Abstract
The conserved central and COOH-terminal regions of troponin T (TnT) interacts with troponin C, troponin I and tropomyosin to regulate striated muscle contraction. Phylogenic data show that the NH2-terminal region has evolved as an addition to the conserved core structure of TnT. This NH2-terminal region does not bind other thin filament proteins and its sequence is hypervariable among fiber type and developmental isoforms. Previous studies have demonstrated that NH2-terminal modifications alter the COOH-terminal conformation of TnT and thin filament Ca2+-activation, yet the functional core structure of TnT and the mechanism of NH2-terminal modulation are not well understood. To define the TnT core structure and investigate the regulatory role of the NH2-terminal variable region, we investigated two classes of model TnT molecules: 1) NH2-terminal truncated cardiac TnT. 2) Chimera proteins consisting of an acidic or basic skeletal muscle TnT NH2-terminus spliced to the cardiac TnT core. Deletion of the TnT hypervariable NH2-terminus preserved binding to troponin I and tropomyosin and sustained cardiac muscle contraction in the heart of transgenic mice. Further deletion of the conserved central region diminished binding to tropomyosin. The re-introduction of differently charged NH2-terminal domains in the chimeric molecules produced long-range conformational changes in the central and COOH-terminal regions to alter troponin I and tropomyosin bindings. Similar NH2-terminal charge effects are demonstrated in naturally occurring cardiac TnT isoforms indicating a physiological significance. These results suggest that the hypervariable NH2-terminal region modulates the conformation and function of the TnT core structure to fine-tune muscle contractility.
Keywords: troponin T core structure, NH2-terminal variable region, long range conformational effect, solid phase protein binding assay, muscle thin filament regulation
The contraction of striated muscle is regulated by Ca2+ via the troponin (Tn) complex that is composed of three subunits: troponin C (TnC, the calcium binding subunit), troponin I (TnI, the inhibitory subunit), and troponin T (TnT, the tropomyosin (Tm)-binding subunit). During the activation of contraction, increased cytosolic Ca2+ binds to TnC to induce a series of conformational changes in Tn and Tm that “opens” the muscle thin filament to allow the interaction of myosin with actin and cross bridge cycling (1). Through direct binding to TnC, TnI, Tm and actin, TnT anchors the Tn complex to the thin filament and transmits the Ca2+ induced conformational changes from Tn to Tm (2–4).
Three homologous TnT genes have evolved in vertebrates to encode fast skeletal, slow skeletal and cardiac muscle TnT isoforms (5–8). These TnT isoforms have highly conserved amino acid sequences in the COOH-terminal region containing binding sites for TnC, TnI and Tm (9–11). The central region of TnT is also highly conserved and contains a second Tm-binding site (2). The NH2-terminal region of the TnT polypeptide chain does not contain any known protein binding sites and phylogenic data show that this region has evolved as an addition to the conserved core structure of TnT (12). In contrast to the central and COOH-terminal regions, the NH2-terminal region of TnT is highly variable among the three muscle fiber type isoforms (13) and is alternatively spliced for additional length and charge variations (2, 14).
The crystal structures of partial cardiac and skeletal Tn complex have revealed the molecular interactions that occur in the globular domain of Tn (15, 16). In agreement with protein binding data (9–11), the high resolution Tn structures demonstrate interactions between the TnT COOH-terminal domain and TnI and TnC. The partial crystal structure of Tn does not include the central region of TnT or the NH2-terminal variable region. This is consistent with the previous observation that TnT is an elongated molecule with its central and NH2-terminal regions forming the asymmetric tail portion of the Tn complex (17–19).
