments. For other posttranslational modifications [e.g., O-linked N-acetylglucosamine (O-GlcNAc) and phosphorylated serine and threonine residues], however, localization is substantially more difficult because the peptides either lose the modification in a charge separation process (O-GlcNAc) (1–3) or by a β-elimination event with a neutral loss of phosphoric acid (e.g., phosphoserine into dehydroalanine). In 1998, Zubarev et al. (4) described a new fragmentation technique called electron capture dissociation (ECD) as a gentler fragmentation technique compared with CID. More recently, Syka et al. (5) and Pitteri et al. (6) demonstrated that peptide cations can also be reduced and converted into radicals by reaction with radical gaseous anions, in an electron transfer process. The reduced peptides show similar fragmentation patterns as observed in ECD experiments, and the process is designated electron transfer dissociation (ETD). Although ETD has been tested in pilot experiments to localize posttranslational modifications, no large-scale analysis using ETD has yet been published.
In this study, we present a global proteomic profiling of phosphopeptides subjected to fragmentation using ETD in an ion trap mass spectrometer. A total of 84,000 ETD and CID tandem MS (MS/MS) spectra from 130 liquid chromatography (LC)-MS/MS runs using three different proteolytic enzymes (Lys-C, trypsin, and Glu-C) allowed us to identify 1,435 unique phosphorylation sites from proteins encoded by 500 genes, of which 1,141 (≈80%) had not been described previously. Using identical samples for ETD and CID analysis, we found that ETD was superior to CID both in the number of phosphopeptides identified as well as amino acid sequence coverage per phosphopeptide. Importantly, from a single sample, our strategy was able to identify 294 of 368 phosphorylation sites (368 phosphorylation sites were previously known in the literature for the phosphopeptides identified in our study, out of which we were able to identify 294) previously reported (80%) in phosphopeptides characterized in our analysis. This number is significant because the phosphorylation sites described in the literature have mostly been discovered through decades of research on individual proteins.
Finally, we took advantage of the large number of phosphorylation sites identified in this study to carry out a bioinformatics analysis to identify novel phosphorylation motifs. Using a hierarchical clustering approach to identify potential phosphorylation motifs, we identified 68 motifs, of which 15 were previously undescribed, illustrating the power of combining experimental and computational approaches for revealing hidden signatures. Overall, our studies establish ETD as an essential component of any comprehensive phosphoproteomics analysis.
Results
Electron Transfer Dissociation (ETD) of Phosphopeptides.
To evaluate ETD for a global analysis of the phosphoproteome, we analyzed phosphopeptides enriched using TiO2 purification on an ion trap equipped with ETD capability. We first decided to use Lys-C generated peptides for this study as it has been reported that multiple charged peptides are fragmented more favorably in ETD experiments (6). Lysates from human embryonic kidney 293T cells treated with serine/threonine and tyrosine phosphatase inhibitors were separated by reversed phase chromatography into 30 fractions, each of which was split into three sets (Fig. 1). One of the sets was subjected to an in-solution digestion by using Lys-C followed by enrichment of phosphopeptides by using TiO2 as described recently (7). The phosphopeptide-enriched fractions were analyzed by using LC-MS/MS (ETD) and 24,622 resulting ETD spectra were searched against the Human RefSeq database. From this experiment, 676 unique peptides were identified of which 606 were phosphopeptides (≈90%), derived from 287 proteins. Of the 606 phosphopeptides, 344 were singly phosphorylated, 208 were doubly phosphorylated, and 54 peptides contained three or more phosphorylated residues. The majority of the phosphorylation sites were localized to serine residues (84%) whereas threonine and tyrosine phosphorylation accounted for 14% and 2.3%, respectively. These values are similar to the results from a recent study using CID (S, 78%; T, 18%; Y, 3.8%) (8). Supporting information (SI) Data Set 1 lists the phosphopeptides that were identified, together with their charge states, peptide identification score, site of phosphorylation, and whether the site of phosphorylation has been previously described. Fig. 2 A–D shows representative ETD MS/MS spectra of phosphorylated peptides carrying 5, 4, 3, or 2 charges, respectively. A clear pattern in the ETD spectra was observed. First, the spectra were dominated by a few very intense ions, which are the result of charge stripping e.g., a peptide bearing 5 positive charges shows an envelope comprising the 5+ precursor along with ions that can be assigned as the 4+, 3+, 2+, and 1+ versions of the precursor ion (Fig. 2A). Secondly, most of the fragment ions can be assigned as c or z ions, and are generally of lower intensity. Nearly equal numbers of fragments were assigned as originating from the C-terminal part of the peptide (48%) as from the N-terminal part (52%). The details of fragmentation for each phosphopeptide are also included in SI Data Set 1. This large data set could be used for a detailed analysis of fragmentation observed with ETD as has already been done for CID-based fragmentation (9).
