Short abstract
A report on the meeting 'Translational Control' at Cold Spring Harbor, New York, 6-10 September 2006.
Abstract
A report on the meeting 'Translational Control' at Cold Spring Harbor, New York, 6-10 September 2006.
Regulation of gene expression at the level of mRNA translation is a fundamental mechanism for moderating cellular events. A recent meeting at Cold Spring Harbor Laboratory served to highlight the modes, mechanisms, and architecture of translational control and of the translational apparatus itself (a full list of presentations is available at http://meetings.cshl.edu/meetings/abstracts/2006transc_absstat.html). This report concentrates on several prominent themes in the regulation of mRNA translation. Central to the topics discussed here is the observation that the translation of single specific mRNAs, subsets, or even a majority of the mRNAs in a cell, is controlled almost exclusively through a multitude of interactions that occur between RNA-binding proteins and regulatory elements embedded throughout the mRNA (Figure 1).
Figure 1.
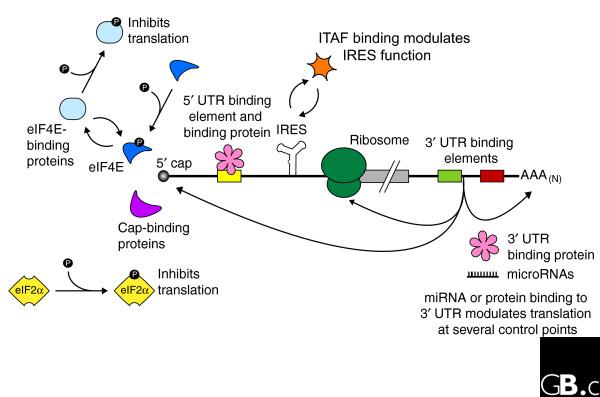
Regulation of eukaryotic mRNA translation occurs at numerous control points. Recognition of 3' UTR sequence or structural elements (green and red boxes) by RNA-binding proteins leads to either activation or repression of translation, often through alteration of the 3' poly(A) tail or through interactions with proteins that bind at the 5' terminal cap structure (that is, the initiation factor eIF4E or cap-binding proteins). Repression of translation by miRNAs can occur through inhibition of translation initiation or elongation, and may also lead to changes in the status of the mRNA 3' poly(A) tail. Elements found within the mRNA 5' UTR (yellow box) can bind regulatory proteins that repress translation by inhibiting 48S ribosome scanning. Global regulation of mRNA translation is commonly achieved through modification of the translational apparatus (that is, by phosphorylation of the translation initiation factors eIF2α and eIF4E) and the ribosome itself, or modulation of protein partner binding affinities (such as the phosphorylation of the eIF4E-binding proteins). Translation can be initiated independent of the mRNA 5' cap through a structured internal ribosome entry site (IRES) in the 5' UTR whose efficiency in initiating translation is, in turn, modulated by trans-acting factors (ITAFs).
A common architecture for translational repression
Binding of proteins to regulatory elements in the 3' untranslated region (3' UTR) of mRNA can facilitate repression of mRNA translation, and evidence suggests that several mRNAs that can be regulated in this way are controlled via a common messenger ribonucleoprotein (mRNP) architecture. The core repression complex consists of proteins that mediate an interaction between the 3' UTR-binding proteins and an mRNA 5' cap-binding protein, either the translation initiation factor eIF4E or a mimic. This arrangement sequesters the mRNA 5' cap in a nonproductive closed-loop conformation and inhibits an obligatory step in translational initiation. An example of this mode of control was presented by Paul Lasko (McGill University, Montréal, Canada), who described the repression of two mRNAs, hunchback and caudal, that are important for axis formation in the Drosophila embryo. An important point to note is that although the recognition elements within the two mRNAs bind distinct proteins, the respective mRNP complexes both lead to repression of translation through an interaction with a unique mRNA cap-binding protein, 4EHP.
