Abstract
The majority of human microRNA (miRNA) loci are located within intronic regions and are transcribed by RNA polymerase II as part of their hosting transcription units. The primary transcripts are cleaved by Drosha to release ∼70 nt pre-miRNAs that are subsequently processed by Dicer to generate mature ∼22 nt miRNAs. It is generally believed that intronic miRNAs are released by Drosha from excised introns after the splicing reaction has occurred. However, our database searches and experiments indicate that intronic miRNAs can be processed from unspliced intronic regions before splicing catalysis. Intriguingly, cleavage of an intron by Drosha does not significantly affect the production of mature mRNA, suggesting that a continuous intron may not be required for splicing and that the exons may be tethered to each other. Hence, Drosha may cleave intronic miRNAs between the splicing commitment step and the excision step, thereby ensuring both miRNA biogenesis and protein synthesis from a single primary transcript. Our study provides a novel example of eukaryotic gene organization and RNA-processing control.
Keywords: Drosha, intron, MicroRNA, processing, splicing
Introduction
MicroRNAs (miRNAs) are noncoding RNA species of ∼22 nt that induce post-transcriptional gene silencing through base-pairing with their target mRNAs (Bartel, 2004; Kim, 2005). This intriguing family of regulatory factors is transcribed by RNA polymerase II (pol II) and regulated both temporally and spatially (Cai et al, 2004; Lee et al, 2004). Like other pol II transcripts, the primary transcripts (pri-miRNAs) are capped and polyadenylated. Pri-miRNAs contain a local stem–loop structure that encodes miRNA sequences in the arm of the stem. This stem–loop structure is cleaved by the nuclear RNase III type enzyme Drosha in a process known as ‘cropping' (Lee et al, 2003). Drosha functions as a complex called ‘Microprocessor' that also contains a double-stranded RNA-binding protein, DGCR8 (also known as Pasha in flies and nematodes) (Denli et al, 2004; Gregory et al, 2004; Han et al, 2004; Landthaler et al, 2004). The processing reaction releases the stem–loop-shaped intermediates that are referred to as pre-microRNAs (pre-miRNAs). Each pre-miRNA is subsequently exported to the cytoplasm by exportin-5 (Yi et al, 2003; Bohnsack et al, 2004; Lund et al, 2004), and in the cytoplasm these precursors are cleaved by the cytoplasmic RNase III Dicer (Bernstein et al, 2001; Grishok et al, 2001; Hutvagner et al, 2001; Ketting et al, 2001; Knight and Bass, 2001). Dicer produces an RNA duplex of ∼22 nt, one strand of which is selected and incorporated into the effector complex (Khvorova et al, 2003; Schwarz et al, 2003) known as the RNA-induced silencing complex (RISC). RISC binds to the 3′ untranslated region (UTR) of the target mRNA in a sequence-specific manner, and induces mRNA cleavage, translational inhibition or cleavage-independent mRNA degradation (Pillai, 2005; Valencia-Sanchez et al, 2006).
It was initially thought that miRNAs are encoded mostly in ‘intergenic regions' (in other words, in their own transcription units) (Kim, 2005). This was because the majority of miRNA loci are located in noncoding transcription units (TUs) or in poorly characterized coding TUs. However, more careful analysis revealed that a significant number of miRNAs are in the introns of TUs that are predicted or annotated to be protein-coding. The numbers may change, however, because many of these TUs have not been experimentally validated for their protein-coding capacity.
Most early studies of miRNA gene structure have been conducted on exonic miRNA loci in noncoding TUs (Cai et al, 2004; Lee et al, 2004; Houbaviy et al, 2005). However, a recent study by Rodriguez et al (2004) showed that the majority of the human miRNA loci are in fact located within the intronic regions of either coding or noncoding TUs (Rodriguez et al, 2004). The expression of these miRNAs largely coincides with the transcription of the hosting TUs, indicating that the intronic miRNAs and their hosting genes may be coregulated and generated from a common precursor transcript (Baskerville and Bartel, 2005). This raises an interesting question as to how these intronic miRNAs get processed.
In an analogous manner to intronic small nucleolar RNAs (snoRNAs) (Filipowicz and Pogacic, 2002), it has been generally assumed that Drosha cleaves the excised introns after splicing reaction and thereby both mRNA and miRNA can be coexpressed from a single primary transcript molecule. However, this hypothesis has not yet been experimentally validated. In our present study, we aimed to understand how miRNA-bearing introns are processed by two separate machineries (the spliceosome and the Microprocessor) to generate both mature mRNA and miRNA from a single TU.
Results
EST analysis of miRNA-containing transcription units
In a previous study, Rodriguez et al (2004) demonstrated that the majority of the miRNA loci reside in intronic regions of hosting TUs. We performed a similar series of analyses using a slightly different set of databases that contain ESTs as well as known genes. By employing the UCSC genome browser, we mapped miRNA-encoding sequences to hosting TUs (Karolchik et al, 2003). From our current analysis of 221 miRNAs in total, we found that 155 miRNAs mapped to at least one EST (Figure 1A and Supplementary Table 1). These data also revealed that the vast majority of miRNA genes reside in intronic regions (127 out of 155 miRNAs, 82%) (Figure 1A). In addition, 19 miRNA loci were found to reside within the exons of hosting TUs (exonic, 12%), whereas nine miRNAs match both introns and exons, depending on the alternative splicing patterns (‘mixed', 6%). Excluding the ‘mixed' examples, the percentages of intronic and exonic miRNAs are therefore 87% (127 out of 146 miRNAs) and 13% (19 out of 146 miRNAs), respectively. Our observations are similar to those of a previous report in which intronic and exonic miRNAs were estimated to be 80 and 20%, respectively (Rodriguez et al, 2004).
