Abstract
ATP-dependent chromatin remodeling complexes play a critical role in chromatin dynamics. A large number of in vitro studies have pointed towards nucleosome sliding as the principal remodeling outcome of SWI/SNF action, whereas few have described histone octamer transfer as the principal outcome. In contrast, recent in vivo studies have linked the activity of SWI/SNF to histone eviction in trans from gene promoters. In this study, we have found that the chimeric transcription factor Gal4-VP16 can enhance SWI/SNF histone octamer transfer activity, resulting in targeted histone eviction from a nucleosome probe. This effect is dependent on the presence of the activation domain. We observed that under conditions mimicking the in vivo relative abundance of SWI/SNF with respect to the total number of nucleosomes in a cell nucleus, the accessibility of the transcription factor binding site is the first determinant in the sequence of events leading to nucleosome remodeling. We propose a model mechanism for this transcription factor-mediated enhancement of SWI/SNF octamer transfer activity.
Keywords: chromatin remodeling, histone eviction, nucleosome, octamer transfer, SWI/SNF
Introduction
A growing number of protein complexes have been implicated in chromatin dynamics. Among them are the ATP-dependent chromatin remodeling complexes (Neely and Workman, 2002), which can be divided into several subfamilies on the basis of the similarities of their ATPase subunits. One of these subfamilies is SWI2/SNF2, where the yeast SWI/SNF (ySWI/SNF) complex is the founding member (Neely and Workman, 2002; Martens and Winston, 2003). The SWI2/SNF2 chromatin remodeling complexes have demonstrated several biochemical activities, including transient exposure of nucleosomal DNA, nucleosome sliding (movement of the histone octamer in cis along the DNA) and histone octamer transfer. Nucleosome sliding (Whitehouse et al, 1999; Jaskelioff et al, 2000; Saha et al, 2002; Kassabov et al, 2003; Zofall et al, 2006) and octamer transfer activity (Lorch et al, 1999; Phelan et al, 2000) have separately appeared to be the principal or exclusive activity in a number of studies. It is thought that these dissimilar outcomes, obtained in different biochemical analyses, rely on the nature of the experimental approaches used in each particular study, and that the underlying mechanisms of activity might be similar (Martens and Winston, 2003), even when comparing to complexes from other subfamilies (Langst and Becker, 2004). Interestingly, although in vitro studies analyzing the activity of SWI2/SNF2 complexes have mostly found sliding as the principal outcome, a growing amount of in vivo data strongly link histone eviction in trans (also referred as nucleosome eviction or nucleosome disassembly) to the activity of these chromatin remodeling complexes (Reinke and Horz, 2003; Boeger et al, 2004; Korber et al, 2004, 2006; Liu et al, 2006).
SWI/SNF complexes can facilitate the access of transcription factors to their cognate binding sites within nucleosomes (Cote et al, 1994; Kwon et al, 1994; Utley et al, 1997). This fact led to the initial concept that in vivo SWI/SNF activity regularly precedes transcription factor binding. However, SWI/SNF interacts with DNA and nucleosomes without sequence specificity (Quinn et al, 1996; Cote et al, 1998). This fact, and the low abundance of ySWI/SNF in the cell nucleus compared with the number of nucleosomes, implies that this complex needs to be targeted to the regions of the genome where its activity is required (Peterson and Workman, 2000). Targeting of SWI/SNF complexes relies principally on transcription factors with which it interacts (Becker and Horz, 2002; Neely et al, 2002; Martens and Winston, 2003). In addition to targeting, however, little is known about the influence that transcription factors could have on the catalytic activity of these complexes. Most of in vitro studies on SWI/SNF remodeling activity have not considered the possible influence of transcription factors.
Our laboratory has previously reported that ySWI/SNF-stimulated transcription on nucleosome arrays requires the presence of transcription factors carrying an activation domain able to target this complex (Neely et al, 1999). Similar results have also been observed for mammalian SWI/SNF (Kadam et al, 2000). In the present study, we show that, upon targeting, nucleosome eviction becomes the principal outcome of ySWI/SNF activity instead of nucleosome sliding. We show that this nucleosome eviction effect relies on the inherent octamer transfer activity of ySWI/SNF, which is enhanced by transcription factors in an activation domain-dependent fashion. We also show that, under conditions mimicking the in vivo low abundance of ySWI/SNF relative to the nucleosomes, accessibility of the transcription factor binding site is a prerequisite to unleash these chromatin remodeling events.
Results
A nucleosome probe accessible for Gal4 derivatives binding upon reconstitution
In order to study the effect of Gal4 derivatives on SWI/SNF remodeling activity, we designed a 216- bp DNA probe containing a single Gal4 binding site close to the 3′ end of this DNA segment (dSHA probe; Figure 1A). We have previously reported that the concerted action of Gal4 derivatives and ySWI/SNF generates nucleosome eviction in an activation domain-independent manner when five Gal4 binding sites are present in the nucleosomal probe (Owen-Hughes et al, 1996). The goal of the present work was to analyze a situation where transcription factor binding itself cannot destabilize the nucleosome, allowing us to analyze the importance of an SWI/SNF-interacting activation domain on chromatin remodeling. A prerequisite for these studies was a nucleosome probe accessible to Gal4 derivative binding.
Figure 1.
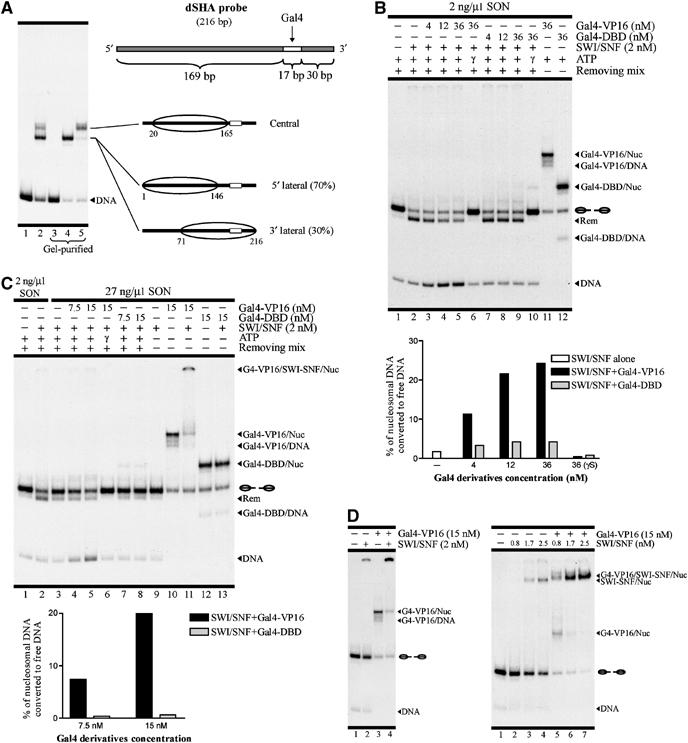
Activation domain-containing Gal4 derivatives stimulate nucleosome eviction catalyzed by ySWI/SNF. (A) Top right: scheme of the dSHA DNA segment. Left: electrophoretic analysis for dSHA probe reconstitution (lane 2), mock reconstitution (lane 1) and gel-purified species (lanes 3–5). The characterized nucleosome populations related to each nucleosomal DNA band are shown schematically at the right side of the gel. Ovals represent histone octamers. (B–D) The denotation SON corresponds to ‘short oligonucleosomes'. The probe used corresponds to gel-purified lateral nucleosome populations. Conditions for each lane are detailed on top of each gel picture. Migration of the different species is indicated on the right of each gel. Nucleosome migration is shown schematically. The denotation ‘Rem' corresponds to remodeled (slid) nucleosome. (B) Remodeling assay performed using standard stringency (2 ng/μl SON). (C) Remodeling assay performed using high stringency (27 ng/μl SON, lanes 3–13). Lanes 1 and 2 correspond to standard stringency. (D) Analysis of targeting effect of Gal4-VP16 under low stringency levels. The assays were performed as in (B) and (C), but the removing mix was omitted in order to analyze only binding. All lanes contain ATP-γ-S to get insight into how binding and targeting are affected before nucleosome remodeling. Left: assay performed under standard stringency. Right: assay performed using 0.33 ng/μl SON and a 4% polyacrylamide gel (60:1 AA:Bis) in the electrophoretic step.