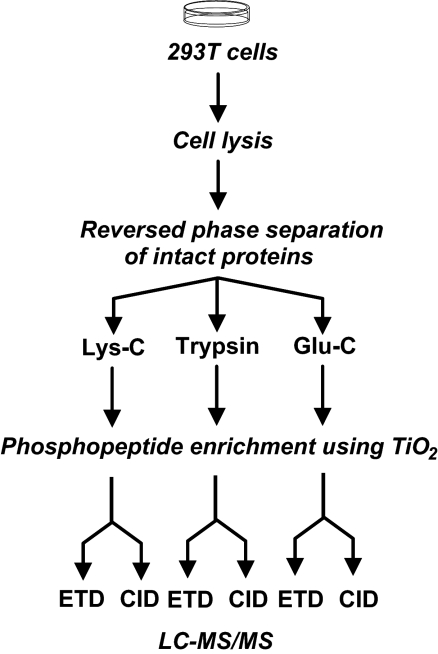
Fig. 1.
A schematic depicting the strategy for analysis of phosphopeptides carried out in this study. All fractions were analyzed by LC-MS/MS. A total of 30 Lys-C digested fractions were analyzed by LC-MS/MS (ETD); 12 of the selected Lys-C fractions (1/3 of each sample was used) were subjected to a replicate analysis, with one part of a sample analyzed by ETD and the other by CID; the remaining 2/3 of the 12 protein fractions were digested by using trypsin or Glu-C, respectively, and analyzed in replicate by LC-MS/MS (CID) and LC-MS/MS (ETD). A fraction (7%) of the digested samples before enrichment of phosphopeptides was also analyzed by CID.
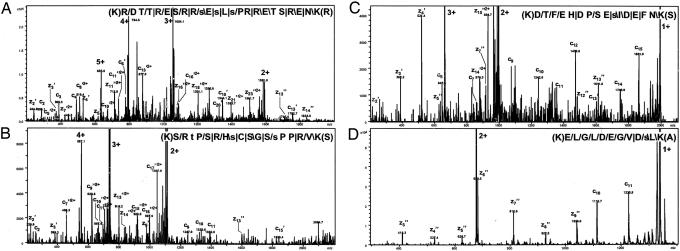
Fig. 2.
MS/MS spectra of four phosphorylated Lys-C peptides identified by ETD. Phosphopeptides with charge states of + 5 (A), +4 (B), +3 (C) and +2 (D) are shown. The four peptides originate from splicing factor, arginine/serine-rich 2 interacting protein, splicing coactivator subunit SRm300, Bcl2-associated transcription factor 1 and DnaJ homolog, subfamily C, member 9, respectively. The peptide sequence with the fragmentation pattern is shown in each panel. The signs: “/”, “ /”, and “|” designate that the C-terminal type fragments, N-terminal fragments, or both types of fragments, respectively, were identified. For all spectra, except A, the intensity axis has been enlarged by a factor of ≈5. Fragment ions resulting from charge stripping of the precursor ion are assigned with charge states (in bold). Small letters indicate phosphorylated residues.
ETD Versus CID for Analysis of Phosphopeptides.