Figure 1.
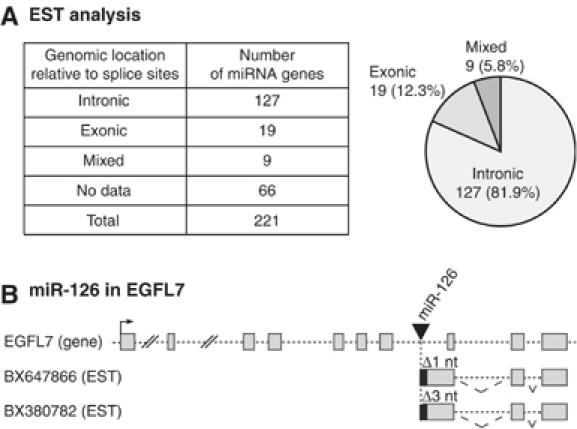
EST analysis of microRNA. (A) The genomic location of miRNAs relative to their hosting genes. In Supplementary Table 1, the hosting gene status for each miRNA is described in a greater detail. (B) ESTs that match EGFL7 gene. The 5′ ends of two ESTs begin at 1 or 3 nt downstream of the 3′ end of mature miR-126.
Interestingly, some EST fragments mapped to a location that is either immediately upstream or downstream of miRNA loci. The 5′ or 3′ ends of these ESTs match the putative Drosha cleavage sites, implicating that they may be the cleavage products of the cropping process. For instance, miR-126 resides in the seventh intron of the EGFL7 gene. The 5′ ends of two EGFL7 ESTs begin at 1 or 3 nt downstream of the 3′ end of the mature miR-126 (Figure 1B), indicating that they may have been derived from the 3′ cleavage products of cropping. Intriguingly, these two ESTs possess the miR-126-encoding intronic sequences but lack the downstream introns. This observation suggests that miRNA-harboring introns may be spliced slowly compared to the downstream introns and that Drosha may cleave the miR-126 hairpin before the miRNA-encoding intron is spliced out.
This observation raised the question of whether Drosha destroys the unexcised intron of the primary transcript before the completion of splicing and thereby prohibits protein synthesis from the given transcript. In other words, is miRNA biogenesis mutually exclusive to protein biogenesis when the miRNA is encoded in the intron of a protein-coding gene?
Processing of miRNA-harboring introns
In order to examine whether the miRNA hairpin is indeed released from an unexcised intron, we depleted Drosha and determined the level of the unspliced transcripts. If Drosha cleaves unexcised introns, it is expected that the unspliced introns would accumulate in Drosha-depleted cells. SiRNA against Drosha was transfected into HeLa cells and total RNA was analyzed by reverse transcription–PCR (RT–PCR). The primer sets had been designed to amplify both the nascent transcripts and the partially spliced transcripts (Figure 2A, left panel). Structures of the genes analyzed in this study are shown in Supplementary Figure 1. The first set of primers (Figure 2A, Puf and Pur, shown in red) amplifies the upstream region. Two distinct transcripts (Un and Up, shown in red) were detected using this upstream primer set. The longer PCR product (Un) amplified by Puf and Pur corresponds to a nascent transcript retaining the upstream intron. The shorter PCR product (Up) corresponds to the partially spliced transcript that lacks the upstream intron. The second set of primers (Figure 2A, Pdf and Pdr, shown in blue) amplifies the downstream region. These primers also detected two kinds of transcripts: nascent (Dn) and partially spliced transcripts (Dp) (Figure 2A, shown in blue). In all of the cases that we analyzed, partially spliced transcripts (Up and Dp) accumulated upon Drosha depletion, whereas the levels of nascent transcripts (Un and Dn) only slightly increased (Figure 2A, right panel).
Figure 2.
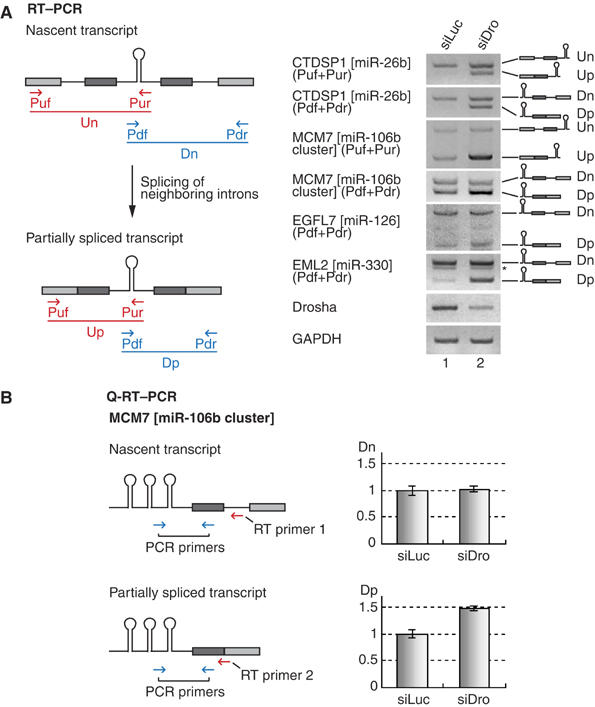
Splicing of microRNA-encoding introns. (A) RT–PCR for the detection of nascent and partially spliced transcripts. The positions of primers (arrows), exons (filled boxes) and introns (solid lines) are indicated. PCR products are depicted as either red or blue solid lines. SiRNA duplexes targeting luciferase (siLuc) or Drosha (siDro) were transfected into HeLa cells and, after 48 h, total RNA was prepared and used for RT–PCR. (B) Real-time PCR of the nascent or partially spliced transcript of MCM7 gene. Different RT primers were used to distinguish these two transcripts, and the same PCR primer set was used for real-time PCR reaction (n=5).