Nucleosome reconstitution of the dSHA probe generated nucleosome populations contained in two major bands on non-denaturing gels (Figure 1A). The slower migrating band correlated with a unique central nucleosome population, whereas the faster migrating band was represented by the histone octamer located to either the 5′ or the 3′ end of this DNA segment (lateral nucleosome populations), as defined by nucleosome mapping (Figure 1A; Supplementary Figure 1; Langst et al, 1999). Restriction enzyme accessibility assays determined that around 70% of the lateral nucleosome populations corresponded to the 5′ population. Consistently, gel-purified lateral nucleosome populations were mostly accessible to Gal4 derivatives binding, as the 5′ lateral nucleosome population contains the Gal4 binding site in the DNA linker region (Figure 1A; Supplementary Figure 1). Binding accessibility of the gel-purified central nucleosome population was even higher, in agreement with the translational location of the histone octamer on this nucleosome population (Figure 1A; Supplementary Figure 1). Gal4 derivatives binding did not disrupt the nucleosome structure (data not shown and see the following text).
The VP16 activation domain stimulates ATP-dependent nucleosome eviction catalyzed by ySWI/SNF
SWI/SNF binds nonspecifically to DNA and nucleosomes. Thus, the interaction of this complex with the nucleosome probe can be restricted by simply altering the cold competitor nucleosome concentration. Accordingly, two different competitor nucleosome concentrations were set for the remodeling assays. Standard stringency corresponds to an oligonucleosome concentration of 2 ng/μl (∼15 nM in nucleosome units), whereas high stringency corresponds to 27 ng/μl (∼200 nM). The ‘cold nucleosomes to probe nucleosome' ratio corresponds to ∼20:1 at standard stringency and ∼280:1 at high stringency, for non-gel-purified probe (∼0.7 nM). These ratios are higher in the assays using gel-purified nucleosome probe whose concentration corresponds to around 0.15 nM. The ratio ‘nucleosomes to SWI/SNF' (2 nM, approximately) correspond to ∼8:1 at standard stringency and ∼100:1 at high stringency. For assays using non-gel-purified nucleosome probe, the oligonucleosomes used as histone donors in the reconstitution process generated the concentration defined as standard. As purification of a mononucleosome probe from non-denaturing gels eliminates the donor oligonucleosomes used in the reconstitution process, in assays using gel-purified probes, generation of both standard and high stringency was accomplished by adding oligonucleosomes from appropriate stocks.
Remodeling assays consisted of a short preincubation with a Gal4 derivative (prebinding), followed by the addition of ySWI/SNF and the remodeling incubation (usually 30 min). In order to observe the fate of nucleosome remodeling, after the two steps indicated above, a mix containing calf thymus DNA, long oligonucleosomes and an oligonucleotide containing the Gal4 binding site (referred to as removing mix) was added, followed by a short incubation. The samples were then analyzed by electrophoresis on non-denaturing gels. The removing mix was added to all the conditions analyzed, except where Gal4 derivatives and/or SWI/SNF binding were to be analyzed.
When ySWI/SNF remodeling activity was analyzed at standard stringency, we observed strong sliding activity (Figure 1B, compare lanes 1 and 2) with a pattern consistent with descriptions given in previous reports studying complexes of the SWI2/SNF2 subfamily. This included the generation of a nucleosome band that migrates faster than the central and lateral nucleosome populations (Rem; Figure 1B), reflecting the movement of the histone octamer beyond the DNA ends (Jaskelioff et al, 2000; Fan et al, 2003; Flaus and Owen-Hughes, 2003). For nucleosome probes bearing linker DNA, such as our 216 bp dSHA probe, this faster migration is believed to be due to wrapping of the free DNA end onto a portion of the histone octamer surface, previously exposed on the opposite end by the sliding activity (Kassabov et al, 2003; Zofall et al, 2006).
Notably, in the presence of Gal4-VP16, an increase in the free DNA signal was observed, indicating DNA dissociation from the histone octamer, accompanied by a reduction in the slid nucleosome signal (Figure 1B, lanes 3–5). The effects of ySWI/SNF on the nucleosome were dependent on ATP hydrolysis, as in the presence of ATP-γ-S, neither sliding nor nucleosome dissociation was observed (Figure 1B, lane 6). In the presence of Gal4-DBD (Gal4 DNA-binding domain), which lacks an activation domain, sliding remained the principal remodeling reaction independent of the increment in Gal4-DBD concentration and no apparent nucleosome dissociation was observed (Figure 1B, lanes 7–9). Gal4-VP16 and Gal4-DBD had a similar binding strength, as observed in Figure 1B (compare lanes 11 and 12). This assay was performed using purified lateral nucleosome populations. Similar results were obtained using purified central nucleosome populations (data not shown). In order to analyze the levels of nucleosome dissociation for each condition in more detail, the percentage of nucleosomal DNA converted to free DNA was calculated. Quantification of the nucleosome dissociation levels consists of subtracting the free DNA signal present in the starting material (nucleosome alone; e.g., lane 1 in Figure 1B) from the free DNA signal obtained in a particular condition. The resulting value is divided by the nucleosomal DNA signal present on the starting material, obtaining the fraction (expressed as percentage on the graphs) of nucleosomal DNA that is converted to free DNA. As observed in the graph of Figure 1B, a low level of nucleosome dissociation was obtained in the presence of Gal4-DBD, slightly higher than in the presence of ySWI/SNF alone and 5–6 times lower than the values obtained when Gal4-VP16 was tested at concentrations of 12 and 36 nM.
