Abstract
The mammalian SWI/SNF chromatin remodeling complex is becoming increasingly recognized for its role in tumor suppression, based on its ability to regulate accessibility of proliferation-associated genes to transcription factors. However, understanding the biological role of the complex is complicated because the same complex seemingly plays both positive and negative roles in gene expression. Work described here reveals that a choice between two independently encoded, closely related variants of a major subunit of the ARID protein family determines whether the SWI/SNF complex forms further associations with activator versus repressor complexes. The choice distinguishes assemblies with opposite effects on cell-cycle activity. The specific complexes control access of factors such as E2F1, Tip60, and HDAC1/2/3 to the promoters of various cell-cycle-specific genes, with c-Myc emerging as a particularly critical target.
Keywords: cell cycle, c-Myc, chromatin remodeling, E2F, SWI/SNF
Introduction
SWI/SNF-type chromatin remodeling complexes are present in all eukaryotic cells, where they act in an ATPase-dependent manner to disrupt chromatin structure and facilitate the binding of transcriptional regulators to nucleosomal sites (reviewed in Vignali et al, 2000; Mohrmann and Verrijzer, 2005). The activity of mammalian SWI/SNF complexes is crucial for proper differentiation and development. The complexes are also essential for negative control of proliferation, and subunits critical to this function are recognized as tumor suppressors (reviewed in Klochendler-Yeivin et al, 2002; Martens and Winston 2003; Roberts and Orkin, 2004; Biegel, 2006). However, very little is known about the role of the complexes in positive control of proliferation.
The entity typically designated the mammalian SWI/SNF complex, sometimes called the BAF complex, is actually a small series of related complexes of variable composition, resulting from the presence in most cells of alternative versions of certain subunits (illustrated schematically in Figure 1A). The complexes contain a core ATPase, either BRG1 or BRM, plus seven or more noncatalytic subunits that contain various DNA-binding and protein-binding motifs, which modulate the targeting and activity of the ATPase. The stably associated noncatalytic components of the complex include the p270/ARID1A subunit, which is a member of the ARID family of DNA-binding proteins (Wilsker et al, 2005). Like the ATPases, ARID-containing subunits of the BAF complex occur as two broadly expressed, independently encoded proteins that are mutually exclusive within the complexes (Inoue et al, 2002; Wang et al, 2004b). The alternative to p270/ARID1A is the closely related protein ARID1B. These products are derived from separate genes that are similar in size and exon structure (Figure 1B). The ARID family subunits are the only other subunits that occur as alternatives within most cells (as opposed to tissue-specific variants); so they are likely to confer specificity of function. In support of this concept, recent results using an siRNA-knockdown approach indicate that p270/ARID1A, which is frequently deficient in tumor tissue samples (Wang et al, 2004a), is required for proper differentiation-associated cell-cycle arrest, whereas ARID1B is dispensable for this function (Nagl et al, 2005, 2006).
Figure 1.
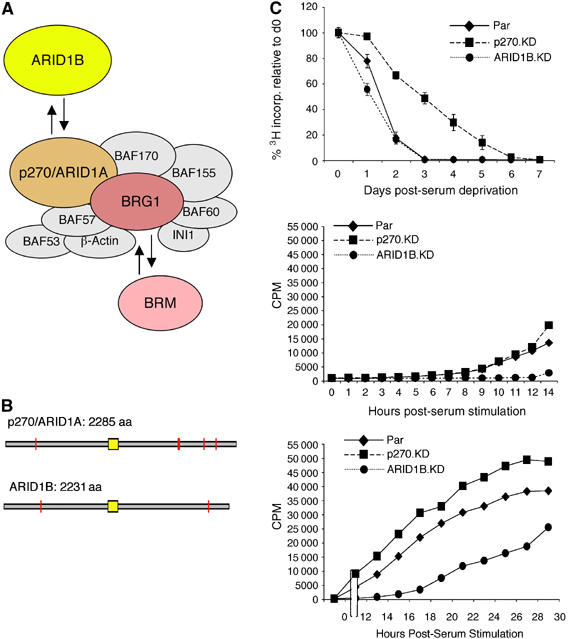
Knockdown phenotypes. (A) The multisubunit SWI/SNF chromatin-remodeling complex contains a core ATPase, either BRG1 or BRM. The ARID family subunits, p270/ARID1A and ARID1B, are also mutually exclusive alternatives. SWI/SNF complexes, therefore, are actually a series of related assemblies whose exact components depend on the choice of ATPase and ARID family subunit. (B) The p270/ARID1A and ARID1B family proteins are illustrated as open gray bars aligned according to the position of the ARID DNA-binding domain (indicated in yellow). Orange bars indicate occurrences of LXXLL motifs, which are implicated in protein–protein interactions. p270/ARID1A and ARID1B are 80% identical within the ARID motif, and approximately 60% identical in the region C-terminal of the ARID domain, with a lesser degree of conservation in the N-terminal region (reviewed in Wilsker et al, 2005). (C) DNA synthesis was measured as the rate of [3H]thymidine incorporation. Each point is an average and the average deviation from three independent cell lines of the respective knockdown series (or a triplicate plating from the parental cells). The average deviation is included at all points, but in most cases is less than the font size of the symbol representing the data point. In the serum stimulation panels, time 0 corresponds to day 3 of serum deprivation for the parental and ARID1B-knockdown lines, and day 7 for the p270-knockdown lines.
Cell-cycle arrest during differentiation is generally considered ‘irreversible' and differs in specific ways from reversible cell-cycle arrest resulting from conditions such as growth factor depletion. The ARID subunit-knockdown lines were engineered in a cell model that permits analysis of both processes. The parental MC3T3-E1 pre-osteoblast cell line consists of non-transformed cells that undergo a tightly regulated and well-characterized progression through cell-cycle arrest and into tissue-specific gene expression when exposed to differentiation medium (e.g., Quarles et al, 1992; Stein et al, 1996; Beck et al, 2001). These cells are also highly sensitive to the presence of serum growth factors, undergoing reversible cell-cycle arrest when deprived of serum.
Analysis of reversible cell-cycle arrest reveals that mammalian SWI/SNF complexes include specialized subsets with reciprocal roles in cell-cycle control. In the absence of p270/ARID1A, arrrest is delayed, but the cells resume cell-cycle activity rapidly once serum is restored. Conversely, arrest occurs normally in ARID1B-depleted cells but cell-cycle re-entry is delayed. Complexes specific for each of the ARID family subunits are present on the promoters of cell-cycle-specific genes in quiescent cells. Upon serum stimulation, the anti-proliferative p270/ARID1A-specific complexes dissociate from each promoter in a sequence correlating tightly with promoter activation. The ARID1B-containing complexes remain associated with the active promoters, in an ARID1B-dependent manner. The SWI/SNF complexes form further associations with activator versus repressor complexes and with specific pro-proliferative or anti-proliferative transcription factors, dependent upon the choice of ARID family subunit. Regulation of c-Myc expression is profoundly dependent on the activity of the SWI/SNF complexes, and the ARID subunits play a critical role in the access that these regulatory factors can gain to the c-Myc promoter.