We also carried out real-time PCR experiments to confirm the above results. Two different RT primers were used to distinguish nascent and partially spliced transcripts. The RT primer that is complementary to the downstream intronic sequence was to detect the nascent transcript (Figure 2B, RT primer 1), whereas the RT primer that anneals to the downstream exon–exon junction could detect the partially spliced transcript (Figure 2B, RT primer 2). Consistent with the above results, only the partially spliced transcripts accumulated upon depletion of Drosha. This indicates that Drosha is required specifically for the turnover of the partially spliced transcripts. There may be at least two explanations to this observation. The first and obvious possibility is that, in Drosha-depleted cells, the partially spliced transcripts accumulated because cropping reaction was slowed down. In a normal condition, Drosha would rapidly remove these transcripts by cleaving away the hairpin in the intron. Second possibility is that Drosha may somehow be required for the splicing of miRNA-containing introns but not of the adjacent introns. The depletion of Drosha may have slowed down the splicing of miRNA-containing introns, resulting in the accumulation of transcripts that retain the miRNA-harboring introns.
To test this possibility, we determined the amount of spliced mRNA in Drosha-depleted cells by RT–PCR (Figure 3). If Drosha is indeed required for splicing, splicing products (mature mRNA) would be reduced in Drosha-depleted cells. Primers that are complementary to the neighboring exons were used to detect spliced mRNAs (Figure 3A). For all the genes that we tested (CTDSP1 containing miR-26b, MCM7 containing miR-106b cluster, EGFL7 containing miR-126 and EML2 containing miR-330), the levels of fully spliced mRNAs remained unaffected in Drosha-depleted cells compared to those in control siRNA-treated cells. We also performed quantitative real-time PCR on MCM7 mRNA (Figure 3B). The MCM7 gene contains miR-106b cluster in its 13th intron. As expected, the level of spliced mRNA in Drosha-depleted HeLa cells was not significantly different from that in control siRNA-treated cells. Therefore, it is unlikely that Drosha is required for the splicing of the miRNA-harboring introns.
Figure 3.
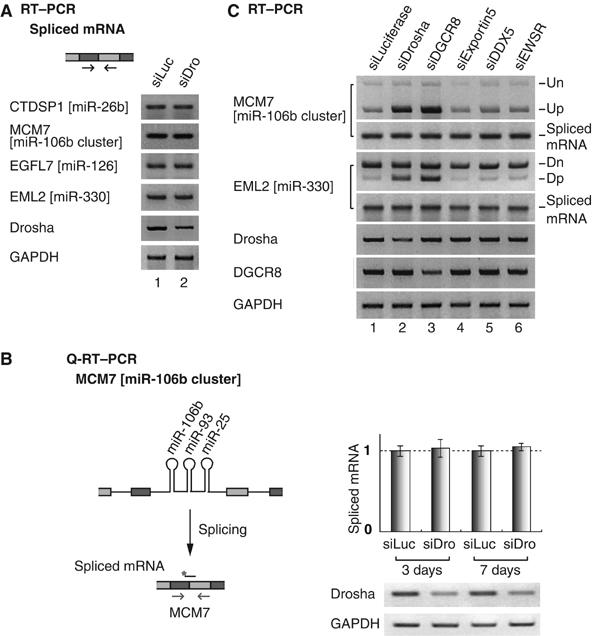
The production of spliced mRNA is unaffected by miRNA cropping. (A) RT–PCR to detect spliced mRNA. SiRNA duplexes targeting luciferase (siLuc) or Drosha (siDro) were transfected into HeLa cells and, after 48 h, total RNA was prepared and used for RT–PCR. (B) Quantitative real-time PCR of MCM7 spliced mRNA. The arrows and the black bar with an asterisk indicate the position of PCR primers and Taqman probe, respectively. Each Ct value of MCM7 was normalized to the Ct value of GAPDH (n=6). (C) Depletion of DGCR8 exerts the same effects as those of Drosha on the processing of miRNA-encoding introns. SiRNA was transfected into HeLa cells as in panel A and Figure 2A.
Comparable results were observed when DGCR8, the essential cofactor for Drosha, was depleted instead of Drosha (Figure 3C). As in Drosha-depleted cells, the partially spliced transcript retaining miRNA-encoding intron (Up and Dp) accumulated, whereas the spliced mRNA level did not significantly change in DGCR8-depleted cells. On the contrary, the knockdown of other genes that are not involved in cropping (exportin-5, DDX5 and EWSR) did not alter the level of these transcripts (Figure 3C).