Next, we evaluated the effect of these Gal4 derivatives on ySWI/SNF remodeling using high stringency. As expected, the higher concentration of cold nucleosomes hindered SWI/SNF sliding of the nucleosome probe on its own (Figure 1C, compare lanes 2 and 3). However, when Gal4-VP16 was present, nucleosome dissociation levels similar to those obtained at standard stringency were observed (Figure 1C, lanes 4 and 5). These results were also obtained when Gal4 derivatives containing the Gcn4 or Gal4 activation domains were assayed (data not shown). In contrast, no remodeling was observed when ySWI/SNF was assayed in the presence of Gal4-DBD (Figure 1C, lanes 7 and 8). At this stringency level, no interaction between ySWI/SNF and the nucleosome probe was detected, but the interaction became strong upon targeting by Gal4-VP16 (Figure 1C, lanes 9 and 11, respectively). As expected, Gal4-DBD could not target ySWI/SNF to the probe (Figure 1C, lane 13). This situation was consistent with remodeling found only in the presence of Gal4-VP16, indicating that under high stringency conditions, the only possibility for ySWI/SNF–nucleosome probe interaction is through Gal4-VP16. More importantly, under these conditions, nucleosome dissociation, and not sliding, was the principal remodeling outcome (Figure 1C, lanes 1 and 5, compare generation of slid nucleosome (Rem) with increment in free DNA).
ySWI/SNF was able to interact with the nucleosome probe at standard stringency, but binding was strongly stimulated by Gal4-VP16 (Figure 1D, left panel, compare lanes 2 and 4), indicating that, to some extent, the targeting effect was also present at standard stringency. Interactions with the nucleosome probe appeared as defined bands in more diluted gels (Figure 1D, right panel). The targeting effect by Gal4-VP16 was also observed at oligonucleosome concentrations below standard stringency (Figure 1D, right panel, 0.33 ng/μl). This situation suggested that the nucleosome eviction effect found in the presence of Gal4-VP16 might depend only on the ability of the transcription factor to favor the interaction of ySWI/SNF and the nucleosome probe. However, time-course assays performed at standard stringency in the absence of Gal4-VP16 (Figure 2) showed that even when ySWI/SNF alone remodels (and therefore interacts with) all the nucleosome probes, this remodeling proceeds almost exclusively by sliding. Nucleosome eviction appeared only in the presence of Gal4-VP16, indicating that in these assays the influence of this transcription factor extends beyond facilitating the interaction of ySWI/SNF with the nucleosome probe.
Figure 2.
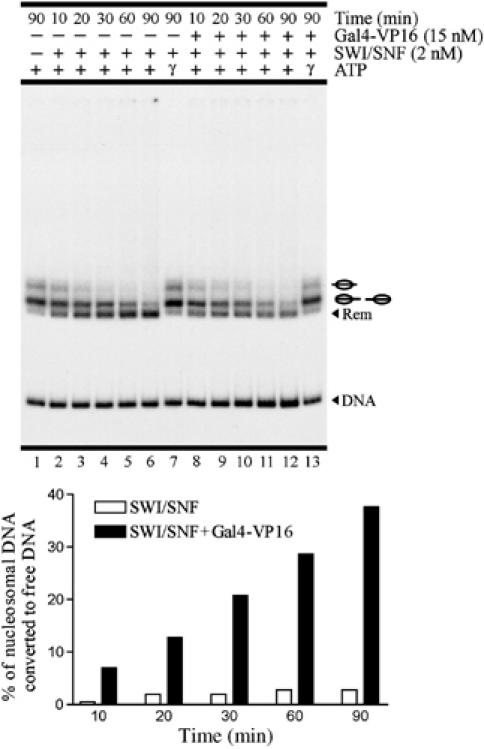
In the absence of Gal4-VP16, complete nucleosome remodeling by ySWI/SNF consists almost entirely of sliding activity. Time-course analysis performed under standard stringency. Non-gel-purified reconstituted probe was used in this assay. See legend in Figure 1 for a general description of remodeling assay figures.
We have also observed that the interaction of Gal4-VP16 with both the nucleosome probe and SWI/SNF appears to be required for obtaining the nucleosome eviction effect, as remodeling assays comparing a nucleosome probe bearing the Gal4 binding site to a probe lacking this site indicate that the eventual VP16–SWI/SNF interaction without transcription factor–nucleosome probe interaction cannot stimulate either nucleosome eviction or sliding activity (Supplementary Figure 2).
Gal4-VP16-stimulated nucleosome eviction occurs by histone octamer transfer
The above experiments indicate that the presence of the activation domain on the transcription factor is required for targeting of SWI/SNF remodeling activity and also for stimulation of its nucleosome eviction activity. Previous studies in our laboratory have indicated that both DNA and oligonucleosomes can act as acceptors of histone octamers released from nucleosome probes during histone octamer transfer (Walter et al, 1995; Owen-Hughes et al, 1996). To determine whether Gal4-VP16-stimulated nucleosome eviction occurred by histone octamer transfer, we tested the importance of the oligonucleosomes as a histone acceptor in these experiments. Thus, we performed a remodeling assay with the gel-purified lateral nucleosome populations without including oligonucleosomes in the reaction mixes. Under these conditions, the level of nucleosome dissociation generated by Gal4-VP16 and ySWI/SNF combined was around three times lower (P<0.001) than the level obtained in the presence of oligonucleosomes and only slightly higher than the nucleosome dissociation generated by ySWI/SNF alone in the absence of oligonucleosomes (Figure 3, compare lanes 2, 4 and 6). This result indicated that the presence of oligonucleosomes was required for nucleosome probe dissociation, consistent with the known features of SWI/SNF octamer transfer activity. Moreover, in the presence of a small amount of cold DNA (previously purified from oligonucleosomes), the nucleosome dissociation generated by Gal4-VP16 and ySWI/SNF combined was two-fold higher (P<0.001) than under the same conditions in the absence of oligonucleosomes and this cold DNA (Figure 3, compare lanes 6 and 7).
Figure 3.
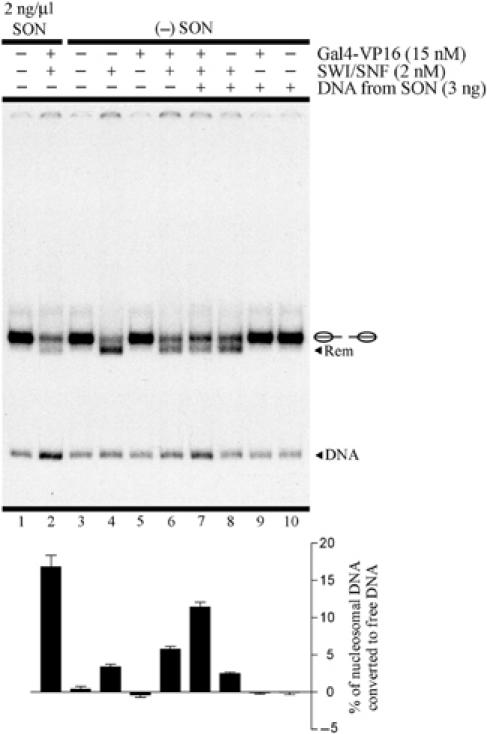
Nucleosome eviction effect relies on SWI/SNF octamer transfer activity. The remodeling assay compared standard stringency (lanes 1 and 2) with the absence of oligonucleosomes in the reaction (lanes 3–6) and the presence of 3 ng (0.2 ng/μl) of DNA purified from SON (lanes 7–10). See legend in Figure 1 for a general description of remodeling assay figures. The probe used in this assay corresponds to gel-purified lateral nucleosome populations. The graph bars are below their corresponding gel lanes. Error bars correspond to one standard deviation from three independent experiments.