The general belief is that ETD preserves posttranslational modifications as well as providing more complete peptide backbone fragmentation. However, as described above, the intensities of c and z backbone fragment ions are most often of low intensity, which can potentially obscure identification of the phosphopeptides. To assess whether ETD is indeed better suited for phosphopeptide analysis than CID, we subjected 12 of 30 samples to CID fragmentation (Fig. 1). The base peak chromatograms of identical samples subjected to both a LC-MS/MS (ETD) and a LC-MS/MS (CID) experiment were quite comparable as seen in SI Data Set 1 (A–D). In the two sample sets, 272 unique peptide ions were identified by ETD versus 190 by using CID. Of 272 peptides identified by ETD, 228 were phosphopeptides, whereas 132 of 190 peptides identified by CID were phosphopeptides. A total of 48 phosphopeptides were identified by both techniques (Fig. 3A). The CID experiment identified more nonphosphorylated peptides than the ETD experiment, the number being 58 versus 44 (Fig. 3B). That a greater number of phosphopeptides was identified by ETD (73%) could arise because of several reasons. (i) In CID, the predominant loss of the phosphate moiety by neutral loss generally precludes further fragmentation of phosphoserine and phosphothreonine containing peptides. This incomplete fragmentation is in sharp contrast to a more complete backbone fragmentation observed in ETD. (ii) Larger peptides carrying three or more positive charges are usually generated by Lys-C. The higher charge state of peptides has been reported to be favorable for ETD experiments, whereas in CID, multiple charged fragment ions generated from such peptides can make it difficult to match MS/MS data. (iii) Although we have established that the LC separation step for samples analyzed by ETD and CID is indeed reproducible, the selection of peptide ions for MS/MS experiments is still somewhat random and only controlled by the intensity of the peptide ions. In the following experiments, these questions will be addressed in greater detail.
Fig. 3.
Phosphopeptide identifications from ETD and CID experiments. (A) The overlap among phosphopeptides identified from Lys-C digested samples. (B) The corresponding data for nonphosphorylated peptides from the same samples.
Combining ETD with CID for Phosphopeptide Analysis.
To ensure that the same peptide is picked for ETD and CID fragmentation, we re-ran selected fractions in alternating CID and ETD mode. Examination of the ETD and CID spectra of which both had identified the exact same phosphopeptide showed that, on average, ETD resulted in ≈40% more back-bone fragment ions transforming into a 23% better sequence coverage, over CID (SI Data Set 1). For the group of peptides where an identical amino acid sequence was identified by both methods, although different residues were assigned as phosphorylated, we found that ETD had 47% higher sequence coverage as compared with CID (data not shown). The higher sequence coverage increases the confidence when assigning the modified residue. In Fig. 4, a direct comparison of the CID (Upper) and ETD (Lower) spectra are shown for four phosphopeptides identified in the alternating CID/ETD experiment. Fig. 4 A and B shows two phosphopeptides for which the two methods identified exactly the same residue as phosphorylated, whereas the identified residue differed for the phosphopeptides shown in Fig. 4 C and D. It is clear that the predominant fragments in the CID experiments arise from the β-elimination of H3PO4, whereas the ETD experiments provide a higher degree of amino acid coverage and a higher signal-to-noise ratio for the fragment ions together with a more uniform fragment distribution.
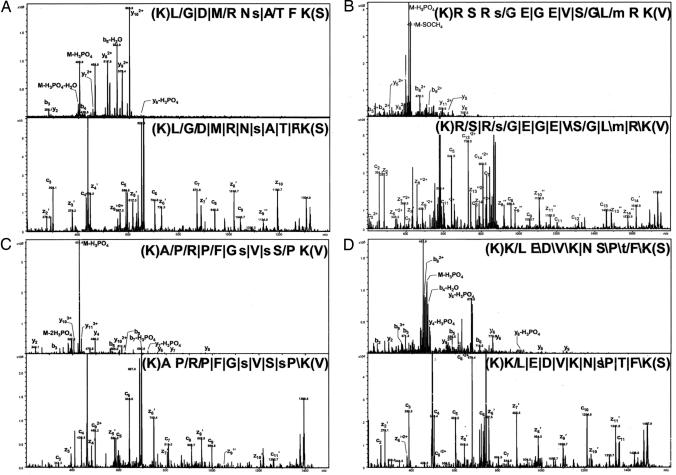
Fig. 4.