Claudia Bagni (University of Rome, Italy) reported that the specificity of translational repression of target mRNAs by the fragile X mental retardation protein (FMRP) in neuron dendrites is achieved not through accessory proteins but through the noncoding RNA, BC1. BC1 acts as a bridging molecule for FMRP, which in turn interacts with the novel neuronal eIF4E-binding protein, Cyf1p. Binding of eIF4E by Cyf1p is believed to underlie a cap-dependent mechanism of translational repression that is important for properly regulated protein synthesis at the synapse and for learning and memory. While sequestration in cis of the mRNA 5' cap has emerged as a fundamental mechanism for preventing translation, it remains to be seen whether differences in composition of the mRNPs are due simply to tissue specificity or whether they provide additional complexity or plasticity to the regulatory event.
MicroRNAs, translational repression, and mRNP aggregates
MicroRNAs (miRNAs) have emerged in recent years as mRNA-specific regulators of translation. Witold Filipowicz (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) discussed evidence that repression of mRNA translation by miRNA is reversible. He presented data from human cell culture studies showing that miR122 represses translation of the arginine/lysine transporter CAT-1 mRNA and enriches the localization of the target mRNA in P bodies, which are cytoplasmic mRNP granules believed to be sites of mRNA storage and/or mRNA degradation. Under conditions of amino acid deprivation or other stress, repression is relieved, and the mRNA re-enters active translation, concomitant with the loss of target mRNA co-localization with P bodies, an effect dependent on the RNA-binding protein HuR. The observation that an mRNA can cycle between repressed and translationally active states, and that this cycling may involve a spatial segregation within the cell cytoplasm, was supported by Roy Parker (University of Arizona, Tucson, USA). His results from yeast studies support the hypothesis that all cellular mRNAs cycle through P bodies, and that this movement represents a fundamental aspect of the mRNA life cycle that has been previously unrecognized but which is important for regulation of mRNA metabolism, including translational control.
The elucidation of a role for P bodies in the control of mRNA translation by miRNA will benefit from new insight that may result from studies presented by Rolf Thermann (EMBL, Heidelberg, Germany). He described an in vitro experimental system to evaluate miRNA-mediated translational control using Drosophila embryo extracts, and presented evidence that this translational repression resulted in a dramatic reduction in the association of 48S translational initiation complex with the mRNA, but an accumulation of mRNP complexes with sedimentation coefficients greater than 80S. These findings are consistent with the hypothesis that the aggregation of mRNAs into large mRNP complexes, which may correspond to P bodies or other cytoplasmic foci, is an important mechanism in mRNA translational repression.
Polyadenylation and translational activation
The relative level of polyadenylation at the 3' end of the message has great impact on the translational status of an mRNA. During early embryonic development, the translation of specific maternal mRNAs is activated through the addition, in the cytoplasm, of a poly(A) tail, and the specificity of this polyadenylation has been ascribed to cis-acting elements in the RNA. Eulalia Belloc (Centre de Regulacio Genomica, Barcelona, Spain) described how mRNA elements that mediate polyadenylation (cytoplasmic polyadenylation elements) can be found alongside recognition elements that lead to mRNA deadenylation (AU-rich elements). The relative arrangement of these elements may have an impact on the polyadenylation profile of the mRNA. These findings suggest that combinatorial binding of proteins to these elements mediates complex and potentially reversible changes to the mRNA poly(A) tail and the translation status of mRNA. Joel Richter (University of Massachusetts Medical School, Worcester, USA) discussed a means of temporal control of mRNA translation through changes in poly(A) tail length by the restructuring of a 3' UTR mRNP. He suggested that, in response to phosphorylation of the cytoplasmic polyadenylation element binding protein (CPEB), poly(A) ribonuclease becomes displaced from the multiprotein mRNP, thereby facilitating poly(A) addition catalyzed by another complex member, Gld-2, a noncanonical poly(A) polymerase.