To generalize our conclusion, we randomly selected several TUs and determined the levels of unspliced transcripts and spliced mRNAs by RT–PCR (Figure 4). In all cases that we tested, the unspliced transcripts accumulated in Drosha-depleted cells, whereas the level of spliced mRNA remained largely unchanged. The positions of these miRNAs inside the corresponding introns are highly variable. Also, the lengths of the introns vary greatly (ranging between ∼2000 and ∼10 000 nt). Thus, the processing of miRNA-harboring introns is unlikely to be affected by the location of miRNA or the length of the intron.
Figure 4.
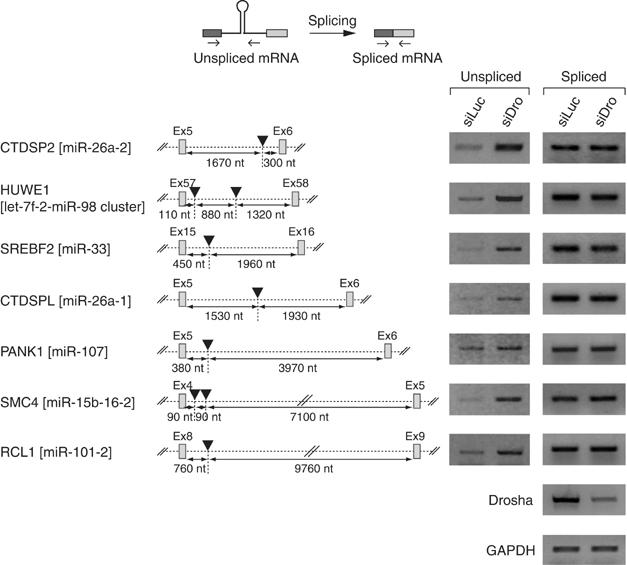
Processing of miRNA-harboring introns is unaffected by the length of the intron and the location of miRNA. The black triangle indicates the location of miRNA.
Splicing is not a prerequisite for cropping
In order to examine if the pre-mRNA splicing and cropping process have any effects on each other, we designed reporter constructs that contain the sequences of the BCL2L2 gene under the control of the human cytomegalovirus promoter (Figure 5A). The RNA transcribed from this construct is expected to contain two exons and one intron. A let-7a-1 miRNA hairpin was inserted into the middle of the intron so as to determine the effects of having an miRNA hairpin upon splicing (Figure 5A, BCL-let-7a). In addition, a splicing-deficient construct was designed by introducing mutations at the 5′ and 3′ consensus splice sites to examine the influence of splicesome assembly on the cropping process (Figure 5A, BCL-m-let-7a). After transfection of these plasmids into HEK293T cells, total RNA was extracted and analyzed by RT–PCR. As controls, the transcripts for GAPDH and the neomycin resistance gene were measured (Figure 5B). The neomycin resistance gene is encoded in the reporter plasmid; so the expression of this gene is expected to reflect the transfection efficiency.
Figure 5.

Pre-mRNA splicing is not required for cropping. (A) Reporter constructs. Upper row: wild-type BCL2L2 mRNA (exon: filled box; intron: solid line). The miRNA stem–loop was inserted into the EcoRI site in the intron. Middle row: miRNA-inserted construct. Lower row: splicing-deficient form of miRNA-inserted construct (red dots indicate the sites of mutation). The mutated sequences are indicated in red. (B, C) RT–PCR and Northern blotting. Reporter constructs were transfected into HEK293T cells and, 2 days later, total RNA was extracted for RT–PCR and Northern analysis. In panel C, the band indicated with an asterisk (*) represents other members of the endogenous let-7a family. 18S rRNA stained with EtBr is shown as loading control.
The insertion of the let-7a-1 hairpin did not notably alter the levels of mature BCL2L2 mRNA (Figure 5B, primers Pf and Pr, lanes 2 and 3), which again demonstrates that cropping of an intronic miRNA hairpin does not significantly affect the splicing efficiency of the given intron. The pre-mRNA with mutated splice sites (BCL-m-let-7a) remained unspliced as expected (Figure 5B, lane 4).
The levels of let-7a miRNA expressed from these reporter constructs were determined by Northern blotting (Figure 5C). Let-7a miRNA was successfully expressed from the reporter constructs (Figure 5C, lanes 3 and 4). Importantly, the level of pre-let-7a-1 from the splicing-deficient construct (BCL-m-let-7a) was only slightly decreased compared with the wild-type construct (BCL-let-7a) (Figure 5C, compare lanes 3 and 4). These results demonstrate that a splicing event is not a prerequisite for pri-miRNA processing.
Cropping does not affect splicing reaction
To further examine the effects of cropping upon splicing, we generated an additional intronic miRNA reporter (CTDSP1-miR-26b) using the sequences of the CTDSP1 gene (Figure 6A and Supplementary Figure S1a). The genomic sequence of CTDSP1 was amplified by PCR from HeLa genomic DNA and inserted at the downstream of the cytomegalovirus promoter. The transcript expressed from this construct contains four exons and three introns. The miR-26b hairpin is present in the second intron. We also generated a mutant reporter (CTDSP1-miR-26b-m) that has altered sequences in the miRNA stem region, which prevents cropping (Figure 6A, lower). These miR-26b reporters were transfected into HEK293T cells and total RNA was subsequently extracted for Northern analysis. The wild-type construct yielded miR-26b, but the mutant construct failed to produce any detectable levels of this miRNA species (Figure 6B). This mutant hairpin RNA can be immunoprecipitated with DGCR8 as efficiently as the wild-type hairpin (Supplementary Figure S2), indicating that this mutation in the hairpin does not affect the affinity to DGCR8. This mutation may instead prevent the catalytic reaction (Han et al, 2006).