Direct comparison of phosphopeptides subjected to alternating CID and ETD experiments. The CID and ETD experiments identified the exact same sites for the two phosphopeptides show in A and B (from tumor protein D52-like 2 isoform E and tripartite motif-containing 28 protein, respectively) whereas C and D show phosphopeptides with identical amino acid sequence but different assignments of the phosphorylated residues (PDZ and LIM domain 5 isoform a and tumor protein D52 isoform 1, respectively). The peptide sequence with the fragmentation pattern is shown in each panel. The signs: “/”, “ /”, and “|” designate that the C-terminal type fragments, N-terminal fragments, or both type of fragments, respectively, were identified. All intensity axes have been enlarged ≈5 times.
Choosing a Protease for ETD-Based Phosphopeptide Analysis.
As shown in the above experiments, for Lys-C generated phosphopeptides, the ETD experiment identified significantly more phosphopeptides and phosphorylation sites than the corresponding CID experiments. To address the effect of the protease selection in a phosphorylation-directed ETD analysis, a subset of our samples (12 of 30) were digested with different proteases (Fig. 1). Trypsin is generally the preferred protease for LC-MS/MS analysis although other cleavage agents have also been used in proteomics experiments (10, 11). With a combined frequency of ≈11% for lysine and arginine residues in humans, the average tryptic peptide is ≈10 aa long with two or three positive charges, whereas the expected length with Lys-C, is twice that of trypsin. Comparing fractions digested with both Lys-C and trypsin, we found to our surprise that trypsin provided a similar number of ETD identified phosphopeptides as Lys-C (Fig. 5). The overlap between the tryptic phosphopeptides identified by ETD and CID was also similar to that observed in the case of the equivalent Lys-C experiment (data not shown). Further analysis of the tryptic phosphopeptides revealed an average length of 16 aa bearing an average of 3.3 positive charges. This result was explained by the finding of a high number of missed proteolytic events (on average 1.1 missed cleavages per phosphopeptide). If the enzymatic digestion were inefficient, one would also expect nonphosphorylated peptides to show the same characteristics. For this purpose, we also carried out a control experiment where a fraction (7%) of the digested samples were analyzed before enrichment of phosphopeptides. We found that the tryptic peptides in this data set had an average length of 12 aa and, on average, 0.3 missed cleavages (from 1,156 unique peptides) (data not shown). Thus, the number of missed cleavages events is significantly higher for phosphorylated peptides (≈4 times greater).
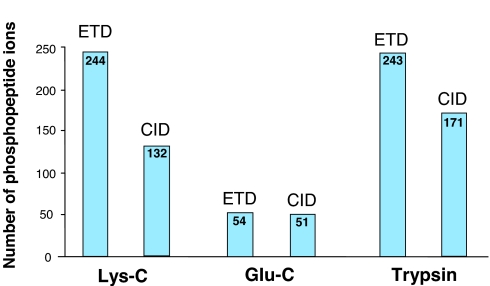
Fig. 5.
A summary of phosphopeptide analysis by CID and ETD. Identical samples digested with three proteases were analyzed by CID and ETD. The number of phosphopeptide ions identified in each of the sample sets digested with trypsin, Glu-C, and Lys-C is shown.
The enrichment of peptides with higher number of missed cleavages must therefore be accounted for by other reasons. One possible explanation is that the proximity of phosphorylated residues to arginine/lysine residues (12, 13) or acidic residues such as aspartic and glutamic acid (14) can hamper the efficiency of digestion by trypsin. An analysis of our data showed that, for 42% of the tryptic phosphopeptides, a phosphorylated residue was 1 or 2 aa away from the cleavage site, suggesting that the phosphorylated residue might have hindered cleavage by trypsin as noted previously (12, 13). Additionally, an analysis of the number of aspartic and glutamic acid residues adjacent to arginine or lysine residues revealed a 3 times higher occurrence in the phosphopeptides as compared with non-phosphopeptides. Similar results were observed for Lys-C peptides. A possible explanation for this phenomenon is that TiO2 used for enrichment not only enriches for phosphorylated residues, but also acidic residues, leading to an increased likelihood of enriching peptides that are phosphorylated and also contain more acidic residues. These phosphopeptides are thus more likely to have missed cleavage sites because of the increased likelihood of aspartic and glutamic acid residues occurring adjacent to a tryptic cleavage sites (14). We conclude that these missed cleavages are due to phosphorylation occurring in close proximity to sites of proteolytic cleavage and recommend that a higher number of missed cleavages events should be allowed during the database searching step of phosphoproteomics experiments.