Results
The ARID family subunits have opposing roles in cell-cycle regulation
The ARID subunit-knockdown lines and the integrity of the complexes incorporating the remaining subunits have been described previously (Nagl et al, 2005). The effect of depletion of the ARID family subunits on cell-cycle activity was evaluated through analysis of DNA synthesis activity in serum-deprived cells (Figure 1C). The upper panel shows that ARID1B-depleted cells arrest with the same kinetics as parental cells, ceasing DNA synthesis by day 3, whereas arrest in p270/ARID1A-depleted cells is delayed by 2–3 days. A strikingly converse result is seen when serum is replenished (middle and lower panels). Resumption of DNA synthesis is markedly delayed in the ARID1B-knockdown lines, indicating a specific role for the ARID1B-containing subset of complexes in cell-cycle activation. The p270/ARID1A-depleted cells consistently re-entered the cell cycle with slightly accelerated kinetics, further supporting the conclusion that these complexes play a specific role in the repression of cell-cycle activity.
Activation of cell-cycle-specific genes is delayed in the absence of ARID1B
The kinetics of activation of a marker panel representing serum-responsive and cell-cycle-specific genes linked with regulation by mammalian SWI/SNF complexes was examined by Western blotting (Figure 2). In the parental cells, c-Myc, cdc2, cyclin E, and cyclin A are induced at increasingly later times, at about 4, 7, 12, and 18 h post-stimulation, respectively. (Induction of c-Myc is not transient in these cells as it is in some fibroblast models of serum response; c-Myc levels are constitutively high in proliferating MC3T3-E1 cells in comparison with NIH3T3 cells; Beck et al, 2001.) Deficiency of p270/ARID1A correlates with accelerated activation of each marker, just as it correlates with accelerated onset of DNA synthesis. For example, cyclin E (Figure 2C) is not detected until 12 h post-stimulation in the parental cells, but is apparent by 10 h in the p270/ARID1A-depleted cells. In an exactly converse pattern, ARID1B deficiency delays activation of the entire marker panel. In the same example (Figure 2C), cyclin E is not detectable in the ARID1B-depleted cells for at least 16 h after stimulation. These results support the conclusion that p270/ARID1A-containing SWI/SNF complexes have an anti-proliferative function, whereas ARID1B-containing SWI/SNF complexes have a pro-proliferative function. Positive regulation by SWI/SNF complexes defines a new aspect of regulation of cell-cycle-specific genes.
Figure 2.
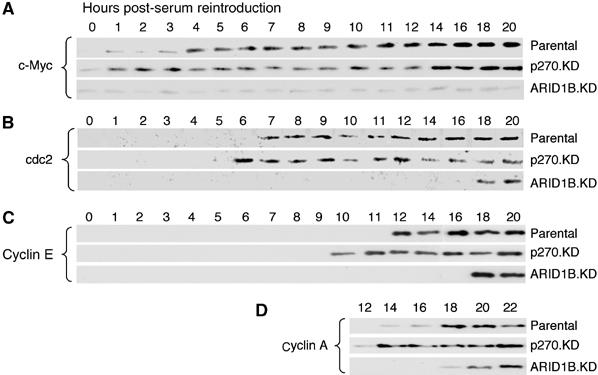
Induction of cell-cycle-specific proteins in the knockdown lines. Cell cultures, serum-depleted and re-stimulated as described in Figure 1, were harvested at hourly intervals up to 12 h post-stimulation, and at two hourly intervals thereafter, as indicated above the panels. Lysates were analyzed by Western blot for c-Myc (A), cdc2 (B), cyclin E (C), and cyclin A (D). For each protein, accumulation is accelerated in p270/ARID1A-depleted cells and delayed in ARID1B-depleted cells, consistent with the DNA synthesis kinetics seen in Figure 1.
Promoter association by p270/ARID1A correlates with repression of cell-cycle-specific genes
Probing parental cells in chromatin immunoprecipitation (ChIP) assays (Figure 3) indicates that each gene in the panel is a direct target for both p270/ARID1A- and ARID1B-defined complexes. Significantly, though, the timing of promoter occupation is different. Complexes containing ARID1B are present on each promoter at all times assayed. In contrast, the presence of p270/ARID1A on each promoter correlates tightly with repression. Complexes defined by the p270/ARID1A subunit dissociate from the promoters in a temporal pattern that coincides closely with the pattern of activation. For example, c-Myc is repressed at time 0 and actively expressed by 4 h after serum stimulation; the p270/ARID1A subunit is present at the c-Myc promoter at time 0, but is dissociated by 4 h after stimulation. At this time, p270/ARID1A is still present on the cdc2, cyclin E, and cyclin A promoters. The p270/ARID1A subunit dissociates from the cdc2 promoter sometime between 4 and 8 h post-stimulation, consistent with the rise in cdc2 levels at about 7 h. p270/ARID1A is present on the cyclin E promoter until at least 8 h post-stimulation, but is dissociated by 12 h when cyclin E levels begin to rise. In the last example, cyclin A expression begins at about 18 h post-stimulation, and complexes containing the p270/ARID1A subunit are still present on the promoter at 12 h after serum restoration. As would be expected from these data, the p270/ARID1A subunit is dissociated from the cyclin A promoter in proliferating cells (seen below in Figure 6).
Figure 3.
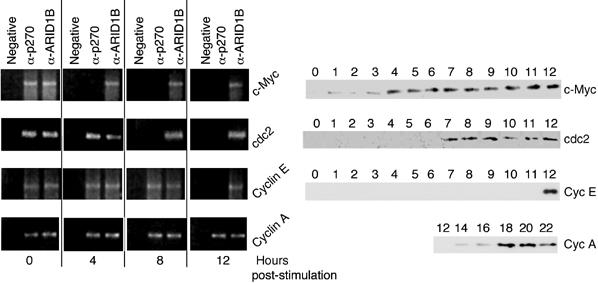
Promoter association analysis. Parental cells were harvested at 0, 4, 8, and 12 h post-stimulation and analyzed in ChIP assays for the presence of p270/ARID1A and ARID1B at the indicated promoters. In each case, ARID1B is present both before and after induction, whereas the presence of p270/ARID1A coincides only with repression. Portions of the parental cell induction patterns from Figure 2 are repeated for ease of comparison with the ChIP results.
Figure 6.