Considering that SWI/SNF-type complexes, such as RSC and SWI/SNF, have been shown to facilitate histone octamer transfer (Lorch et al, 1999; Phelan et al, 2000) and that in our nucleosome remodeling assays the free DNA probe is in fact a piece of DNA able to act as a histone acceptor, a certain extent of octamer transfer from oligonucleosomes towards the naked DNA probe was expected to be present in these assays. In this instance, the presence of Gal4-VP16 or Gal4-VP16-SWI/SNF on the DNA probe might preclude transfer of histone octamers from oligonucleosomes towards this probe, but not the transfer from nucleosome probe towards oligonucleosomes, resulting in an increased percentage of naked DNA probe in these remodeling reactions. By incubating dSHA DNA probe with oligonucleosomes and ySWI/SNF, under our assay conditions, we observed that the extent of octamer transfer from oligonucleosomes towards the DNA probe (Lorch et al, 1999; Phelan et al, 2000) was minimal compared with the extent of nucleosome probe dissociation that Gal4-VP16 and ySWI/SNF generated under the same conditions (Supplementary Figure 3). Thus, blocking of the minimal extent of nucleosome formation on the probe cannot account for the levels of nucleosome dissociation obtained for the nucleosome probe, indicating that the increment of free DNA observed in our remodeling assays corresponds to a direct effect of Gal4-VP16 and ySWI/SNF on the nucleosome probe.
Accessibility of the Gal4 binding site is the first determinant leading to nucleosome eviction
Under high stringency conditions, the appearance of nucleosome remodeling by SWI/SNF required targeting mediated by Gal4-VP16. Considering the low abundance of the SWI/SNF complex in yeast (100–500 copies per nucleus; Peterson and Workman, 2000) and the few number of nucleosomes in a particular gene promoter compared with the total number of nucleosomes on the whole yeast genome, it is clear that our analyses performed at high stringency are closer to the in vivo situation than those performed using standard stringency. This suggests that accessibility of the transcription factor binding site should play a primary role in the sequence of events leading to nucleosome remodeling. As mentioned above, upon reconstitution of the dSHA probe, the proportion of the resulting lateral nucleosome populations is 70–30% (5′ lateral to 3′ lateral). As the Gal4 binding site is located 30 bp inside the nucleosome in the case of the 3′ lateral nucleosome population (see schemes in Figure 1A), we wanted to assess the capability of Gal4-VP16 to target SWI/SNF nucleosome remodeling activity on this nucleosome population in comparison with 5′ population, where the Gal4 binding site is located in a free DNA region (linker DNA).
To date, no methods have been described for separation of 5′ and 3′ lateral reconstituted nucleosome populations from each other. To perform this, we developed an approach that combines digestion with a particular restriction enzyme for a whole reconstitution, followed by purification of the non-digested nucleosomal DNA from a non-denaturing gel. As schematized in Figure 4A, the HinfI site is in a free DNA region of the 5′ population. On the other hand, the EcoRI site locates on linker DNA in the case of the 3′ population. As in the electrophoretic separation, the digested (shorter) nucleosome probe runs faster than the non-digested (full length) nucleosome probe, we were able to obtain the 5′ population by gel-purifying the full-length EcoRI-treated nucleosomal DNA (3′ population digested by EcoRI). Analogously, the 3′ population was obtained by gel-purifying the non-digested HinfI-treated nucleosome probe.
Figure 4.
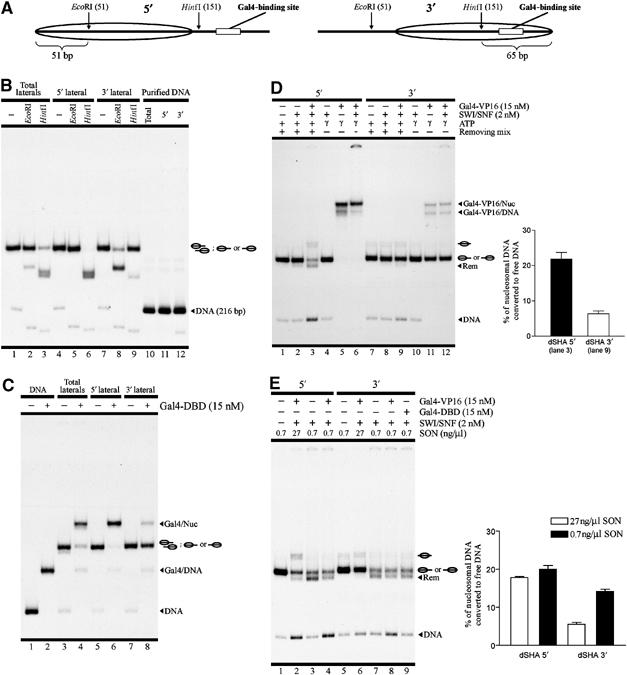
Under high stringency levels, Gal4 binding site accessibility determines the occurrence of nucleosome remodeling. (A) Schematic representation of 5′ and 3′ lateral nucleosome populations, indicating the position of the restriction sites used for the differential purification of these populations. (B) Restriction enzyme accessibility assay (lanes 1–9) and DNA integrity analysis (lanes 10–12) of the gel-purified nucleosome populations. The description of nucleosome populations analyzed and restriction enzymes used is on the top of the gel. ‘Total laterals' refers to the gel-purified mixed lateral nucleosome populations. Migration of the non-digested nucleosome probes is shown schematically on the right of the gel, where ‘DNA (216 bp)' refers to non-digested naked DNA migration. (C) Analysis of Gal4 binding site accessibility to Gal4-DBD on different versions of dSHA probe. Description of the probes used and presence of Gal4-DBD is shown on top. The identity of the different bands is detailed on the right side of the gel picture. (D) Remodeling assay comparing 5′ lateral nucleosome population with 3′ population, performed under high stringency. Error bars correspond to one standard deviation from three independent experiments. (E) Assay comparing 5′ nucleosome population with 3′ population under high and low (0.7 ng/μl SON) stringency levels. The graph details the eviction levels for the ‘Gal4-VP16+SWI/SNF' lanes. Error bars correspond to one standard deviation from three independent assays. For (D) and (E), see legend in Figure 1 for a general description of remodeling assay figures.
It is widely known that DNA accessibility is significantly reduced inside a nucleosome. Accessibility of a DNA stretch on a particular translational location in the nucleosome may vary depending on DNA sequence, but it has been observed that in general nucleosomal DNA sites located 50 bp or deeper into the nucleosome show an accessibility of one order of magnitude lower than sites located closer to the nucleosome edge (roughly, 20 bp or less) (Anderson and Widom, 2000; Anderson et al, 2002). Thus, the location of EcoRI and HifI sites relative to the nucleosome edge of 5′ and 3′ lateral nucleosomes, respectively (see Figure 4A), makes these restriction enzymes highly reliable for purification and differential analysis of these two nucleosome populations. Standard DNA purification from the purified 5′ and 3′ lateral populations showed intact DNA content for these nucleosome probes (Figure 4B, lanes 11 and 12, respectively). The 5′ population could be purified to homogeneity, as this purified nucleosome probe was not digested by EcoRI, but completely digested by HinfI (Figure 4B, lanes 5 and 6, respectively). On the other hand, a significant enrichment of the 3′ population was reproducibly obtained, as indicated by the strong EcoRI digestion of this purified nucleosome probe and the minimal digestion with HinfI (Figure 4B, lanes 8 and 9, respectively). Consistent with the differential pattern of restriction enzyme digestions, binding by Gal4-DBD further corroborated the identity of these lateral nucleosome populations (Figure 4C). For the mix of 5′ and 3′ populations, Gal4-DBD binds to most of the nucleosomal probe (Figure 4C, lane 4), but binding was complete when the purified 5′ lateral nucleosome population was tested (Figure 4C, lane 6). A weak binding signal was observed for the 3′ population (Figure 4C, lane 8). We attribute this binding signal to contaminant 5′ population, as other more drastic purifications of the 3′ population (stronger digestions with the corresponding restriction enzyme) correlated with no binding by Gal4-DBD. However, these 3′ probe purifications contained more significant levels of nicked nucleosomal DNA (fastest migrating band in Figure 4B, lane 12) and were discarded for the remodeling assays (data not shown).