Another protease, Glu-C, cleaving proteins at the C terminus of glutamic acid was also tested (see SI Fig. 6 E and F for base peak chromatograms) and resulted in dramatically fewer identified peptides in comparison with Lys-C and trypsin (Fig. 5). We attribute the poor outcome of Glu-C digests for identifying phosphopeptides to the fact that although the average length of peptides generated is indeed in a favorable range (between trypsin and Lys-C), the fragmentation in ETD is impaired, as noted previously for CID (15).
Discovery of Phosphorylation Motifs Through a Bioinformatics Analysis.
We have previously developed the Human Protein Reference Database (www.hprd.org) as a proteomic resource, which includes posttranslational modifications of proteins (16). Currently, there are 6,297 known unique phosphorylation sites annotated in this database. A comparison of the phosphorylation sites identified in our current study against the known sites revealed that ≈80% of the identified sites were novel, i.e., previously unreported (78% serine; 18% threonine; 5.6% tyrosine) (SI Data Set 1). We also carried out a comparison of the converse i.e., the number of instances where a known phosphorylation site on a sequenced phosphopeptide was identified or not identified in our study. We were able to identify 294 of 368 reported phosphorylation sites from a single sample in our study. All of those 368 phosphorylation sites were collected from 103 separate publications. This result has important implications for systems biology approaches because it shows that, from a single sample, we were able to capture 80% of phosphorylation events on these peptides ever recorded. Further, it also proves the importance of an in-depth analysis using complementary and partially overlapping technologies to obtain a comprehensive picture of the phosphoproteome. When we compared the gene ontology distribution of proteins corresponding to phosphopeptide data set to that of nonphosphorylated peptide data set, we found that certain biological processes were overrepresented in the phosphopeptide data set (regulation of nucleobase and nucleic acid metabolism, cell growth and/or maintenance) whereas others (protein metabolism, energy pathways) were overrepresented in the nonphosphorylated data set. It is important to note that regulation of nucleobase and nucleic acid metabolism is not generally associated with phosphorylation. This finding shows how global analyses such as the current study can serve to highlight unanticipated results, which can then be systematically investigated.
Because a large majority of the phosphorylation sites that we identified were previously undescribed, we reasoned that it is possible that they contain short conserved amino acid sequences around the phosphorylation site, or motifs, that have not been previously described. To test this hypothesis, we used a hierarchical clustering strategy in which sequences surrounding the phosphorylation sites were aligned and grouped to create small clusters (groups of peptides containing certain conserved residues). If a consensus pattern i.e., a similar stretch of amino acids, was present in ≥5 phosphopeptide sequences, we considered it as a potential motif. This analysis led to identification of a total of 68 motifs. To determine which of these motifs have not been previously described, we took advantage of a compendium of known motifs from the literature that we have recently created (17). We observed a large number of motifs that are known to be substrate motifs for proline-directed kinases ([pS/pT]P; PxpSP) such as MAP kinases, CDK5, and GSK3 kinases (18). In addition, a number of motifs corresponding to casein kinase 2, protein kinase C (PKC), and protein kinase A (PKA) were observed. Overall, of the 1,114 previously undescribed sites, 85% matched at least one known motif. Importantly, this approach also led to the identification of 15 motifs that had not been previously described (Table 1) highlighting the importance of coupling global experimental studies with computational approaches for refining our understanding of biological signals such as motifs.
Table 1.