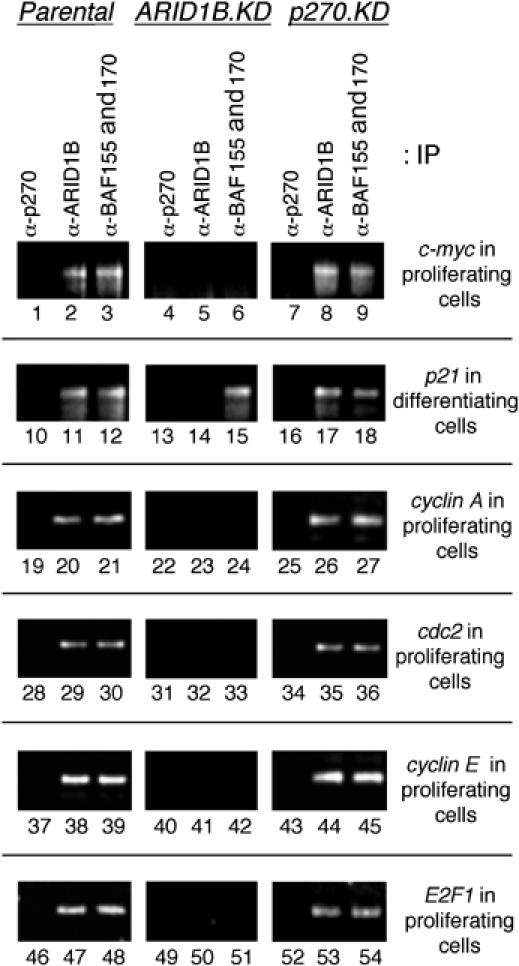
Requirement for ARID1B in SWI/SNF complex association with cell-cycle-specific promoters. Parental cells, or ARID subunit-depleted knockdown (KD) lines, were subjected to ChIP assays to monitor the presence of p270/ARID1A, ARID1B, or the ubiquitous subunits BAF155/170 at selected promoters. Cells were either actively proliferating, or at day 4 of differentiation-associated cell-cycle arrest, as indicated to the right or each panel. Loss of BAF155/170 reactivity on the c-myc, cyclin A, cdc2, cyclin E, and E2F1 promoters in ARID1B-depleted cells (middle panels) indicates a requirement for ARID1B in SWI/SNF association with each of these promoters. In contrast, ARID1B is not required for SWI/SNF association with the p21 promoter.
Interaction with pro-proliferative versus anti-proliferative E2F transcription factors
The target genes considered here are regulated in large part by the cell-cycle-specific E2F transcription factor family. This collective term refers to a series of related transcription factors, of which the best characterized are E2F1–E2F3, which are regarded as activating, and E2F4–E2F5, which are regarded as repressing. These are the predominant species associated with cell-cycle-regulated promoters in proliferating and quiescent cells, respectively (reviewed in Bracken et al, 2004; Frolov and Dyson, 2004; Blais and Dynlacht, 2004; Zhu et al, 2005). To determine whether E2F family members interact differentially with the SWI/SNF complexes distinguished here, ARID subunit-specific complexes were separated by immune precipitation and probed for the presence of E2F1, E2F4, and E2F5 (Figure 4A). Consistent with the pro-proliferative function of ARID1B-containing complexes, E2F1 (an activator E2F) was detected only in the ARID1B-containing subset (lanes 3 and 9). E2F1 was not detected in p270/ARID1A-specific complexes or in a negative control reaction performed on lysates from ARID1B-depleted cells (lanes 2 and 6). The repressor E2Fs (E2F4 and E2F5) associate with both sets of complexes (lanes 2, 3, 5, and 9). This supports the conclusion that p270/ARID1A-containing complexes have an anti-proliferative function, but reveals that ARID1B-containing complexes also associate with repressor E2Fs. This actually accords with the pattern of promoter occupancy exhibited by the two different sets of SWI/SNF complexes seen in Figure 3, and suggests that ARID1B-containing complexes contribute somewhat to repression of proliferation even though the net effect of ARID1B depletion is impaired cell-cycle activation. Activation involves dissociation of the p270/ARID1A-containing complexes from target promoters, whereas the ARID1B complexes appear to remain in place (or rapidly dissociate and re-associate) during the switch from repressor to activator E2F. A schematic representation of potential promoter interactions, based on the knockdown phenotypes, the kinetics of ARID subunit association with G1-activated promoters, and the association of ARID subunit-specific complexes with specific E2F factors in vivo is presented in Figure 4B.
Figure 4.
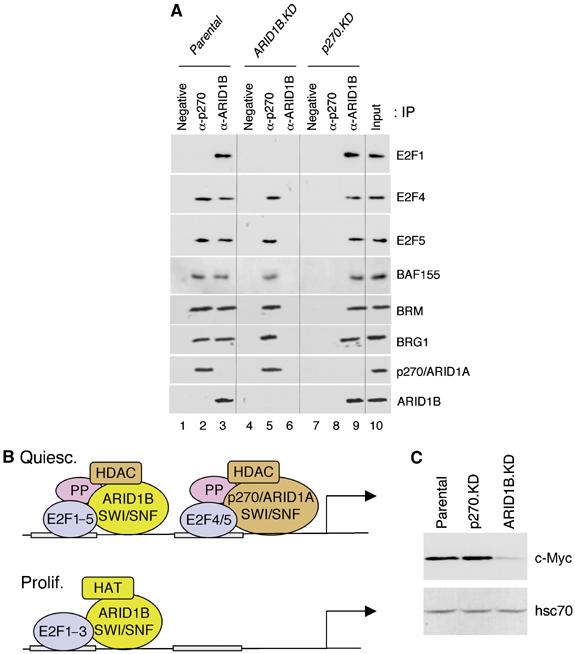
Stable association of E2F factors with SWI/SNF complexes. (A) p270/ARID1A-specific complexes and ARID1B-specific complexes were separated by immune precipitation from lysates of exponentially growing MC3T3-E1 cells, and probed for the presence of E2F1, E2F4, and E2F5. The negative control is a mAb of non-mammalian specificity (to SV-40 virus T-antigen). Antibodies reactive to SWI/SNF subunits BAF155, BRM, BRG1, p270/ARID1A, and ARID1B were used as positive controls. We and others have shown previously that p270/ARId1A and ARID1B each associate with both ATPases (Inoue et al, 2002; Wang et al, 2004b). (B) The presence of separate positive and negative E2F-binding sites has been demonstrated in multiple G1/S-phase genes including cyclin B1 and cdc2 (Zhu et al, 2004). A hypothetical G1-regulated promoter is depicted here with two E2F-binding sites: a distal positive site and a proximal negative site. The E2F factor family interacts with a family of ‘pocket proteins' (consisting of pRb, p107, and p130) that act as repressors. Repression in quiescent cells is linked with association of the repressor E2Fs, E2F4/5, pocket proteins (PP), and HDAC activity, and with pocket protein-mediated repression of activator E2Fs such as E2F1–3. Serum stimulation initiates a series of events that results in phosphorylation-mediated dissociation of pocket proteins and repressor E2Fs4/5, release of repression on activator E2F1–3, and recruitment of HAT activity. Data presented here indicate that p270/ARID1A-specific SWI/SNF complexes associate with E2F4 and E2F5 and contribute to repression at target promoters, dissociating at the time of activation. Association of ARID1B-specific SWI/SNF complexes with E2F4 and E2F5, and with repressed promoters in quiescent cells, suggests that ARID1B-specific SWI/SNF complexes play an auxiliary role in repression. Nevertheless, the phenotype of ARID1B-depleted cells indicates that ARID1B-specific complexes are required primarily for efficient activation of cell-cycle-specific promoters. ARID1B-specific complexes remain on the promoter (or rapidly dissociate and re-associate) as the promoter becomes activated and bound by an activator E2F. The postulated association of HAT and HDAC activities with specific SWI/SNF complexes is explored further below. (C) Western blot analysis of exponentially growing cells indicates that c-Myc levels are sharply reduced in ARID1B-deficient cells (ARID1B.KD).