Figure 6.
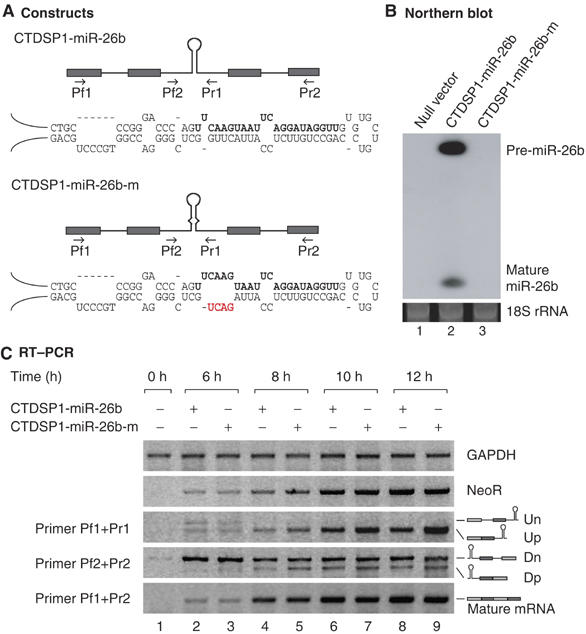
Splicing and cropping of four exons–three introns reporters. (A) Schematic depiction of the constructs used in this experiment. The hairpin represents the miR-26b stem–loop and the arrows indicate the position of primers. The mature miRNA sequences are written in bold whereas the mutated sequences are shown in red. (B) Northern blot analysis of miR-26b reporter expression. Two days after transfection into HEK293T cells, total RNA was extracted and analyzed by Northern blotting. (C) Time-course experiment of the reporter constructs. After transfection of the reporter plasmid into HEK293T cells, total RNA was prepared at the indicated time. The neomycin resistance gene was used as transfection efficiency control. All experiments were performed at least three times and representative results are shown.
We performed RT–PCR to detect splicing products at different time points (Figure 6C). Compared with the wild-type construct, the cropping mutant yielded higher steady-state levels of partially spliced transcripts (Up and Dp) (Figure 6C). In contrast, the steady-state levels of nascent transcripts (Un and Dn) from the mutant were not significantly different from that of wild type (Figure 6C). Consistent with our initial results (Figures 3, 4 and 5), both constructs generated comparable levels of fully spliced mature mRNA regardless of their susceptibility to Drosha-mediated cropping (Figure 6C, primer Pf1+Pr2). This indicates that Drosha may not significantly affect splicing reaction. From these observations, we conclude that partially spliced transcripts may be the major substrates for Drosha in vivo and that intron cleavage by Drosha may not affect the production of mature mRNA.
Discussion
It has been suggested previously that spliced introns may be debranched and be used as the substrate for Drosha (Ying and Lin, 2006), with analogy to the intronic small nucleolar RNAs (snoRNAs) (Filipowicz and Pogacic, 2002). However, our results strongly indicate that splicing is not a prerequisite for miRNA production and that an unspliced intron may serve as the substrate for the Microprocessor. If Drosha indeed cleaves unexcised introns before splicing and releases pre-miRNA, what happens to the rest of the transcript? Does cropping preclude the production of mature mRNA? Surprisingly, our data suggested otherwise: spliced mRNA is produced at a normal rate.
It may seem paradoxical that an unspliced intron is cleaved by Drosha before splicing reaction and yet this cleavage does not reduce the level of the splicing product. Recently, it has been shown by Dye et al (2006) that a continuous intron is unnecessary for pre-mRNA splicing in vivo. The authors engineered an intron by inserting either a cotranscriptional cleavage element or a ribozyme so that the intron would be cotranscriptionally cleaved. The exons franking this intron were accurately and efficiently spliced, suggesting that the preceding exon may be tethered to the elongating pol II complex until the next exon emerges from pol II (Dye et al, 2006). Tethering of exons to the elongating pol II complex may optimize the process of splice site selection. Our current study supports and extends this ‘exon-tethering' model.
Our results imply that the miRNA-harboring intron may be cotranscriptionally defined and the splice sites may be physically tied to each other in a relatively stable complex (Figure 7). It is conceivable that the 5′ and 3′ splice sites are linked by U1 snRNP, the U2AF heterodimer and SR proteins in a form of splicing commitment complex (CC, also known as E complex) (Figure 7). The Microprocessor cleaves the intron to release pre-miRNA. Because the exons have already been paired and tethered to each other, splicing catalysis would still occur efficiently regardless of the discontinuity of the intron (Figure 7). This model would explain why mature mRNA levels remain largely unchanged when cropping has been blocked (Figures 3 and 6). Thus, exon pairing/tethering may be an elegant and efficient way to coordinate the two separate processes of miRNA biogenesis and protein synthesis from a single TU. This provides a novel example of multiple gene organization and RNA-processing control.
Figure 7.