Novel motifs that were identified by hierarchical clustering
| Novel motif | Occurrence in phosphopeptides | |
|---|---|---|
| 1 | pS[E/D]X[E/D][E/D] | 55 |
| 2 | pSPXXXP | 31 |
| 3 | pSPXXXT | 27 |
| 4 | DXXXp[S/T]P | 14 |
| 5 | GGpS | 13 |
| 6 | p[S/T]PPP | 12 |
| 7 | QXp[S/T]P | 12 |
| 8 | PSp[S/T]P | 11 |
| 9 | PPXp[S/T]P | 9 |
| 10 | PPp[S/T]P | 9 |
| 11 | EXSXp[S/T]P | 9 |
| 12 | PXpSPX[R/K] | 8 |
| 13 | PpSXL | 7 |
| 14 | PLp[S/T]P | 6 |
| 15 | TpTP | 5 |
We mapped the phosphorylation sites identified in this study to 500 unique genes (See Table 2 for a summary of the data). SI Table 3 shows the top 20 proteins from which phosphorylation sites were identified in this study. The protein with the largest number of identified phosphorylation sites (119 sites) was a splicing coactivator subunit of predicted molecular mass 300 kDa, SRm300 (also referred to as serine/arginine repetitive matrix 2). Interestingly, two related proteins, SRm160 and SRm75, which exist in a complex with the SRm300 protein, also featured on this list with 25 and 12 sites, respectively, making this probably the most phosphorylated protein complex yet described.
Table 2.
A summary of the phosphopeptide dataset reported in this study
| Category | No. of examples |
|---|---|
| No. of unique phosphorylation sites | 1,435 |
| Novel phosphorylation sites identified | 1,141 |
| No. of unique phosphorylated proteins | 500 |
| Phosphopeptides with 1, 2, 3, or 4 phosphorylation sites | 868; 374; 93; 24 |
| Phosphorylated (serine:threonine:tyrosine) residues | 1,096:266:73 |
| Phosphopeptides identified by both CID and ETD | 129 |
| Protein with the most phosphorylation sites identified: SRm300 (serine/arginine repetitive matrix 2) | 119 total (71 novel, 48 known) |
Discussion
The success of a phosphoproteomics experiment is highly dependent on the use of particular enrichment methods and sample processing steps. Phosphopeptide enrichments protocols employing TiO2 and immobilized metal affinity chromatography are biased toward peptides containing negatively charged residues. This bias implies that it is possible that certain phosphorylation motifs are overrepresented whereas others are underrepresented in the enriched phosphopeptide data set. Another parameter that is affected by the presence of acidic residues is cleavage by proteases. It is known that trypsin does not cleave efficiently when an acidic amino acid is located adjacent to arginine or lysine residues. Because of this behavior of trypsin, as demonstrated in this study, the identified phosphorylated tryptic peptides are longer when reagents such as TiO2 is used for enrichment, resulting in a higher number of positive charges per peptide. As a result, tryptic peptides are almost as good as Lys-C for phosphorylation analysis by ETD. This finding also highlights the need for allowing greater flexibility in terms of missed cleavage events during a database search. Steps can be taken to minimize this bias toward enrichments of peptides with a number of acidic residues by derivatization (19) or charge based fractionation (20).
In this study, we have demonstrated the advantages of using ETD as a fragmentation method for global phosphoproteomics analysis. In all, when comparing identical fractions we found 60% more phosphopeptides with ETD than with CID. Most studies thus far have used CID as a fragmentation method for phosphorylation analysis in conjunction with tryptic digests. Although, a large number of phosphorylation sites can undoubtedly be discovered by using the conventional CID-based approach, we believe that incorporation of ETD as a fragmentation method will be necessary to obtain more comprehensive coverage as well as to improve the confidence of phosphorylation sites because ETD is a complementary technique to CID. As we have demonstrated, alternating CID and ETD experiments can be easily carried out on existing mass spectrometers and provide an excellent platform to carry out phosphoproteomics analysis. Finally, it is likely that coupling of CID to ETD for peptide identification will be fruitful especially in cases where the peptide coverage is not high such as proteins based on “single peptide identification.” Given the central role of mass spectrometric analysis in systems biology approaches today, we believe that inclusion of ETD as a fragmentation method will be advantageous for any proteomic analysis.