Activation of c-Myc is dependent on ARID1B
The activating role of ARID1B-containing SWI/SNF complexes is not strictly required for induction of cdc2, cyclin E, and cyclin A. Expression of these gene products is markedly delayed, but each is expressed at levels comparable to those in parental cells by 18–20 h post-stimulation (Figure 2B–D). In contrast, c-Myc is detectable at a low level but is not induced further in ARID1B-depleted cells at any time tested (Figure 2A), even though its activation normally precedes each of the other genes, and even though c-Myc is normally maintained at a relatively high level in these cells (Beck et al, 2001). A probe of levels of c-Myc in exponentially growing cells (Figure 4C) reveals that c-Myc is constitutively low in ARID1B-deficient cells, although a basal level of expression is detectable. The basal level is apparently sufficient to maintain normal cell-cycle functions, as the ARID1B-depleted cells do not show proliferation defects during exponential growth, but these data indicate a critical role for ARID1B-containing SWI/SNF complexes in the activation of c-Myc in response to serum, and an inability to induce or maintain high levels of c-Myc in the absence of ARID1B.
Constitutive expression of c-Myc largely alleviates the effects of ARID1B depletion
E2F1 expression is activated in early G1 partly in response to c-Myc (Leone et al, 1997; Fernandez et al, 2003). Therefore, the dependence of c-Myc induction on ARID1B raises a question of the extent to which the delay in expression of later G1 genes is a consequence of the lack of c-Myc rather than a direct consequence of the deficiency of ARID1B. To address this question, c-Myc was expressed ectopically from an adenovirus expression vector, Ad-cMyc, before serum restoration (Figure 5A, blots 2 and 4). Parental cells infected with a negative control virus (Ad-9S) show the same kinetics of c-Myc expression seen in uninfected cells; that is, expression of c-Myc becomes detectable by 4 h post-stimulation (Figure 5A, blot 1). E2F1 becomes detectable in parental control-infected cells about 2 h after c-Myc (Figure 5B, blot 1); constitutive expression of c-Myc accelerates this event by about an hour (Figure 5B, blot 2). In ARID1B-depleted cells infected with the negative control virus, E2F1 expression is delayed to about 18 h post-stimulation (Figure 5B, blot 3). Constitutive expression of c-Myc is sufficient to restore nearly normal kinetics to E2F1 expression in the ARID1B-depleted cells, but accumulation of E2F1 is still delayed about 2 h compared to what is seen in parental cells (Figure 5B, compare blot 4 with blot 2). Constitutive expression of c-Myc with the concomitant accelerated expression of E2F1 also accelerates the activation of cdc2, cyclin E, and cyclin A in ARID1B-depleted cells. Nevertheless, in each case, expression in the ARID1B-depleted cells lags an hour or two behind expression in parental cells comparably infected with Ad-cMyc (Figure 5C–E, blot 4 versus blot 2 in each panel). We conclude that the most severely rate-limiting step in cell-cycle activation in ARID1B-depleted cells is upstream of E2F1, and affects the expression of c-Myc, a critical direct target of ARID1B-specific complexes during cell-cycle activation. However, the promoters of genes such as cdc2, cyclin E, and cyclin A are also direct targets for ARID1B-specific SWI/SNF complexes.
Figure 5.
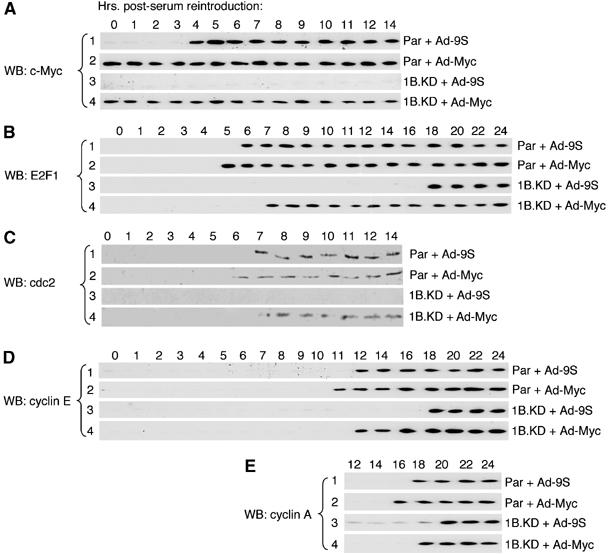
Constitutive expression of c-Myc in ARID1B-depleted cells. Parental (Par) or ARID1B-knockdown (1B.KD) cells, serum-depleted as described in Figure 1, were infected with either an Ad-cMyc expression virus or a negative control virus (Ad-9S), as indicated to the right of each panel. Twenty-four hours post-infection, cells were given fresh serum (time 0). Individual cell cultures were lysed at hourly intervals up to 12 h post-stimulation, and at two hourly intervals thereafter as indicated, and analyzed by Western blot (WB) for expression of c-Myc (A), E2F1 (B), cdc2 (C), cyclin E (D) and cyclin A (E).
ARID1B is required for SWI/SNF association with cell-cycle-specific promoters in proliferating cells
The p270/ARID1A-containing complexes dissociate from target promoters before the respective gene products are expressed. This situation affords the opportunity to determine whether ARID1B is required for promoter association by the complexes in proliferating cells, because the assay will not be complicated by the presence of the alternative version of the complexes. Association of the complexes was tracked with antibodies to the respective ARID family subunits, and also with an antibody reactive against two closely related ubiquitous subunits, BAF155 and BAF170, which are present together and have been identified in all known versions of the complexes. The absence of BAF155 and BAF170 from a promoter therefore indicates that no version of the complex is present. In proliferating parental cells, p270/ARID1A is not detectable at the c-Myc promoter, whereas ARID1B and the BAF155/170 subunits are present (Figure 6, lanes 1–3). In ARID1B-depleted cells, ARID1B is not detected at the promoter and neither are the BAF155 or BAF170 subunits (lanes 4–6), indicating that SWI/SNF complex is unable to associate with the c-Myc promoter in the absence of the ARID1B subunit. This is not due to an overall loss of integrity of the complexes, as the complexes remain otherwise intact in the absence of either ARID family subunit (Nagl et al, 2005). Moreover, the pattern at the c-Myc promoter can be compared with the association pattern at the p21WAF1/CIP1 promoter. p21 is induced during differentiation, and ARID1B is normally present at the promoter at this time (lane 11), but activation is not dependent on ARID1B (Nagl et al, 2005). Association of BAF155/BAF170 continues unimpaired on the p21 promoter in ARID1B-depleted cells (lane 15), indicating that association of the complex with this promoter is independent of ARID1B. Analysis of cyclin A, cdc2, cyclin E, and E2F1 shows the same pattern seen at the c-Myc promoter; ARID1B is required for SWI/SNF complex association with each of these cell-cycle-specific promoters. Overall, the results indicate that ARID1B-specific SWI/SNF complexes participate directly, in an ARID1B-dependent manner, at each step in a series that extends from induction of c-Myc, through induction of E2F1, to induction of E2F-responsive genes.
Association of histone acetyltransferase and deacetyltransferase activities with specific subsets of mammalian SWI/SNF complexes
The dissociation of activator and repressor functions suggests that histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities may interact with distinct SWI/SNF complexes (model illustrated in Figure 4B). Stable association of SWI/SNF components with the HDAC-containing Sin3 or N-CoR repressor complexes has been reported previously (Underhill et al, 2000; Sif et al, 2001; Battaglioli et al, 2002; Kuzmichev et al, 2002), but these observations were made before it was possible to distinguish the ARID subunits; so this question was re-examined here. Direct biochemical assay of immune complexes specific for either p270/ARID1A or ARID1B shows HDAC activity present in both sets of complexes (Figure 7A, upper panel), in agreement with the finding that both sets of complexes are present on repressed promoters. HAT activity has not been reported to date in association with mammalian SWI/SNF complexes, but analysis here reveals its presence, and that it is limited to ARID1B-containing complexes (lower panel), consistent with the finding that only the ARID1B-containing complexes are associated with active promoters. Neither activity is detected in immuno-reactive material isolated from the respective knockdown lines, verifying the specificity of the interactions.