Phylogenic lineage of TnT shows a short to long, simple to complex evolution of the NH2-terminal variable region (12). Fast skeletal muscle TnT with the NH2-terminal variable region removed retains activity in the calcium activation of actomyosin ATPase (20–22). Similarly, the exchange of NH2-terminal truncated cardiac TnT into myofibrils retains calcium activated force development (23). On the other hand, removing the NH2-terminal region from both skeletal and cardiac TnT decreased the maximal force (20, 23) and increased the binding affinity to Tm compared to that of the intact TnT molecule (20, 23, 24). Further suggesting a regulatory function of the TnT NH2-terminal variation, alternative splicing within the NH2-terminal region is responsible for the developmental isoform switches of both cardiac and fast skeletal muscle TnT (14). We have previously demonstrated that structural alterations within the TnT NH2-terminal region alters the molecular conformation and flexibility of the central and COOH-terminal regions of TnT to affect the binding affinity to TnI, TnC and Tm (19, 25, 26), Ca2+ activation of ATPase activity (27), shortening velocity of isolated cardiomyocytes (28), and Ca2+ sensitivity of force development (29). Its variable structure and the resulting functional effects suggest a hypothesis that the NH2-terminal region of TnT is a modulatory addition to the TnT central and COOH-terminal core structure, which functions to fine-tune muscle contractility.
There are three variables in the TnT NH2-terminal structure: Amino acid sequence, length (mass) and charge. The NH2-terminal sequence of TnT isoforms shows very little conservation and therefore provides little information for a functional correlation. Furthermore, the size of vertebrate TnT varies by as many as 70 amino acid residues, almost completely resulting from variations in the length of the NH2-terminal region. A striking observation is that although the sizes of embryonic and adult fast skeletal TnT isoforms overlap, they fall into two distinct and non-overlapping acidic and basic charge classes, resulting from developmentally regulated alternative splicing of 6 exons that encode the NH2-terminal region (14, 26). Therefore, charge variation may be a determinant for the TnT hypervariable NH2-terminal region to modulate the conformation and function of the core structure.
In the present study, we investigated representative model proteins to define the TnT core structure and investigate the regulatory effects of NH2-terminal charge. The results demonstrate preserved biochemical and physiological activity of the TnT core corresponding to the conserved central and COOH-terminal regions. The addition of differently charged exogenous NH2-terminal segments alters the molecular conformation of the TnT core structure to affect the interactions with TnI and Tm. Similar NH2-terminal effects are demonstrated in naturally occurring cardiac TnT isoforms, suggesting that the NH2-terminal variable region plays a role in modulating the conformation and function of the TnT core to fine-tune muscle contractility.
EXPERIEMNTAL PROCEDURES
Construction of cDNAs encoding NH2-terminal truncated and chimeric TnT proteins
cDNAs encoding two cTnT proteins with different NH2-terminal truncations were designed to remove either only the NH2-terminal variable region (McTnT-ND72) or extend the deletion to include a portion of the conserved central region (McTnT-ND91). The two cDNA coding templates were constructed using polymerase chain reaction (PCR) to place a methionine codon prior to leucine 72 (McTnT-ND72) or aspartic acid 91 (McTnT-ND91) of the adult mouse cardiac TnT-4 ((30), Fig. 1). The truncated cDNA was amplified from an intact adult mouse cardiac TnT cDNA cloned in pBluescript SK(−) (31) using mutagenesis primers (McTnT-ND-F72 containing an ATG codon prior to the codon for Leu72 and an NdeI restriction endonuclease cloning site (5’-AGCCCCATATGCTCTTCATGCCCAACTT-3’)) or (McTnT-ND-F91 containing an ATG codon prior to the codon for Asp91 and an NdeI cloning site (5’- GACCATATGGACATCCACAGGAA -3’)) paired with a vector primer next to the cDNA 3’ end. The resultant PCR fragments were modified by NdeI and XhoI at the ends and cloned into the pAED4 prokaryotic expression plasmid (32). Purified plasmid DNA was verified by dideoxy chain termination sequencing.
Figure 1. TnT molecules studied.