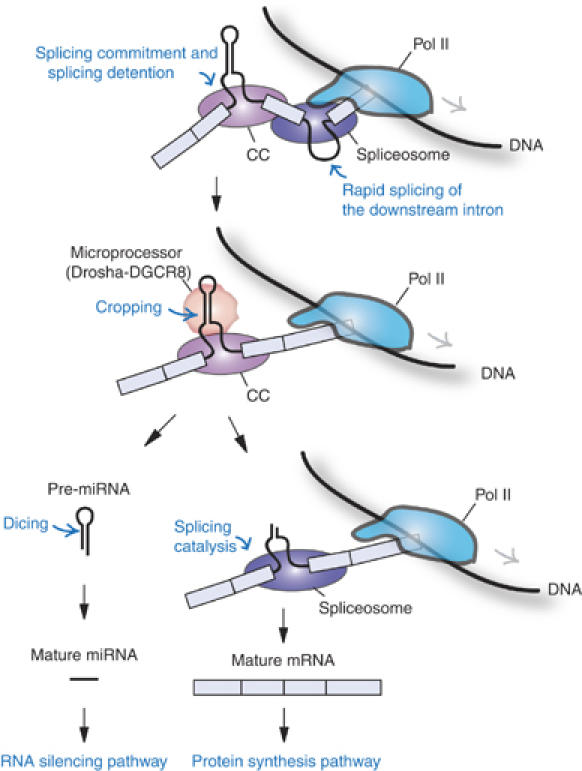
A model for the processing of intronic microRNA. The exons flanking miRNA are cotranscriptionally defined and physically bound to each other by a ‘splicing commitment complex' (CC). Downstream introns may be spliced out rapidly, whereas splicing of the miRNA-harboring intron is detained. The Microprocessor cleaves the intron to release pre-miRNA that is subsequently processed by Dicer. Because the exons had already been paired and tethered to each other, splicing catalysis would still occur efficiently regardless of the discontinuity of the intron. Details are described in the text.
We note that we cannot formally exclude the possibility that excised introns may also participate in miRNA production. To address this possibility, we have tried exhaustively to determine whether spliced introns (in both lariat form and debranched form) accumulate when Drosha is depleted. Thus far, we have not been able to detect excised introns by any methods that we tried (RT–PCR and RNase protection assay), indicating that excised introns are extremely unstable in mammalian cells (data not shown). It is also noted that there remains a possibility that there are multiple overlapping transcription units and/or that an miRNA-producing precursor may not be the same molecule as the mRNA-producing one. These possibilities are technically not feasible to be addressed at this point.
It is interesting that the adjacent introns seem to be spliced more rapidly than miRNA-encoding intron. This suggests that the binding of Microprocessor may interfere with the splicing of miRNA-harboring intron to some extent. In fact, Drosha knockdown resulted in small but reproducible increases in spliced mRNA levels (Figure 3B).
It is also noteworthy that the pre-miRNA level expressed from the BCL construct was slightly reduced when splicing sites were mutated (Figure 5C). This suggests that splicing may facilitate cropping to some extent. It is tempting to speculate that the splicing commitment complex may facilitate the recruitment of the Microprocessor to the miRNA hairpin. The recruited Microprocessor (or its associated factor(s)) may briefly interfere with the splicing reaction. Once the hairpin is cleaved away by Drosha, the interfering factor(s) may be removed, allowing the completion of splicing reaction. In fact, an immunoprecipitation experiment has shown that Drosha interacts with proteins that are known to be involved in splicing (Gregory et al, 2004). It would be interesting in the future to investigate whether Drosha or DGCR8 interacts with any of the splicing factor(s) so as to coordinate the splicing and cropping processes.
Materials and methods
MicroRNA database
The total human miRNA list was retrieved from the microRNA registry release 4.0 (http://microrna.sanger.ac.uk/sequences/; Griffiths-Jones, 2004). The number of total human miRNAs used in this study was 221. Sense and antisense miRNAs from the same stem were counted independently.
EST analysis
A BLAT search (Kent, 2002) was initially carried out to identify the position of the miRNA locus. After identifying the miRNA site using BLAT, its position was compared with gene loci using RefSeq, Known gene, human mRNA and human EST database. When each miRNA was matched with a gene that was verified as having protein-coding capabilities, the hosting gene status was marked with ‘Known gene' (see Supplementary Table 1). If the miRNA species was matched to a full-length mRNA, but not matched to known gene or with RefSeq, that is, its protein-coding capability was only predicted, a ‘Predicted' hosting gene status was assigned. Finally, when the miRNA was matched only with an EST, the hosting gene status was designated as ‘EST only'. We used spliced ESTs only, as their unspliced counterparts cannot distinguish whether the sequence is exonic or intronic.
The cleavage products of Drosha were analyzed using all types of EST, both spliced and unspliced EST. If either the 5′ or 3′ ends of the mature miRNA duplex were not known exactly, the Drosha and Dicer cleavage sites were estimated based on the assumption that all miRNA duplexes contain 2-nt 3′ overhangs at both ends.
Cell culture and transfection
HeLa and HEK293T cells were cultured in DMEM (WelGENE) supplemented with 10% FBS (WelGENE). Transfection to HEK293T cells was carried out using the calcium-phosphate method. Lipofectamine 2000 (Invitrogen) was used for transfection of HeLa cells.