Figure 7.
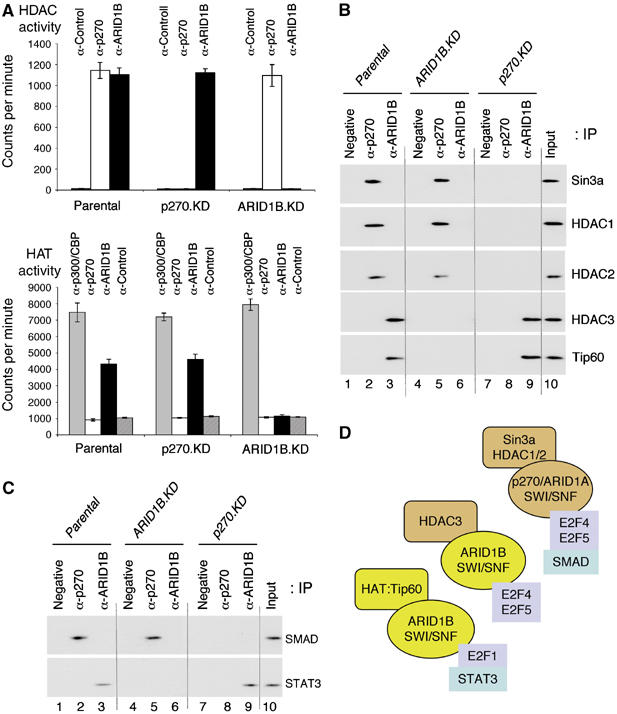
Association of HAT and HDAC activities with SWI/SNF complexes. (A) HDAC assays were performed on Sepharose-immobilized immune complexes specific for p270/ARID1A or ARID1B, as indicated. The negative control antibody is described in Figure 4. The respective knockdown lines were included as further negative controls. Assays were performed in triplicate, using three independently isolated knockdown lines from each series, or a triplicate plating of parental cells. Results are represented as average counts per minute and average deviation. HAT assays were performed on Sepharose-immobilized immune complexes prepared with controls as described above. As an additional control, a parallel reaction with a mAb reactive with the acetyltransferases p300/CBP was included. Reactions were performed in the presence of [3H]acetyl coenzyme A and scored by detecting acetylation of the histone substrates in a filter binding assay. Results, performed in triplicate as described above, are represented as the average and average deviation of counts per minute per filter. (B) Immune complexes, as described in Figure 4A, were probed for the presence of various components of repressor (HDAC-containing) or activator (HAT-containing) complexes. (C) The immune complexes in panel B were additionally probed for the presence of SMAD and STAT3. (D) The figure is a schematic representation of the HAT and HDAC activities associated with ARID subunit-specific subsets of SWI/SNF complexes. Also shown are identified associations with specific transcription factors.
The nature of the associated activities was probed in Western blots of the isolated immune complexes (Figure 7B). Because the Sin3 repressor complex has been detected repeatedly in association with mammalian SWI/SNF complexes, and is associated with E2F-mediated repression (Lai et al, 2001; Rayman et al, 2002), we probed for the presence of the Sin3a subunit determinant of this complex. Sin3a was readily detectable in association with the p270/ARID1A-containing complexes (lanes 2 and 5), but not with the ARID1B-containing complexes (lanes 3 and 9), suggesting that HDAC activities associated with these complexes are of different origin. The Sin3 complex contains two HDAC subunits, HDAC1 and HDAC2 (reviewed in Silverstein and Ekwall, 2005), both of which can also be seen in Figure 7B in association with p270/ARID1A, but not with ARID1B. The HDAC activities commonly present in transcription regulatory complexes extend to a third species, HDAC3, which is not a stable member of the Sin3 complex but is associated with other repressor complexes including the N-CoR complex (reviewed in Yang and Seto, 2003). Probing for HDAC3 shows a pattern converse to that seen with Sin3a and HDAC1/2. HDAC3 is associated with ARID1B-containing SWI/SNF complexes (lanes 3 and 9), but not with p270/ARID1A-containing complexes (lanes 2 and 5). Thus, the HDAC activities associated with the ARID subunit-defined subsets of SWI/SNF complexes are indeed derived from different sources. Considering the basis of the ARID1B-associated HAT activity, we reasoned it would likely be one associated with E2F-responsive promoters during G1/S-phase activation. The HAT activity most clearly linked with E2F in this context is the Tip60 HAT subunit of the TRRAP activator complex (Taubert et al, 2004). A probe for Tip60 reveals that it is indeed stably and specifically associated with ARID1B-containing complexes (Figure 7B). This does not imply that this is the only associated HAT, but it is consistent with the behavior of the complexes on promoters.
Current models of activation of E2F-responsive promoters in G1/S-phase cells recognize, in addition to a requirement for dissociation of pocket proteins and a switch in E2F family members, a requirement for dissociation of a repressor activity and a further requirement for recruitment of an activating function. The present studies link separate anti-proliferative and pro-proliferative subsets of mammalian SWI/SNF complexes with these roles. The secondary repressor function linked with ARID1B-containing complexes is presumably not essential, as its loss is not reflected in the biological phenotype of ARID1B-depleted cells, where the net effect is deficiency of the pro-proliferative function.
Role of ARID subunits in association of other factors at the c-Myc promoter
The different roles of the ARID subunit-determined complexes were explored directly in the context of the c-Myc promoter. Repression of c-myc is linked with association of SMAD family members in an interaction that involves E2F4 (Chen et al, 2002; Frederick et al, 2004), whereas activation is associated with promoter binding by the STAT3 transcriptional activator and activator E2Fs including E2F1 (Hiebert et al, 1989; Roussel et al, 1994; Kiuchi et al, 1999; Albert et al, 2001; Bowman et al, 2001). Probing the separate complexes with antibodies specific for SMADs and STAT3 showed a pattern consistent with the emerging model, that is, antibodies specific for the SMAD family of negative regulators reacted only with the p270/ARID1A-specific complexes, whereas antibodies specific for the positive transactivator STAT3 reacted only with the ARID1B-specific complexes (Figure 7C). The interactions identified with the specific complexes are illustrated schematically in Figure 7D.
To determine whether binding of any of these factors to the c-Myc promoter is affected by deficiency of the ARID family subunits, promoter association was assayed in quiescent and proliferating cells, and in each of the knockdown lines. These assays show that SMAD and E2F4 and the co-repressing factors Sin3a, HDAC1, and HDAC2 are each associated with the c-Myc promoter in quiescent, but not proliferating, cells (Figure 8C–G, lane 1 compared with lane 4). Promoter binding by these factors is not affected by deficiency of ARID1B (Figure 8C–G, lane 3), but binding by all except E2F4 is dependent on the presence of p270/ARID1A (C–G, lane 2).
Figure 8.