Materials and Methods
Sample Preparation and Processing.
Human embryonic kidney 293T cells were treated with the phosphatase inhibitors pervanadate and calyculin A (Sigma, St. Louis, MO) for 25 min. The cells were then lysed by ultrasonic treatment followed by nitrogen cavitation (Parr Instrument Company Moline, IL). Cell debris was removed by centrifugation at 16,000 × g for 10 min. Urea was added to the protein sample (final concentration: 6 M), followed by reduction, alkylation, and acidification. Five hundred microliters (≈500 μg) of the protein sample was injected by using a loop onto a reversed phase column (mRP-C18 High-Recovery Protein Column; Agilent Technologies, Santa Clara, CA) maintained at 80°C. Proteins were separated by using a 1%/min gradient of 0.1% TFA (buffer A) and acetonitrile/0.08% TFA (buffer B) delivered by a binary pump at 800 μl/min (1100 series; Agilent Technologies) and detected at 280 nm (UV detector 1100 series; Santa Clara, CA). Protein fractions were collected every 30 s, and the resulting fractions were divided into three sets. The fractions were dried down and redissolved in 40 μl of 0.1 M NH4HCO3 containing either 0.15 μg or 0.3 μg of either trypsin (Promega, Madison, WI), lysyl endopeptidase/Lys-C (Wako Chemicals, Richmond, VA), or endoproteinase Glu-C (Roche Diagnostics GmbH, Penzberg, Germany). Protein fractions were digested overnight at room temperature. For quality control purposes, 7% of selected digested fractions were taken before the phosphopeptide enrichment step and analyzed by LC-MS/MS (CID).
Phosphopeptide Enrichment using TiO2.
The peptides obtained from the digestion step were dried down, and selected fractions were redissolved in 20 μl of 80% acetonitrile, 66 mg/ml 2,5-dihydrobenzoic acid, and 1% TFA and loaded onto a home-built TiO2 (Glygen Corp., Columbia, MD) microcolumn (1–2 mm) packed in a GelLoader tip, as described by Larsen et al. (7). After washing twice with 50 μl of 80% acetonitrile containing 1% TFA, the bound peptides were eluted in 20 μl of 3% NH4OH, pH 10.5, dried, and redissolved in 5 μl of 0.1% formic acid before LC-MS/MS analysis.
LC-MS/MS and Data Analysis.
All LC-MS/MS analyses were performed on an Agilent 1100 Series HPLC-Chip/MS system interfaced to a 3-D ion trap mass spectrometer (Agilent 6340; Agilent Technologies) equipped with an ETD source. Peptides were separated by reversed-phase LC with a precolumn/analytical column nano-flow setup (HPLC-Chip cube; Agilent Technologies) and analyzed in either CID only, ETD only, or alternating CID/ETD modes.
Protein identification was performed by using Spectrum Mill Proteomics Workbench Version A.03.03 (Agilent Technologies) searching the human subset of the NCBI RefSeq database. An initial search was performed by using two missed cleavages with complete proteolytic specificity (Trypsin, Lys-C, or Glu-C), +/−4 Da for the precursor mass, +/−0.7 Da for the fragment masses, 40% minimum scored peak intensity, and 5+ for the maximum ambiguous charge state for the spectra with precursors of unassigned charge state. Phosphorylation of serine, threonine, and tyrosine residues, together with oxidation of methionines, was allowed as variable modifications. After this first search, spectra were manually validated (see below for details), and a smaller database was created by using the proteins identified by these validated spectra. Further searches were performed against this database, allowing for semienzymatic cleavages, and up to four missed cleavages for spectra that contained a sequence tag >4.
To validate that a protein was actually present in the sample, at least one of the peptide identifications must exhibit a confident score (>14–16 depending on charge state) and a large δ-forward reverse score (>5) along with at least three complementary c and z cleavages within the matched sequence. For peptides with a lower score from the same protein, the peptides must still exhibit a large sequence tags of matched c and/or z ions but do not require a large score and/or delta-reverse score. Further, peptide matches that did not meet the above criteria were allowed if a confident identification of the same peptide was found in the same data set. In a second step, peptides were validated in “peptide” mode where the remaining peptide identifications were evaluated individually. First, any peptide with a score >16 was automatically validated. For the remaining peptides, each spectrum was inspected independently. In general, 5+ identifications required a score >14, 4+ a score >12, 3+ a score >9, and 2+ with a score >7 to be validated as an individual peptide. The counting of peptides and phosphopeptides was carried out in accordance with recently published guidelines (21) to avoid redundancy.