Next, we analyzed the influence of Gal4-VP16 on ySWI/SNF remodeling, comparing 5′ and 3′ lateral nucleosome populations under high stringency conditions. In the case of the 5′ population, Gal4-VP16 was able to target ySWI/SNF to the nucleosome probe (Figure 4D, lane 6) and consequently it was readily remodeled (Figure 4D, lane 3). In contrast, a minimal level of targeting was observed while assaying the 3′ population (Figure 4D, lane 12). Accordingly, a significantly lower level of sliding and nucleosome eviction was obtained for this nucleosome probe (Figure 4D, lane 9). As observed in the graph in Figure 4D, nucleosome dissociation was more than three-fold higher (P<0.01) for the 5′ lateral nucleosome population than for the 3′ population. Differences were significant, despite the mentioned contamination of the 3′ population with the 5′ population. Thus, under conditions resembling the in vivo proportions of total nucleosomes to ySWI/SNF complex (high stringency), the accessibility of the Gal4 binding site determines whether nucleosome remodeling will proceed or not.
These results may appear to contradict early work stating that SWI/SNF facilitates the access of transcription factors to nucleosomal DNA (Cote et al, 1994; Kwon et al, 1994; Utley et al, 1997). However, the difference may be because those studies used relatively low stringency levels (low concentrations of cold nucleosomes or DNA), allowing interaction of SWI/SNF with the nucleosome probe, without the need of targeting and thereafter exposure of nucleosomal DNA for transcription factor access. Then, at low stringency, ySWI/SNF would be able to facilitate the access of Gal4-VP16 to the 3′ lateral nucleosome population and eviction levels closer to the observed for the 5′ population would be observed. Indeed, this is the result obtained when the assay was performed using low stringency (0.7 ng/μl cold oligonucleosomes), reconciling our results with the previously published data. Under these conditions, the nucleosome eviction effect was still dependent on the presence of the activation domain VP-16 (Figure 4E).
Under high stringency levels, remodeling by nucleosome eviction predominated over sliding when analyzing the 5′ population (Figure 4D), as previously observed for the probe containing the mix of both lateral nucleosome populations (Figure 1C). Interestingly, for the 5′ population, the remodeling observed by sliding not only consisted of a faster migrating nucleosome band, but also resulted in the generation of a central nucleosome population (Figure 4D, lane 3; Figure 4E, lane 2). This phenomenon was restricted to the use of high stringency and was also observed, to a lower extent, in assays performed with the probe containing the mix of lateral nucleosome populations (Figure 1C, lanes 4 and 5) or even with probe that was not gel-purified (Supplementary Figure 3). We also observed this result when using a probe with an unrelated DNA sequence (Supplementary Figure 2). To analyze whether these central nucleosome populations correspond to a final product or an intermediate in the remodeling reaction, we performed a time-course analysis under high stringency for the 5′ probe. As observed in Figure 5, the central nucleosome population did not accumulate with time. Rather, it appeared early and was gradually reduced with longer incubations, concurrent with the increase of the free DNA signal.
Figure 5.
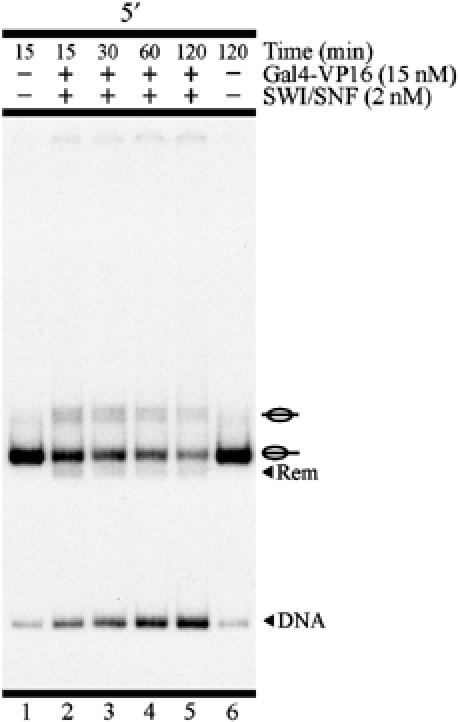
Central nucleosome populations do not correspond to a final product of targeted nucleosome remodeling. The figure corresponds to a time-course analysis for targeted ySWI/SNF remodeling on the 5′ lateral nucleosome population. All lanes correspond to high stringency. See legend in Figure 1 for a general description of remodeling assay figures.
The remodeling assays shown until this point were performed on reconstituted mononucleosomes. It has been proposed that sliding beyond the DNA ends on mononucleosome templates, with the concurrent exposure of a portion of the histone octamer surface, may lead to destabilization of the nucleosome structure and/or facilitation of histone displacement in trans (Flaus and Owen-Hughes, 2003). This opens the possibility for a nucleosome eviction effect limited to the use of mononucleosomes as templates for the remodeling assays, in contrast to the in vivo situation where the nucleosomes are not surrounded by DNA ends. To test this possibility, we performed remodeling assays on nucleosome arrays. A dinucleosome-length segment of the adenovirus E4 promoter containing a single Gal4 binding site (Carey et al, 1990) was placed in the center of a tandem array of the Lytechinus variegatus 5S rDNA sequence in such a way that the Gal4 binding site is found in a linker region upon nucleosome reconstitution. This is determined by the nucleosome phasing imposed by the 5S array (Figure 6A; Horn et al, 2002). Remodeling assays on nucleosome arrays were performed under high stringency, employing conditions similar to those utilized with the mononucleosome probes, including the use of prebinding, remodeling and removing incubations previously mentioned, before the analyses described in Figure 6. As observed in Figure 6B, micrococcal nuclease (MNase) digestion of the end-labeled nucleosome array shows a strong disruption of a nucleosome adjacent to the Gal4 binding site, in the presence of Gal4-VP16 and SWI/SNF (Figure 6B, lanes 9 and 10). Similar to the results obtained with mononucleosomes, this disruption was not observed when combining Gal4-DBD and SWI/SNF (Figure 6B, lanes 7 and 8). To confirm that disruption in this region of the nucleosome array was reflecting nucleosome disassembly, we performed the nucleosome cut-out assay (Owen-Hughes and Workman, 1996). This included placing an internal label in the nucleosome array in a particular position close to the nucleosome located upstream of the Gal4 binding site. After the remodeling assay incubations, the region encompassing this segment of the array was released by incubating with a pair of restriction enzymes surrounding this region (Figure 6A), followed by electrophoretic analysis of the sample. As observed in Figure 6C, Gal4-VP16 stimulated SWI/SNF-mediated nucleosome disassembly, resulting in a larger fraction of the excised segment migrating as naked DNA. Taken together, these assays confirm that the nucleosome eviction effect obtained in the presence of Gal4-VP16 and SWI/SNF is not restricted to the use of mononucleosome probes and can occur within an array of nucleosomes.