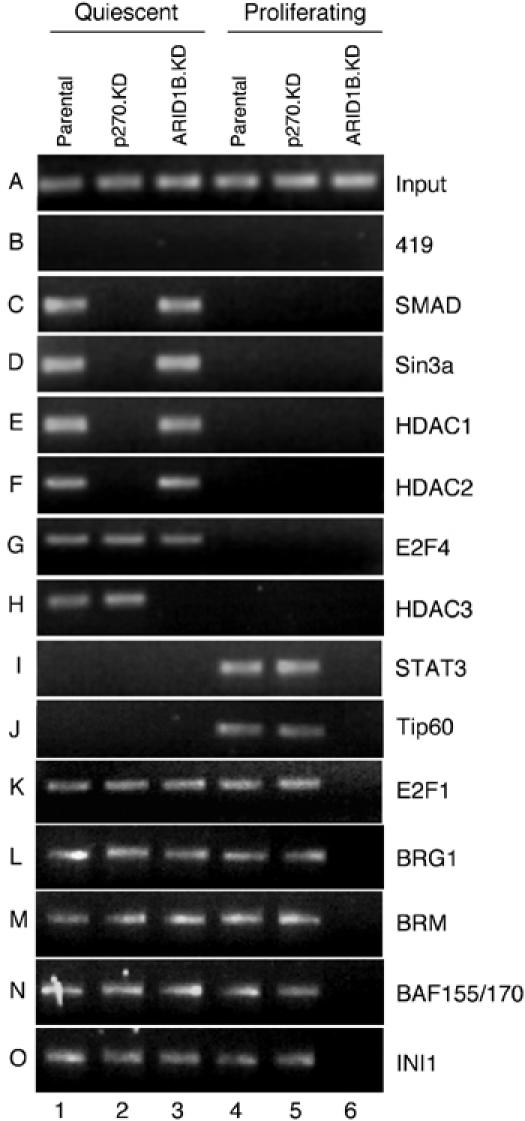
Protein binding at the c-myc promoter in ARID subunit-depleted cells. Association of a panel of regulatory proteins with the c-myc promoter was assessed by ChIP assay in parental cells or knockdown (KD) lines depleted for p270/ARID1A or ARID1B, as indicated above each lane. Cells were either actively proliferating or made quiescent by serum deprivation as described in Figure 1. The negative control mAb is described in Figure 4.
Association of HDAC3 (Figure 8H) shows a different pattern. HDAC3 is associated with the c-Myc promoter only in quiescent cells as would be expected (Figure 8H, lane 1 compared with lane 4), but its presence is dependent on the presence of ARID1B, not p270/ARID1 (Figure 8H, lane 3 compared with lane 2), in agreement with the protein–protein interaction pattern seen in Figure 7. The striking inability of cells to assemble key repressor proteins on the c-Myc promoter in the absence of p270/ARID1A underscores the severity of the cell-cycle arrest phenotype associated with loss of this subunit. That c-Myc levels are eventually repressed in serum-depleted p270/ARID1A-knockdown cells is apparently due to surviving secondary mechanisms of repression, such as the ARID1B-dependent recruitment of HDAC3 to the promoter.
In proliferating cells, the transactivator STAT3 and the coactivating acetyltransferase Tip60 are each present on the c-Myc promoter (Figure 8I and J, lane 4 compared with lane 1), and association of both factors is dependent on ARID1B (lane 6 compared with lane 4). E2F1 is also present (Figure 8K, lane 4), and is unique among the factors assayed in that it is associated with the promoter in both proliferating and quiescent cells (lanes 1 and 4). This accords with accepted models of E2F1 function, which posit repression of the activation function of E2F1 without dissociation from the promoter, by binding of a pRB family member and subsequent recruitment of HDAC activity (reviewed in Blais and Dynlacht, 2004; Bracken et al, 2004; Frolov and Dyson, 2004).
In quiescent cells, promoter association by E2F1 is not dependent on either ARID family protein (Figure 8K, lanes 2 and 3). This is the same pattern seen with E2F4 (Figure 8G). E2F1 association may occur as long as either ARID subunit is present, a possibility that will be addressed in future studies using double knockdowns. Alternatively, there is precedent for the concept that E2F proteins can access promoters independently of the ATPase-dependent chromatin remodeling complexes. E2F4 is able to bind to the cell-cycle-regulated Plk1 promoter in SW13 cells (Gunawardena et al, 2004). Because these cells are deficient in both the BRG1 and BRM ATPases, the result implies that the activity of all subsets of the complexes is dispensable for E2F4 binding to the promoter. SWI/SNF complex activity was, however, necessary for histone deacetylation in this study, consistent with our observations that binding of specific HDACs is dependent on individual subsets of the complexes.
Association of E2F1 with the repressed c-Myc promoter is independent of either p270/ARID1A or ARID1B, but association with the active promoter is dependent on ARID1B (Figure 8K, lane 6). Thus, upon release from repression, E2F1 dissociates from the promoter and re-associates during activation in a manner dependent on the ARID1B-containing subset of complexes. A recent study indicates that binding of activator E2Fs to a panel of cell-cycle-regulated genes normally precedes binding of the acetyltransferase Tip60 to these promoters (Taubert et al, 2004). Results described here indicate that the integrity of the ARID1B-containing complexes is specifically required for both events.
Additional subunits of the SWI/SNF complexes were also assayed for promoter association (Figure 8L–O). BRG1 and BRM both associate with both ARID family subunits (Inoue et al, 2002; Wang et al, 2004b), and both are present on the c-Myc promoter when either ARID subunit is present in the cells and at the promoter. The same behavior is seen with BAF155/BAF170 and INI1, and is consistent with the results shown in Figure 6.
Discussion
These studies reveal a new layer of control in the regulation of cell-cycle-specific genes. The existence of distinct subsets of SWI/SNF complexes with pro-proliferative versus anti-proliferative roles has not been recognized until now. Moreover, recognition that separate SWI/SNF complexes facilitate promoter localization by individual co-activators and co-repressors including HDAC1, HDAC2 and HDAC3, Sin3a, and Tip60 offers significant new insight into the mechanism of promoter assembly by these factors. On still another level, recognition that separate SWI/SNF complexes play required roles in promoter localization by SMAD versus STAT3 offers important insight into the mechanisms of promoter assembly by sequence-specific transcription factors.
The molecular mechanism of action of the ARID subunits is not completely defined. Their DNA-binding domains make sequence-independent DNA contacts (Patsialou et al, 2005). Whether this interaction affects DNA conformation is an open question to be addressed in further studies. Most of the remainder of the protein sequence is apparently devoted to protein–protein interactions, both within and outside the SWI/SNF complex. Encoding two different members of the ARID1 subfamily is characteristic only of higher eukaryotes. The single Drosophila equivalent of ARID1 (termed Osa) is involved in both activation and repression (e.g., Milán et al, 2004). The emergence of alternative related subunits, like the evolution of multiple E2F family members, permits more finely tuned regulation, which may be particularly important in maintaining the temporal order of gene activation and repression during the cell cycle.