Bioinformatics Analysis of Phosphorylation Sites.
For hierarchical clustering, the phosphorylated serine/threonine/tyrosine residues identified in this study and the flanking 7 aa on both sides were used to generate a library of 1,435 15-mer peptide sequences. A hierarchical clustering algorithm was developed by using Python shell scripting (version 2.3) for identifying conserved amino acid sequences, or motifs, present among the phosphopeptides. The algorithm was based on agglomerative hierarchical clustering in which each peptide sequence is initially placed into its own group. Each of these groups contains only a single peptide, referred to as a singleton. All groups were compared with each other and the closest groups were merged into a single new group if the distance between a specific pair of group was less than the threshold distance. The threshold for the distance function was provided by BLOSUM62 matrix, based on a likelihood method estimating the occurrence of each possible pair wise substitution, which assigns a score to every identity or substitution based on the observed frequencies of such occurrences in alignments of related proteins (22). Clustering is continued until the distance between the closest pair is greater than this threshold.
Supplementary Material
Acknowledgments
We thank Ramars Amanchy, Balamurugan Periaswamy, Jakob Bunkenborg, and Jens Andersen for fruitful discussions. This work was supported by National Heart Lung and Blood Institute Contract N01-HV-28180 and by National Institutes of Health Grants U54RR020839 and R01CA106424. A.P. is also supported by a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation.
Abbreviations
- CID
collision-induced dissociation
- ETD
electron transfer dissociation
- MS/MS
tandem MS
- LC
liquid chromatography.
Footnotes
Conflict of interest statement: D.M.H. and N.T. are employees of Agilent Technologies. All other authors have declared no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611217104/DC1.
References
- 1.Haynes PA, Aebersold R. Anal Chem. 2000;72:5402–5410. doi: 10.1021/ac000512w. [DOI] [PubMed] [Google Scholar]
- 2.Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, Hart GW. Anal Biochem. 1996;234:38–49. doi: 10.1006/abio.1996.0047. [DOI] [PubMed] [Google Scholar]
- 3.Chalkley RJ, Burlingame AL. J Am Soc Mass Spectrom. 2001;12:1106–1113. doi: 10.1016/s1044-0305(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 4.Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 5.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Anal Chem. 2005;77:1831–1839. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O'Hair RA, Speed TP, Simpson RJ. Anal Chem. 2003;75:6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 10.Wu CC, Yates JR., III Nat Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 11.Wu CC, MacCoss MJ, Howell KE, Yates JR., III Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 12.Benore-Parsons M, Seidah NG, Wennogle LP. Arch Biochem Biophys. 1989;272:274–280. doi: 10.1016/0003-9861(89)90220-8. [DOI] [PubMed] [Google Scholar]
- 13.Schlosser A, Pipkorn R, Bossemeyer D, Lehmann WD. Anal Chem. 2001;73:170–176. doi: 10.1021/ac000826j. [DOI] [PubMed] [Google Scholar]
- 14.Thiede B, Lamer S, Mattow J, Siejak F, Dimmler C, Rudel T, Jungblut PR. Rapid Commun Mass Spectrom. 2000;14:496–502. doi: 10.1002/(SICI)1097-0231(20000331)14:6<496::AID-RCM899>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Lab Invest. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 16.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikkota SG, Pandey A. Nat Biotechnol. 2007 doi: 10.1038/nbt0307-285. in press. [DOI] [PubMed] [Google Scholar]
- 18.Pelech SL. Neurobiol Aging. 1995;16:247–256. doi: 10.1016/0197-4580(94)00187-6. [DOI] [PubMed] [Google Scholar]
- 19.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 20.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Henikoff S, Henikoff JG. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.