Figure 6.
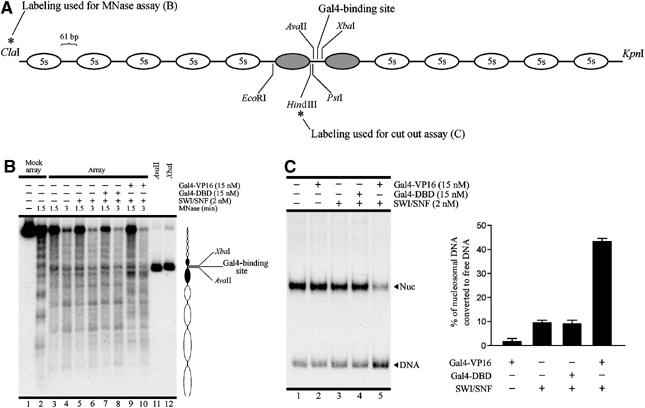
Activation domain-dependent nucleosome eviction on nucleosome arrays. (A) Schematic representation of the nucleosome array used in these assays, showing the relevant restriction sites used in the probe preparations and the assays itself. Ovals represent each of the 12 nucleosomes of the array, where the two gray ovals correspond to nucleosomes covering the E4 promoter insert, which contains a single Gal4 binding site. ‘5s' correspond to the principal translational position adopted for the tandem repeats of the 5s rDNA sequence. (B) MNase digestion of end-labeled array performed after the three incubation steps of the remodeling assays. The picture corresponds to electrophoresis on a 1.5% agarose gel. Conditions for each lane are depicted on top of the gel picture. Digestion of mock-reconstituted array with AvaII and XbaI (lanes 11 and 12) was used to mark the position of the Gal4 binding site. (C) Cut-out assay. The probe was internally labeled at the unique HindIII site (schematized in (A)). After the three incubations of the remodeling assay, a 170-bp region upstream of the Gal4 binding site was released from the array by digestion with EcoRI and PstI (see scheme in (A)) and the fate of this region was analyzed electrophoretically. Error bars in the graph correspond to one standard deviation from three independent experiments.
Discussion
In our present work, we describe the influence of transcription factors on SWI/SNF catalytic activity. Here, we demonstrate that the low level of SWI/SNF octamer transfer activity can be enhanced by the presence of a transcription factor carrying a functional activation domain, favoring nucleosome eviction. Moreover, under stringency levels emulating the relative ySWI/SNF abundance, with respect to nucleosomes in the nucleus, nucleosome eviction predominates over sliding and the accessibility of the transcription factor binding site is the first determinant in the sequence of events leading to nucleosome eviction by SWI/SNF.
In vitro study of SWI/SNF chromatin remodeling activity
Most in vitro studies analyzing SWI/SNF activity have used a total nucleosomes concentration similar to or only slightly higher than the concentration of the complex (Utley et al, 1997; Cote et al, 1998; Jaskelioff et al, 2000; Flaus and Owen-Hughes, 2003; Kassabov et al, 2003; Zofall et al, 2006). Although these studies have given important insights into the catalytic features of SWI/SNF activity, under these conditions there is no targeting requirement for the interaction of SWI/SNF with the nucleosome probes, a situation that is unlikely to occur in vivo. As shown in the present work, under high stringency, a transcription factor able to target SWI/SNF to the nucleosome probe is a prerequisite for remodeling. Importantly, the outcome of SWI/SNF remodeling under these conditions is primarily nucleosome eviction (Figures 1C, 4D, E and 5; Supplementary Figure 2).
The ability of SWI/SNF to change the translational position of the histone octamer on a mononucleosome from central to lateral positions, moving the octamer beyond the DNA end, has been demonstrated in a number of studies. It has been also shown that, in general, when the starting nucleosome is located at one lateral position the octamer is mobilized towards the opposite end. In this case, the absence of central nucleosome populations as intermediates in the remodeling reaction is thought to be due to a high sliding processivity (Jaskelioff et al, 2000; Fan et al, 2003; Flaus and Owen-Hughes, 2003; Kassabov et al, 2003; Zofall et al, 2006). However, here we have found that when SWI/SNF reaches the nucleosome probe through targeting by a transcription factor (high stringency), central nucleosome populations can be observed as an intermediate state, especially when analyzing the purified 5′ lateral nucleosome population. For this probe, generation of central nucleosome populations can only involve mobilization of the histone octamer from 5′ to 3′ towards the Gal4 binding site. This places the Gal4 binding site at the nucleosome edge (Figures 4A, D, E and 5). The appearance of these transient central nucleosome populations suggests that eviction might occur once the histone octamer gets close to the Gal4 binding site, rather than occurring as a consequence of sliding beyond the DNA ends. The analyses performed on nucleosome arrays further support this idea. Thus, our observations may be explained by the bulging mechanism proposed for SWI/SNF remodeling activity, consisting of continuous unpeeling of the DNA from the histone octamer surface at the nucleosome edges (see illustration in Supplementary Figure 4; Becker and Horz, 2002; Langst and Becker, 2004). The long latency of central nucleosome populations suggests that the transcription factor might enhance the access of acceptor DNA to the octamer by blocking or delaying the reassociation of the targeted DNA chain to the histone octamer surface. Alternatively, this effect could also be obtained by a larger extent of DNA unpeeling from the nucleosome, stimulated by the presence of the transcription factor interacting with its target sequence and SWI/SNF (Supplementary Figure 4). Nonetheless, either mechanism or a combination of both would lead to the same effect of enhanced octamer transfer activity from the target nucleosome towards histone acceptors.
Although in the analyses performed on nucleosome arrays a similar level of disruption was expected to occur on the nucleosomes located immediately upstream and downstream of the Gal4 binding site (as this is a site for binding of a Gal4 homodimer), we found a significantly stronger disruption on the upstream nucleosome (Figure 6A and B, data not shown). This phenomenon is maybe due to positioning of the upstream and downstream nucleosomes at different distances from the Gal4 binding site, obtained upon nucleosome reconstitution, and/or due to the influence of the underlying DNA sequences on remodeling itself (Vicent et al, 2004).
Nucleosome eviction on gene promoters
Here, we have demonstrated that, under stringency conditions reflecting the in vivo relative ySWI/SNF abundance with respect to the total number of nucleosomes in the nucleus, the accessibility of the transcription factor binding site is the first determinant leading to targeted remodeling. This fact indicates that in vivo, the position of the cognate binding sites for certain transcription factors, relative to the translational location of nucleosomes, is a key element for regulation of gene expression. It has been proposed that particular DNA sequences with a significant effect on nucleosome positioning are strategically distributed throughout the eukaryotic genome, influencing genome function (Anderson and Widom, 2001; Kiyama and Trifonov, 2002; Minsky, 2004; Yuan et al, 2005). Recently, Rando and colleagues described the positions of nucleosomes in yeast at a genome-wide level, observing that most of the occupied transcription factor binding sites were located in nucleosome-free regions and establishing a strong correlation between nucleosome-excluding sequences and these nucleosome-free regions (Yuan et al, 2005). However, this correlation cannot define whether these nucleosome-free regions are established before or after the binding of particular transcription factors, although one example is given for the first situation.