Using polyadenylation state array (PASTA) analysis to separate and identify mRNA populations from Saccharomyces cerevisiae on the basis of poly(A) tail length, Thomas Preiss (Victor Change Cardiac Research Institute, Sydney, Australia) reported positive correlations between poly(A) tail length and ribosome density (representing active translation), and the possible co-regulation of functionally related mRNAs through poly(A) tail status. Interestingly, an overall correlation between poly(A) tail length and mRNA stability was not observed; however, the association of poly(A)-binding protein (PAB) with the mRNA was shown to correlate with tail status. Supporting the popular theory that PAB association with mRNA, rather than the poly(A) tail itself, acts as the overall modulator of translation, Glover Martin (Massachusetts Institute of Technology, Cambridge, USA) described the efficient expression of a non-adenylated mRNA from dengue virus through binding of PAB to the mRNA 3' UTR despite absence of a poly (A) tail.
Cell signals and translational control of mRNA subpopulations
Under certain cellular conditions, the regulation of large numbers of different mRNAs may be necessary to respond to stimuli or maintain homeostasis. David Stokoe (University of California, San Francisco, USA) described the identification of a class of cellular mRNA targets whose translation is boosted by the mammalian target of rapamycin (mTORC1) signaling pathway, an important regulator of cell growth that acts by stimulating the initiation of translation. This analysis capitalized on the observation that, in conditions of limiting growth factors or nutrient withdrawal, protein synthesis from this class of mRNAs is prevented through the inhibitory action of two mTORC1 pathway members, tuberous sclerosis complex (TSC) proteins 1 and 2. Mutant cells defective in TSC function cannot prevent target mRNA translation in such conditions, and Stokoe carried out a microarray analysis comparing RNA isolated from polysome and non-polysome fractions from wild-type and Tsc1- or Tsc2-deficient mouse embryo fibroblasts in conditions of serum withdrawal. The results show that translation from mRNAs harboring a 5'-terminal oligopyrimidine (TOP) tract within their 5' UTRs is normally dramatically up-regulated in wild-type cells. Stokoe also showed that the 5' UTRs of many of the regulated mRNAs were sufficient to confer translational control on reporter genes in cis. The observation that TOP-containing mRNAs encode predominantly ribosomal proteins and components of the translation machinery suggests that this mechanism of mRNA class-specific modulation of translation in response to environmental changes can control global protein synthesis. Indeed, Oded Meyuhas (Hebrew University-Hadassah Medical School, Jerusalem, Israel) discussed the hypothesis that the unrestrained cell proliferation associated with a deficiency in TSC1 or TSC2 in cultured mammalian cells may result from the derepression of TOP mRNA translation.
In regard to the role of miRNA in controlling translation, Ulf Ørom (University of Copenhagen, Denmark) reported that miR10a can interact with the 5' UTR of mRNAs harboring the TOP regulatory motif and impact, both negatively and positively, on their translation. As well as showing that a miRNA may play a role in enhancing mRNA translation, these findings also suggest that miRNAs might be able to affect global protein production by regulating the levels of the translational apparatus within the cell.
It is generally accepted that mRNA translation is down-regulated at the onset of mitosis. Current hypotheses for the underlying mechanism propose that it is the initiation of translation that is inhibited. Gilad Sivan (Tel-Aviv University, Israel) reported, in contrast, that in cells whose cycle is synchronized in the absence of drugs, the broad decrease in mRNA expression at mitosis is associated with a persisting association of mRNA with ribosomes. This indicates that control of mRNA translation during mitosis can occur at multiple points, including inhibition of ribosome elongation or translation termination.