The ability to distinguish the complexes based on their ARID subunits has greatly facilitated identification and clarification of interactions between SWI/SNF complexes and histone-modifying activities. Sin3 complex members associate specifically with the p270/ARID1A-containing subset, whereas ARID1B-associated HDAC activity is linked with HDAC3, suggesting that it derives from an associated N-CoR or SMRT complex. Underhill et al (2000) isolated an N-CoR complex that contained HDAC3 and approximately 20 other associated proteins, including the SWI/SNF complex members, BRG1, BAF155, BAF170, and INI1/hSNF5; species of a size appropriate to ARID1B were also present. Thus, our results are consistent with previous findings, but reveal that the SWI/SNF subunits associated with N-CoR versus Sin3 derive from distinct complexes defined by the choice of ARID family subunit. Our data strongly support the concept that histone-modifying co-repressor complexes are physically coupled with ATPase-powered chromatin remodeling complexes, and that the ability of the HDAC complexes to access target promoters such as c-Myc is dependent on the functional presence of the ARID family subunits.
HAT activity has not been reported before in association with mammalian SWI/SNF complexes. Our earlier attempts to detect such an activity using a BAF155-specific antibody did not yield detectable signal (Dallas et al, 1998), presumably because the HAT-associated ARID1B complexes were a minor part of the BAF155-specific immune complex. The ability to look at ARID1B complexes separately provides the sensitivity needed to reveal the presence of HAT activity. Analysis of endogenous protein–protein interactions links the presence of this activity with the presence of the Tip60 HAT in ARID1B complexes. This does not exclude the possibility that other HATs also associate with ARID1B complexes, but the identification of Tip60 is significant as this HAT is already strongly implicated in the activation of E2F-responsive promoters (Taubert et al, 2004). Among the various questions under further investigation is whether specificity for binding the co-activator/co-repressor complexes lies directly within the ARID family subunits. A related question concerns interactions between the ARID subunits and specific transcription factors such as SMADs, STAT3, and the E2F family.
The role of p270/ARID1A in cell-cycle repression implies it contributes significantly to the tumor suppression activities of SWI/SNF complexes. Although ARID1A has not yet been characterized directly as a tumor susceptibility gene, it is notable that deficiency of p270 has been detected among human tumor tissue samples (Wang et al, 2004a), and that the chromosomal locus of ARID1A, 1p35.3 (Entrez), lies in a region strongly predicted to harbor as yet unidentified tumor susceptibility genes (reviewed in Schwab et al, 1996). Conversely, ARID1B is a specificity determinant of SWI/SNF complexes with a wide-ranging role in promoting proliferation and an apparently non-essential role in repressing cell-cycle activity, making ARID1B an attractive target for anti-cancer therapy. High-level expression of c-Myc is particularly dependent on ARID1B, and elevated expression of c-Myc has been detected in a wide range of human cancers, often linked with the presence of constitutive upstream signals such as activated forms of STAT3 (reviewed in Inghirami et al, 2005). The effect on the proliferative rate of depleting ARID1B in such cells is under investigation.
Materials and methods
Cell lines and cell culture
Culture and differentiation (by exposure to ascorbic acid and β-glycerol phosphate) of low passage murine calvarial MC3T3-E1 cells, the generation of the knockdown lines, and the integrity of the complexes in the absence of either ARID family subunit have been described previously (Beck et al, 2001; Nagl et al, 2005). Originally, three independent lines in each knockdown series were described with reproducible phenotypes. Representative lines from each series were used here. For serum depletion, monolayers at 90% confluency were washed in PBS and incubated in DMEM plus 0.1% FBS for indicated times. Serum-stimulated cells received fresh medium plus 10% FBS (Summit Biotech, Fort Collins, CO) at time 0.
ChIP assays
ChIP assays were performed with the EZ ChIP™ system (Upstate Cell Signaling Solutions, Lake Placid, NY), according to the manufacturer's directions, modified to include pre-clearing of lysates with 60 μl of 50% slurry of protein G/salmon sperm DNA for 1 h at 4°C, and again overnight. Primers sequences used are as follows:
cdc2 For: CACACAGAAAGGTAGCTGGAG, cdc2 Rev: CAATCAGAGCTGAGCTACGCT;
p21 For: TGCGTGACAAGAGAATAGCCCAG, p21 Rev: TGCAGTTGGCGTCGAGCTGC;
c-myc For: AGCAAGAGAAAATGGTCGGG, c-myc Rev: GCTGCAATGGGCAAAGTTTG;
cyclin E For: GAAACAACAAAGCCTGGTGG, cyclin E Rev: ACAGCCACTCCGGTCTGCGA;
cyclin A For: TAATCGCCCAGACAGAGAAG, cyclin A Rev: ATTGATTTAGGGCAGAGGGA;
E2F1 For: ACGCCGCAACCAATGCTCGC, E2F1 Rev: ATGAGTGGCAGGCCGCGGCG;
Neg control For: TTGACTTCGTCACGGAGACG, Neg control Rev: GAGTGCAAGACAGCGACAAG.
DNA synthesis
Cells were labeled with [3H]thymidine (Perkin-Elmer, Boston, MA) in 1-h pulses at the times indicated in the text, as described previously (Nagl et al, 2005).
Virus infection
The E1A-inactivated (negative control) 9S adenovirus Ad-9S and the c-Myc expression virus Ad-cMyc have been described previously (Zerler et al, 1987; Mitchell and El-Deiry, 1999). Infection was performed at a multiplicity of 25 plaque-forming units per cell.
HAT assay
Immune complexes were isolated from 2 mg of cell lysate, washed three times with p300 lysis buffer (Nagl et al, 2005), and resuspended in HAT buffer (50 mM Tris, pH 8.0, 10% glycerol, 1 mM DTT, 0.1 mM EDTA). The HAT assay was performed in the presence of 0.2 mg calf thymus histone mixture (type II-A) (Roche), 0.113 M n-butyric acid, and 100 nCi [3H]acetyl coenzyme A (specific activity 4.5 Ci/mmol, Amersham Life Science) at 30°C for 10 min. HAT activities were scored by detecting acetylation of histone substrates in a filter binding assay (Brownell and Allis, 1995).
HDAC assay
Assays were performed using an HDAC assay kit (Upstate Biotechnology, cat. no. 12-347) supplemented with [3H]CH3COONa (specific activity 2–5 Ci/mmol; Perkin-Elmer).
Protein assays and antibodies
Protein assays were performed as described previously (Wang et al, 2004b; Nagl et al, 2005). Monoclonal antibodies (mAb) specific for p270/ARID1A (PSG3), ARID1B (KMN1), BAF155/BAF170 (DXD12), p300/CBP (NM11), SV-40 Tag (419), and the cdc2-G6 polyclonal antibody have been described previously (Dallas et al, 1998; Wang et al, 2004b, 2005; Nagl et al, 2005). Antibodies of the following specificities were obtained from commercial sources: p21WAF1/CIP1 (556431) and INI1 (612110) (BD Biosciences, San Jose, CA), hsc70 (Stressgen, San Diego, CA), c-Myc (9E10, sc-40), cyclin A (C-19, sc-596), cyclin B2 (N-20, sc-5235), cyclin E (M-20, sc-481), cyclin A (C-19, sc-596), E2F1 (C-20, sc-193), E2F4 (C-20, sc-1082), E2F5 (MH-5, sc-968), Sin3a (AK-11, sc-767), HDAC2 (H-54, sc-7899), SMAD (H-465, sc-7153), STAT3 (C-20, sc-482), BRM (N-19, sc-6450), and BRG1 (H-88 sc-10768) (Santa Cruz Biotechnology Inc., Santa Cruz, CA), HDAC1 (2062) and HDAC3 (2632) (Cell Signaling Technology, Beverly, MA), and Tip60 (07-038) (Upstate Cell Signaling Solutions, Lake Placid, NY).