Activation of the yeast PHO5 gene is preceded by the eviction of four promoter nucleosomes in a process initiated by binding of the transcriptional activator Pho4 to its cognate binding site UASp1, which is positioned in a linker region of the inactive gene (Reinke and Horz, 2003; Boeger et al, 2004; Korber et al, 2004). Promoter remodeling on this gene has been shown to be dependent on the Pho4 activation domain (Svaren et al, 1994). Additionally, Pho4 initiates the PHO8 gene promoter remodeling, with SWI/SNF participating in a step subsequent to Pho4 binding to this promoter (Gregory et al, 1999). The participation of SWI/SNF at a step subsequent to activator binding, as well as the activation domain dependence for in vivo chromatin remodeling, has also been observed in other yeast genes (Stafford and Morse, 1997; Ryan et al, 1998). Thus, binding of a transcription factor able to target a chromatin remodeling complex may lead to the generation of a nucleosome-free region, allowing the binding of other specific transcription factors and the basal transcription machinery. The influence of DNA sequence on chromatin organization of some gene promoters could be more subtle, limited to the accommodation of particular transcription factor binding sites on linker regions or nucleosome edges. Alternatively, the activity of other more abundant chromatin remodeling complexes may, in a non-targeted fashion, generate windows of opportunity for binding of transcription factors, which in turn target SWI/SNF, leading to generation of nucleosome-free regions.
In the analyses performed on nucleosome arrays, we observed that the combined action of Gal4-VP16 and SWI/SNF generated a certain level of disruption on the surrounding 5S nucleosomes, although at a lower extent than the disruption observed on the nucleosome adjacent to the Gal4 binding site. This disruption could reflect some level of histone eviction. Nevertheless, we have recently observed that targeted histone acetylation in a particular region of a nucleosome array enhances SWI/SNF nucleosome eviction activity in that region of the array (Chandy et al, 2006). Thus, in vivo, the modification status of a particular nucleosome might influence the outcome of ATP-dependent remodeling on that nucleosome relative to the neighboring nucleosomes.
Taken together, our results support the concept that in vivo SWI/SNF nucleosome eviction activity corresponds to ‘transcription factor-enhanced octamer transfer activity of this complex'. Once SWI/SNF is targeted, the transcription factor becomes a forced partner for this complex, at least during part of the remodeling process, leading to nucleosome eviction. Consequently, the otherwise modest octamer transfer activity of this complex becomes dominant only when it is specifically targeted to a particular region of the genome.
Materials and methods
DNA probes, recombinant proteins and protein complexes
pGUB-dSH plasmid was obtained by deletion of the SalI–HindIII region of pGUB plasmid (Juan et al, 1997). dSHA probe was generated by PCR amplification of a 216-bp region of pGUB-dSH, in the presence of [α-32P] dCTP. The dSHA sequence is 5′-ACATTAACCTATAAAAATAGGCGTATCACGAGGC CCTTTCGTCTTCAAGAATTCACGCGTAGATCTGC TAGCATCGATCCATGGACTAGTCTCGAGTTTAAA GATATCCAGCTGCCCGGGAGGCCTTCGCGAAATA TTGGTACCCCATGGAATCGAGGGATCCTCTAGA CGGAGGACAGTCCTCCGGTTACC TTCGAACCACGTGGCCGTCTAGAT-3′ (Gal4 binding site is underlined). Probes for the assays performed on nucleosome arrays were prepared as described in the Results section and in Supplementary data. Recombinant proteins Gal4-VP16 and Gal4_1–94 (Gal4-DBD) were purified as described previously (Utley et al, 1998). Dilutions of the protein stocks were made using Gal4 buffer (200 mM NaCl, 20 mM Hepes–KOH (pH 7.5), 1 mM β-mercaptoethanol, 20 μM ZnSO4, 1 mM EDTA and 10% glycerol). The ySWI/SNF complex was obtained by tandem affinity purification as described previously (Rigaut et al, 1999; Chandy et al, 2006). A portion of the purified complex was extensively concentrated (Amicon Ultra-4 100 000 MWCO, Millipore), quantified by SDS–PAGE/Coomassie staining and used as standard for Western blot quantifications.
Nucleosome reconstitution and purification
All reconstitutions were carried out by the octamer transfer method (Utley et al, 1996). For mononucleosome reconstitutions, 1 pmol of 32P-body-labeled probe was mixed with 3 μg (DNA content) of oligonucleosomes (purified from HeLa cells; Utley et al, 1996) at 1 M NaCl concentration. Dilutions to 0.8, 0.6, 0.4 and 0.2 M NaCl were made using buffer containing 10 mM Tris–Cl pH 7.4, 1 mM EDTA pH 8.0, 5 mM DTT and 0.5 mM PMSF (Buffer DR). Final dilution to 0.1 M NaCl was made using 10 mM Tris–Cl pH 7.4, 1 mM EDTA pH 8.0, 5 mM DTT, 0.5 mM PMSF, 0.1% NP-40, 20% glycerol and 200 μg/ml BSA (Buffer FDR). Reconstitutions prepared for direct use had a starting volume of 25 μl (250 μl final volume) and those for gel purification of the nucleosome used 5 μl as the starting volume (50 μl final). For this purification, the reconstituted material was loaded on a non-denaturing polyacrylamide gel (5%, 37.5:1 AA:Bis, 0.3 × TBE; conditions used throughout the present work, unless specifically indicated). After electrophoresis, the wet gel was exposed to a film and the bands corresponding to nucleosomal DNA were cut from the gel, incubating each gel piece overnight with the elution buffer (100 mM NaCl, 10 mM Tris–Cl pH 7.4, 1 mM EDTA pH 8.0, 5 mM DTT, 0.5 mM PMSF and 100 μg/ml BSA) at 4°C on an orbital shaker. After spinning, the supernatant was transferred to a clean tube and 0.25 volumes of the elution buffer containing 50% glycerol and 0.25% NP-40 were added. Purified nucleosome populations with a low relative concentration were concentrated around three times using Microcon YM-10 (Millipore, 42407) before addition of this buffer. The concentration of all purified probes was further adjusted to the same level using elution buffer containing 10% glycerol and 0.05% NP-40 (buffer R). For purification of 5′ and 3′ lateral nucleosome populations, the same gel purification protocol was used with the following modifications: for reconstitution, the final dilution was carried out using buffer FDR containing only 10% glycerol. After nucleosome reconstitution, the sample was supplemented with 10 mM MgCl2 and 40 U of EcoRI or HinfI was added, incubating at 30°C for 30 min (EcoRI) or 40 min (HinfI). Digestion was stopped with 20 mM EDTA, followed by gel purification of the non-digested nucleosomal probe.
For reconstitution of nucleosome arrays, 0.1 pmol of end-labeled or internally labeled probe was mixed with 4 μg of oligonucleosomes, using 25 μl as the starting volume and 250 μl as the final volume.
Binding assays
A mix containing 8.5 μl remodeling buffer (70 mM KCl, 20 mM Hepes–KOH pH 7.9, 2 mM DTT, 0.5 mM PMSF, 10% glycerol, 0.05% NP-40, 10 mM MgCl2 and 100 μg/ml BSA), 0.5 μl of 60 ng/μl oligonucleosomes, 3 μl SWI/SNF buffer (150 mM NaCl, 10 mM Tris–Cl pH 8.0, 1 mM Mg(CH3COO)2, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10% glycerol, 1 mM DTT and 0.5 mM PMSF), 0.5 μl 450 nM Gal4-DBD (or Gal4 buffer) and 2.5 μl probe was incubated for 30 min at 30°C, followed by gel electrophoresis, gel drying and scanning using phosphor screen and Typhoon 9400 (GE Healthcare). Autoradiography was also performed.