Control of global mRNA translation by alterations in the translation apparatus
The translation of mRNA can be repressed globally in response to cellular perturbations, either as a mechanism for short-term adaptation or to ensure long-term survival. One conserved mechanism involves phosphorylation of the initiation factor eIF2α, which inhibits ternary complex formation and precludes translation of the majority of mRNAs in the cell (that is, those requiring the 5' cap for their expression and many harboring internal ribsome entry sites (IRES)), and numerous protein kinases specifically target eIF2α at differing cell stages or levels of stress. Douglas Cavener (Pennsylvania State University, University Park, USA) and Donalyn Scheuner (University of Michigan, Ann Arbor, USA) both discussed the importance of eIF2α phosphorylation by pancreatic endoplasmic reticulum eIF2α kinase (PERK) in maintaining insulin-secreting beta-cell function. Using a series of tissue- and cell-specific Perk knockout mice, Cavener presented data suggesting that defects in beta-cell differentiation and proliferation during embryonic development underlie the deficiency in glucose-stimulated insulin secretion and proinsulin trafficking in postnatal mice. In contrast, Scheuner presented data from mice harboring homozygous mutations at the regulatory phosphorylation site of eIF2α, indicating that, in the absence of eIF2α phosphorylation, beta-cells underwent apoptotic cell death. It therefore remains under debate how regulation of eIF2α by PERK prevents the symptoms of diabetes; however, the need to regulate global protein synthesis to prevent manifestations of human disease is certain. A role for eIF2α phosphorylation in behavioral learning was described by Mauro Costa-Mattioli (McGill University, Montreal, Canada) in experiments with mice in which the phosphorylation status of eIF2α was modulated. Mutations that decrease eIF2α phosphorylation led to enhanced long-term memory, whereas treatment with a drug that prevents eIF2α dephosphorylation impaired long-term memory. Together, these studies highlight eIF2α and global translational control as key effectors of cellular function.
A wide range of cellular stresses lead to the dampening of overall protein synthesis through the functional activation, by dephosphorylation, of the inhibitory eIF4E-binding protein 4E-BP1. In contrast, modification of eIF4E once it is incorporated into the eIF4F translational initiation complex leads to both positive and negative effects on cap-dependent mRNA translation. Carolina Arias (New York University School of Medicine, New York, USA) unveiled a biological role for the 'nonessential' phosphorylation of eIF4E catalyzed by the protein kinases Mnk1 and Mnk2, which are activated by the mitogen-activated protein kinase (MAPK) pathway. When stressed by infection with vaccinia virus, mouse cells deficient in Mnk1 displayed reduced eIF4E phosphorylation and impairment of virus replication and protein synthesis, despite the presence of Mnk2.
An emerging but poorly understood control point for the regulation of mRNA translation is through modification of the ribosome itself. Chemical modifications of ribosomal proteins or rRNA, or changes in the association of ribosomal or nonribosomal proteins, are postulated to lead to adjustments in the affinity of the ribosome for particular classes of mRNAs. Vivian MacKay (University of Washington, Seattle, USA) discussed a genetic screen in Saccharomyces cerevisiae that revealed a relationship between the protein composition of the 60S ribosomal subunit and aging. Deletion of any of several genes encoding proteins of the large ribosomal subunit led to a significantly longer life span, and deletion of various genes involved in 60S processing or maturation also resulted in longer lived cells. Further research will be needed to understand this intriguing observation.
To evaluate whether ribosome modifications have a role in adjusting translation, Camille Diges (Stanford University School of Medicine, Palo Alto, USA) presented mass spectrophotometric analysis of ribosomes purified from uninfected and poliovirus-infected HeLa cells. Global changes to the posttranslational modifications of the ribosomal proteins were observed, including an overall gain in ribosomal subunit di-methylation and either a decrease or alteration in protein acetylation in 40S and 60S subunits, respectively, upon viral infection. On the basis of these preliminary observations, Diges proposes that post-translational modification of the ribosome fine tunes the binding of non-ribosomal proteins and, in turn, modulates the affinity of the ribosome for specific mRNAs, thereby enabling the ribosome to act directly as a regulator of translation. It remains unclear, however, if the observed changes in ribosome modifications described by Diges do indeed lead to increased efficiency of viral RNA translation; further experiments will be needed to validate this exciting and novel form of translational control.
Further elucidation of the molecular events underlying the regulation of mRNA translation, the discovery of new mechanisms, and the identification of the mRNA targets at each control point, will serve to complement our already broad understanding of translational control within the cell. It is already clear that careful balancing of protein synthesis is essential for critical processes such as embryonic development and cellular adaptation, and learning and memory.