Acknowledgments
We thank Peter Yaciuk for help with hybridoma development, Wafik El-Deiry, Scott Shore, Sal Rusello, E Premkumar Reddy, Xavier Graña-Amat, Judit Garriga, Bayar Thimmapaya, and Ed Harlow for generous gifts of reagents, and members of our laboratory and other colleagues for advice and critical comments. This work was supported by PHS grant RO1GM073257 (EM), DOD BCRP grant DAMD17-01-1-0406 (EM), a Research Incentive Award from Temple University (EM), and by a Shared Resources grant to the Fels Institute (1R24CA88261). NGN Jr. XW, and AP were recipients of DOD BCRP fellowships DAMD-17-02-1-0577, DAMD17-00-1-0453, and DAMD-17-02-1-0578, respectively.
References
- Albert T, Wells J, Funk J, Pullner A, Raschke EE, Stelzer G, Meisterernst M, Farnham PJ, Eick D (2001) The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J Biol Chem 276: 20482–20490 [DOI] [PubMed] [Google Scholar]
- Battaglioli E, Andres ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G (2002) REST repression of neuronal genes requires components of the hSWI. SNF complex. J Biol Chem 277: 41038–41045 [DOI] [PubMed] [Google Scholar]
- Beck GR Jr, Zerler B, Moran E (2001) Gene array analysis of osteoblast differentiation. Cell Growth Differ 12: 61–83 [PubMed] [Google Scholar]
- Biegel JA (2006) Molecular genetics of atypical teratoid/rhabdoid tumor. Neurosurg Focus 20: E11. [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD (2004) Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev 14: 527–532 [DOI] [PubMed] [Google Scholar]
- Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, Irby R, Yeatman T, Courtneidge SA, Jove R (2001) Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci USA 98: 7319–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K (2004) E2F target genes: unraveling the biology. Trends Biochem Sci 29: 409–417 [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD (1995) An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA 92: 6364–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massague J (2002) E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110: 19–32 [DOI] [PubMed] [Google Scholar]
- Dallas PB, Cheney IW, Liao DW, Bowrin V, Byam W, Pacchione S, Kobayashi R, Yaciuk P, Moran E (1998) p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol 18: 3596–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B (2003) Genomic targets of the human c-Myc protein. Genes Dev 17: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF (2004) Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol Cell Biol 24: 2546–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ (2004) Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Gunawardena RW, Siddiqui H, Solomon DA, Mayhew CN, Held J, Angus SP, Knudsen ES (2004) Hierarchical requirement of SWI/SNF in retinoblastoma tumor suppressor-mediated repression of Plk1. J Biol Chem 279: 29278–29285 [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Lipp M, Nevins JR (1989) E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci USA 86: 3594–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inghirami G, Chiarle R, Simmons WJ, Piva R, Schlessinger K, Levy DE (2005) New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle 4: 1131–1133 [DOI] [PubMed] [Google Scholar]
- Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N (2002) Largest subunits of the human SWI/SNF chromatin remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem 277: 41674–41685 [DOI] [PubMed] [Google Scholar]
- Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, Hibi M, Hirano T (1999) STAT3 is required for gp130-mediated full activation of the c-myc gene. J Exp Med 189: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Muchardt C, Yaniv M (2002) SWI/SNF chromatin remodeling and cancer. Curr Opin Genet Dev 12: 73–79 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol 22: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, Tsai SC, Seto E, Zhang Y, Kuzmichev A, Lane WS, Reinberg D, Harlow E, Branton PE. (2001) RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol 21: 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR (1997) Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387: 422–426, Erratum in: Nature 387: 932 [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Milán M, Pham TT, Cohen SM (2004) Osa modulates the expression of Apterous target genes in the Drosophila wing. Mech Dev 121: 491–497 [DOI] [PubMed] [Google Scholar]
- Mitchell KO, El-Deiry WS (1999) Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-Phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ 10: 223–230 [PubMed] [Google Scholar]
- Mohrmann L, Verrijzer CP (2005) Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta 1681: 59–73 [DOI] [PubMed] [Google Scholar]
- Nagl G Jr, Patsialou A, Haines DS, Dallas PB, Beck GR Jr, Moran E (2005) The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle regulation. Cancer Res 65: 9236–9244 [DOI] [PubMed] [Google Scholar]
- Nagl G Jr, Zweitzig DR, Thimmapaya B, Beck GR Jr, Moran E (2006) The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res 66: 1289–1293 [DOI] [PubMed] [Google Scholar]
- Patsialou A, Wilsker D, Moran E (2005) DNA-binding properties of ARID family proteins. Nucleic Acids Res 33: 66–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarles LD, Yohay DA, Lever LW, Caton R, Wenstrup RJ (1992) Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. J Bone Miner Res 7: 683–692 [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD (2002) E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev 16: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH (2004) The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer 4: 133–142 [DOI] [PubMed] [Google Scholar]
- Roussel MF, Davis JN, Cleveland JL, Ghysdael J, Hiebert SW (1994) Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene 9: 405–415 [PubMed] [Google Scholar]
- Schwab M, Praml C, Amler LC (1996) Genomic instability in 1p and human malignancies. Genes Chromosomes Cancer 16: 211–229 [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE (2001) Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev 15: 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet 47: 1–17 [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, Stein JL, Van Wijnen AJ, Montecino M (1996) Transcriptional control of osteoblast growth and differentiation. Physiol Rev 76: 593–629 [DOI] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B (2004) E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol 24: 4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C, Qutob MS, Yee SP, Torchia J (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 275: 40463–40470 [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL (2000) ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nagl NG Jr, Flowers S, Zweitzig DR, Dallas PB, Moran E (2004a) Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer 112: 636–642 [DOI] [PubMed] [Google Scholar]
- Wang X, Liao DW, Van Scoy M, Pacchione S, Yaciuk P, Moran E (2005) Monoclonal antibodies reactive with the BAF155 (SMARCC1) and BAF170 (SMARCC2) components of human SWI/SNF-related complexes. Hybridoma 24: 55–57 [DOI] [PubMed] [Google Scholar]
- Wang X, Nagl G Jr, Van Scoy M, Pacchione S, Yaciuk P, Dallas PB, Moran E (2004b) Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J 383: 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsker D, Probst L, Wain HM, Maltais L, Tucker PW, Moran E (2005) Nomenclature of the ARID family of DNA binding proteins. Genomics 86: 242–251 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2003) Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev 13: 143–153 [DOI] [PubMed] [Google Scholar]
- Zerler B, Roberts RJ, Mathews MB, Moran E (1987) Different functional domains of the adenovirus E1A gene are involved in regulation of host cell cycle products. Mol Cell Biol 7: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR (2004) E2Fs link the control of G1/S and G2/M transcription. EMBO J 23: 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR (2005) Temporal control of cell cycle gene expression mediated by E2F transcription factors. Cell Cycle 4: 633–636 [DOI] [PubMed] [Google Scholar]