Nucleosome remodeling assays
Assays using mononucleosome probes were performed by mixing 2.5 μl probe, 0.5 μl oligonucleosomes (or buffer R), 0.6 μl 100 mM ATP (Roche, 1140965; ATP-γ-S 1162306), 0.5 μl Gal4-DBD or Gal4-VP16 (or Gal4 buffer) and 7.9 μl remodeling buffer. Gal4 derivatives stock concentration varied according to each particular assay. The same applies for oligonucleosome stocks. The mix was incubated for 20 min at 30°C (prebinding step). Then, 3 μl of SWI/SNF complex (or SWI/SNF buffer) was added, incubating for 30 min at 30°C (remodeling step). Then, a mix (1.2 μl) containing 750 ng calf thymus DNA, 500 ng long oligonucleosomes and 65 pmol of Gal4-oligo (removing mix) was added, incubating for 20 min at 30°C. Where Gal4 derivatives and/or ySWI/SNF binding activity was to be assessed, buffer DR was added instead of the removing mix. The samples were then subjected to electrophoresis on non-denaturing polyacrylamide gel. The dried gel was scanned using phosphor screen and Typhoon 9400. Quantitative analysis was performed using ImageQuant TL software (GE Healthcare). The total amount of probe loaded in each lane was normalized with respect to the amount in the starting material lane (nucleosome alone), before proceeding to the calculation described in the Results section. Autoradiography was also performed in each case. For Figures 2 and 5, duration of the remodeling incubation is as indicated in Results.
Remodeling assays using nucleosome arrays were performed as indicated for the mononucleosome probes, with the following modifications: in the remodeling buffer, KCl was raised to 82 mM and MgCl2 reduced to 7.6 mM; the remodeling step proceeded for 1 h; ATP stock was 25 mM. For MNase digestions, after the removing step, 2 μl of 120 mU/μl MNase (90 mU/μl for mock reconstituted array) in 10 mM Tris–Cl (pH 7.4), 15 mM CaCl2, 10 mM NaCl and 100 μg/ml BSA was added. MNase digestion proceeded for 1.5–3 min at 30°C. Further treatment of the samples was performed as described previously (Steger and Workman, 1997). In the cut-out assay, after the Removing step, 0.5 μl of EcoRI (100 U/μl) and 0.5 μl of PstI (100 U/μl) were added to the samples, incubating for 30 min at 37°C. Then, samples were analyzed electrophoretically as performed for the mononucleosome remodeling assays.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Bing Li and the other members of the Workman Laboratory for useful discussions. JLG was supported by the Pew Latin American Fellows Program in the Biomedical Sciences. This work was supported by the NIGMS Grant R37 GM047867 to JLW.
References
- Anderson JD, Thastrom A, Widom J (2002) Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol Cell Biol 22: 7147–7157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JD, Widom J (2000) Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 296: 979–987 [DOI] [PubMed] [Google Scholar]
- Anderson JD, Widom J (2001) Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol 21: 3830–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD (2004) Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell 14: 667–673 [DOI] [PubMed] [Google Scholar]
- Carey M, Lin YS, Green MR, Ptashne M (1990) A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345: 361–364 [DOI] [PubMed] [Google Scholar]
- Chandy M, Gutierrez JL, Prochasson P, Workman JL (2006) SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot Cell 5: 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Peterson CL, Workman JL (1998) Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA 95: 4947–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL (1994) Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 [DOI] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ (2003) Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2003) Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol Cell Biol 23: 7767–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W (1999) Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J 18: 6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Crowley KA, Carruthers LM, Hansen JC, Peterson CL (2002) The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat Struct Biol 9: 167–171 [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Gavin IM, Peterson CL, Logie C (2000) SWI–SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol 20: 3058–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan LJ, Utley RT, Vignali M, Bohm L, Workman JL (1997) H1-mediated repression of transcription factor binding to a stably positioned nucleosome. J Biol Chem 272: 3635–3640 [DOI] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM (2000) Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev 14: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B (2003) SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell 11: 391–403 [DOI] [PubMed] [Google Scholar]
- Kiyama R, Trifonov EN (2002) What positions nucleosomes? A model. FEBS Lett 523: 7–11 [DOI] [PubMed] [Google Scholar]
- Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W (2006) The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J Biol Chem 281: 5539–5545 [DOI] [PubMed] [Google Scholar]
- Korber P, Luckenbach T, Blaschke D, Horz W (2004) Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol Cell Biol 24: 10965–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370: 477–481 [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB (2004) Nucleosome remodeling: one mechanism, many phenomena? Biochim Biophys Acta 1677: 58–63 [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97: 843–852 [DOI] [PubMed] [Google Scholar]
- Liu H, Mulholland N, Fu H, Zhao K (2006) Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol 26: 2550–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD (1999) Histone octamer transfer by a chromatin-remodeling complex. Cell 96: 389–392 [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Minsky A (2004) Information content and complexity in the high-order organization of DNA. Annu Rev Biophys Biomol Struct 33: 317–342 [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Brown CE, Howe L, Workman JL (2002) Transcription activator interactions with multiple SWI/SNF subunits. Mol Cell Biol 22: 1615–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL (1999) Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell 4: 649–655 [DOI] [PubMed] [Google Scholar]
- Neely KE, Workman JL (2002) The complexity of chromatin remodeling and its links to cancer. Biochim Biophys Acta 1603: 19–29 [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Utley RT, Cote J, Peterson CL, Workman JL (1996) Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273: 513–516 [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL (1996) Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J 15: 4702–4712 [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Workman JL (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev 10: 187–192 [DOI] [PubMed] [Google Scholar]
- Phelan ML, Schnitzler GR, Kingston RE (2000) Octamer transfer and creation of stably remodeled nucleosomes by human SWI–SNF and its isolated ATPases. Mol Cell Biol 20: 6380–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL (1996) DNA-binding properties of the yeast SWI/SNF complex. Nature 379: 844–847 [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Ryan MP, Jones R, Morse RH (1998) SWI–SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol Cell Biol 18: 1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2002) Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev 16: 2120–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford GA, Morse RH (1997) Chromatin remodeling by transcriptional activation domains in a yeast episome. J Biol Chem 272: 11526–11534 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Workman JL (1997) Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J 16: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Schmitz J, Horz W (1994) The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J 13: 4856–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Cote J, Owen-Hughes T, Workman JL (1997) SWI/SNF stimulates the formation of disparate activator–nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem 272: 12642–12649 [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394: 498–502 [DOI] [PubMed] [Google Scholar]
- Utley RT, Owen-Hughes TA, Juan LJ, Cote J, Adams CC, Workman JL (1996) In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol 274: 276–291 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M (2004) DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell 16: 439–452 [DOI] [PubMed] [Google Scholar]
- Walter PP, Owen-Hughes TA, Cote J, Workman JL (1995) Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol Cell Biol 15: 6178–6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400: 784–787 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13: 339–346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