Plasmid construction
The BCL2L2 and CTDSP1 genes were PCR-amplified using HeLa genomic DNA. The primers for the BCL2L2 reporter were 5′-GAA TTC TGA TGG CGA CCC CAG CCT CGG CC-3′ (forward) and 5′-GCG GCC GCT CAC TTG CTA GCA AAA AAG GCC C-3′ (reverse). The primers for the miR-26b reporter were 5′-GAC CCC AGT CCA ATA CCT GCT-3′ (forward) and 5′-AGC ATT GTC TGG ATG GAA GAC-3′ (reverse). The PCR product was inserted into the pGEM-T easy vector (Promega).
In addition, the let-7a-1 miRNA sequence was amplified by PCR, cloned into pGEM-T easy and verified by sequencing. The primers for let-7a-1 cloning were 5′-GAT TCC TTT TCA CCA TTC ACC CTG GAT GTT-3′ (forward) and 5′-TTT CTA TCA GAC CGC CTG GAT GCA GAC TTT-3′ (reverse). The BCL2L2 gene was subcloned into a FLAG-tagged-pcDNA3 vector (Invitrogen). And the miRNA sequence was then inserted at the EcoRI site in the intron of BCL2L2.
Site-directed mutagenesis was carried out using the QuickChange Site-Directed Mutagenesis kit (Stratagene) to generate the splicing-deficient construct. The primer sets were 5′-AGT GGG GGC TGG GTA CAA AGC TTC TCA ATT GC-3′ (forward) and 5′-GCA ATT GAG AAG CTT TGT ACC CAG CCC CCA CT-3′ (reverse) for the 5′ intronic consensus sequence. The primers used for the 3′ intronic consensus sequence were 5′-TCC CTT CTC TCC CAC CTG CGG AGT TCA CAG CT-3′ (forward) and 5′-AGC TGT GAA CTC CGC AGG TGG GAG AGA AGG GA-3′ (reverse). The cropping-deficient mutation of miR-26b stem–loop was generated by site-directed mutagenesis using the following primers. Forward:
5′-GCC TGT TCT CCA TTA GAC TGC TCG GGG ACC GGT G-3′, and reverse: 5′-CAC CGG TCC CCG AGC AGT CTA ATG GAG AAC AGG C-3′.
RNA interference
SiRNA duplex was transfected into HeLa cell using RNAiFect transfection reagent (Qiagen) according to the manufacturer's instructions (Lee et al, 2003; Han et al, 2004).
RT–PCR
RT was carried out using the Superscript II kit (Invitrogen) according to the manufacturer's protocol. Total mRNA was prepared using Trizol (Invitrogen), treated with DNase I (Takara) and then reverse-transcribed using an oligo-dT primer (Invitrogen). After RT, the RNA was digested using RNase A (Ambion). The primers used in these analyses were miR-26a-2_exon_forward:
5′-ATC AAC AAT GCT GAC TTC ATA GTG-3′,
miR-26a-2_exon_reverse: 5′-CTT GGC CAG GCT GGC AGT GAA GAG-3′,
miR-26a-2_intron_reverse: 5′-CTT GGC CAG GCT GGC AGT GAA GAG-3′,
miR-26b_intron_forward (Pdf): 5′-CCG GGA CCC AGT TCA AGT AAT TCA-3′,
miR-26b_intron_reverse (Pur): 5′-TCA GCC TAG TGC AGG AAT CCA G-3′,
miR-26b_exon_forward: 5′-AGT GAA CAA CGC GGA CTT CAT C-3′,
miR-26b_exon_reverse: 5′-CTT GGC GAG GCT AGC AGT GAA CAG-3′,
miR-26b_upexon_forward (Puf): 5′-AAG ATC TGC GTG GTC ATC GAC-3′,
miR-26b-downexon_reverse (Pdr): 5′-TGT CCA GCA GGT CAG CTA CTG-3′,
miR-126_exon_forward: 5′-CAG TTA CTG GTG CCA GTG TTG GGA-3′,
miR-126_exon_reverse: 5′-CAC CCT GGA CTG CAG CCT CTG-3′,
miR-126_intron_forward (Pdf):
5′-CAG CGC AGC ATT CTG GAA GAC-3′,
miR-126_downexon_reverse (Pdr): 5′-GAG CTG CTG GAA GGA GTG CAC-3′,
miR-330_intron_forward (Pdf): 5′-GGT GAC TCC CTT CTT CCA GGA TCG-3′,
miR-330_exon_forward: 5′-CCA CCC CCG GCC CCG AAG CTC CGC-3′,
miR-330_exon_reverse: 5′-CAC TGA AGA TAA CTT CTT TGG-3′,
miR-330_downexon_reverse (Pdr):
5′-GAG CGT GTG TCC AGG CTG TAG-3′,
miR-106b-93-25_upexon_forward (Puf): 5′-TTG AAC CTC TGG ACA TGA AGC TC-3′,
miR-106b-93-25_downexon_reverse (Pdr): 5′-TAT CAC ATC TGC TGG TCT CTG AGT C-3′,
miR-106b-93-25_intron_forward (Pdf): 5′-CCT CAC AGG ACA GCT GAA CTC C-3′,
miR-106b-93-25_intron_reverse (Pur): 5′-ACT CAC AAA ACA GGA GTG GAA TCC-3′,
miR-106b-93-25_exon_forward: 5′-GCA TAC GTG GAG ATG AGG CGA G-3′,
miR-106b-93-25_exon_reverse: 5′-CCC CTT GTC TCC TAG AAG AGA G-3′,
miR-15b-16-2_intron_reverse: 5′-TTT CAG TAG TAG TGG TTC CAC CAA G-3′,
miR-15b-16-2_exon_forward: 5′-ATG TAT CCA GAA CGG CCT GCA-3′,
miR-15b-16-2_exon_reverse: 5′-CAT ACC CTC ATC GTG TTC AGT CTG-3′,
miR-26a-1_intron_reverse: 5′-CTT TGC CCA CAA GAC TCC TCG T-3′,
miR-26a-1_exon_forward: 5′-CGG TTG AAA TCG ATG GAA CTA TAC A-3′,
miR-26a-1_exon_reverse: 5′-AAG CTG GCA GTA AAG AGC ACA CAT-3′,
miR-33_intron_reverse: 5′-CGT AGA CCA GGT GAT GCT CCA C-3′,
miR-33_exon_forward: 5′-GTT CCT TCT GCC ATT GCG AGA G-3′,
miR-33_exon_reverse: 5′-GCT GTC CGT AGC GAC AGT AGC AG-3′,
miR-98_intron_reverse: 5′-ACA TCT GCC TCA CCC TAA ACA CC-3′,
miR-98_exon_forward: 5′-GAC TCC ACT GAA CAG AAT CTC TCA G-3′,
miR-98_exon_reverse: 5′-TGC TGT TGT GAG GAT TGC AGA GT-3′,
miR-101-2_intron_reverse: 5′-TGC TTA TCA GTC TTC CCT CTC TGC-3′,
miR-101-2_exon_forward: 5′-TGA CCC TTG GAC AGC AGG ATG-3′,
miR-101-2_exon_reverse: 5′-ATG CCA ACA CAG GTC ATC AGC A-3′,
miR-107_intron_reverse: 5′-CAA CTT TGT ATG TCA CCA GCT CCA C-3′,
miR-107_exon_forward: 5′-CGA GAT TCC ATC AGC AAG GAA GA-3′,
miR-107_exon_reverse: 5′-TAT GCC AGC AGC TTC ATG GAG A,
BCL2L2_Pf: 5′-GAA TTC TGA TGG CGA CCC CAG CCT CGG CC-3′,
BCL2L2_Pr: 5′-GCG GCC GCT CAC TTG CTA GCA AAA AAG GCC C-3′, neomycin-resistance gene: forward: 5′-ATG ATT GAA CAA GAT GGA TTG CAC-3′, and reverse: 5′-TCA GAA GAA CTC GTC AAG AAG GC-3′.
Real-time PCR
Real-time PCR was conducted using the comparative Ct method with the Applied Biosystems 7300 Real-Time PCR system. The primers used for RT were 5′-TTC ACT AAA CCA CTC ACC TAG-3′ (for nascent mRNA, RT primer 1) and 5′-TCT CTG AGT CCT AGC TGT CT-3′ (for partially spliced mRNA, RT primer 2). Real-time PCR reaction was conducted by using the SyBR green method, and the PCR primers were 5′-CCT CAC AGG ACA GCT GAA CTC C-3′ (forward) and 5′-CCC CTT GTC TCC TAG AAG AGA G-3′ (reverse).
To measure the level of the MCM7 spliced mRNA, the TaqMan probe was designed to bind the splice junction, and primers were designed to anneal at adjacent positions to the probe. The probe sequence is 5′-6FAM-TGC TCT GGC ACG TCT GAG AAT GGT G-TAMRA-3′. The PCR primers were 5′-GCT ATC CTG CGC CTT TCC A-3′ (forward) and 5′-TCA CAT CTT CTT TCT CCA CCA CAT-3′ (reverse). The TaqMan GAPDH endogenous control kit was purchased from Applied Biosystems.
Northern blot analysis for miRNA
Thirty micrograms of HEK293T total RNA was resolved on 12.5% urea–polyacrylamide gel and transferred to a Zetaprobe-GT-membrane (Bio-Rad). Oligonucleotides complementary to let-7a or miR-26b were end-labeled using T4 polynucleotide kinase (Takara) and used as probes.
Supplementary Material
Supplementary Table
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank Dr Dong-Ki Lee, Sun Woo Hong, and Yoontae Lee for critical comments and technical help, Drs David Bartel and Christopher Burge for discussions on splicing and SeungYoon Nam and Dr Sang Hyuk Lee for their advice on database searches. We are also grateful to Dr Sang Hyun Park, Sung Hee Baek, Deog Su Hwang and to the members of our laboratory for their assistance with designing these experiments, and YKK thanks The Bioneers, the research group of undergraduates at Seoul National University. This work was supported by grants from the National Research Laboratory Program (M1050000010905J000010910), the Molecular and Cellular BioDiscovery Research Program (2004-01749), the Korean Research Foundation Grant (KRF-2005-005-J16001) and the BK21 Research Studentship.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ (2006) Exon tethering in transcription by RNA polymerase II. Mol Cell 21: 849–859 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 14: 319–327 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S (2004) The microRNA Registry. Nucleic Acids Res 32: D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004) The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN (2006) Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Dennis L, Jaenisch R, Sharp PA (2005) Characterization of a highly variable eutherian microRNA gene. RNA 11: 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ (2003) The UCSC Genome Browser Database. Nucleic Acids Res 31: 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Pillai RS (2005) MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11: 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SY, Lin SL (2006) Current perspectives in intronic micro RNAs (miRNAs). J Biomed Sci 13: 5–15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table
Supplementary Figure 1
Supplementary Figure 2