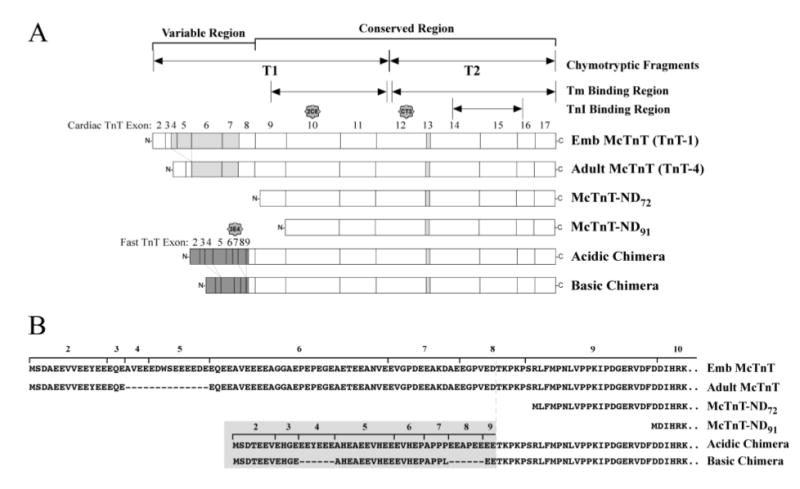
(A) Linear maps show the two naturally occurring mouse cardiac TnT isoforms and the engineered model TnT molecules used in this study. The adult mouse cardiac TnT (TnT-4) differs from the embryonic cardiac TnT (TnT-1) by the exclusion of exons 4 and 5 from the NH2-terminal variable region. The truncated mouse cardiac TnT molecules deleted only the NH2-terminal variable region encoded by exons 2-8 (McTnT-ND72) or further including a conserved segment encoded by exon 9 (McTnT-ND91). The two chimeric TnT molecules are identical to the mouse cardiac TnT in their central and COOH-terminal regions (exons 9-17) but contain the NH2-terminal domain of an acidic chicken fast skeletal TnT (encoded by exons 2-8 from chicken fast skeletal TnT-3) (Acidic chimera) or a basic chicken fast skeletal TnT (encoded by exons 2-3, 5-7 from chicken fast skeletal TnT-4) (Basic chimera). The TnI binding region, tropomyosin binding regions, T1 and T2 chymotryptic fragments and the mAb 3E4, CT3 and 2C8 epitopes are designated. Light grey exons indicate alternately spliced exons in cardiac TnT; dark grey exons indicate chicken fast skeletal muscle TnT exons. (B) The NH2-terminal amino acid sequences of the six TnT molecules are aligned to demonstrate the structural relationships. Residues originated from chicken fast skeletal TnT are highlighted in grey.
The most variable NH2-terminal charge exists among alternative spliced fast skeletal muscle TnT isoforms (33, 34). The isoelectric point (pI) of embryonic versus adult mouse fast skeletal muscle TnT isoforms ranges from 5.04 to 9.19 (33). To build model molecules for testing the NH2-terminal charge effect, two recombinant chimera cDNAs were constructed by joining the 5’ variable region of embryonic or adult chicken fast skeletal muscle TnT cDNA (fsTnT-3 or fsTnT-4; respectively (35)) to cDNA encoding the conserved core structure of mouse cardiac TnT. As illustrated in Fig. 1, the Acidic chimeric TnT consists of chicken fast skeletal TnT exons 2-8 spliced to mouse cardiac TnT exons 9–17. The Basic chimeric TnT consists of chicken fast skeletal muscle TnT exons 2, 3, 5, 6 and 7 spliced to mouse cardiac TnT exons 9–17.
The chimeric cDNA was constructed using a three-step recombinant PCR method. First, the chicken NH2-terminal region containing exons 2–8 of TnT-3 or exons 2–3 and 5–7 of TnT-4 was amplified from intact cDNA templates cloned in M13mp18 phagemid (kindly provided by Dr. Larry Smillie, University of Alberta (35)). The PCR used primer CfTnTe7R (5’-GGGTTTGGTCTCCTCTGGAGG -3’) with the 3’ region complimentary to the exon 7 of chicken fsTnT and the 5’ region complimentary to the exon 9 of mouse cardiac TnT paired with a vector primer M13R. In a separate PCR reaction, the exon 9-17 region of mouse cardiac TnT cDNA was amplified from intact TnT-4 cDNA cloned in pBluescript SK(−) (31) using primer McTnTe7F (5’-AGAGGAGACCAAACCCAAGCCCA-3’) with the 3’ region complimentary to the exon 9 of mouse cardiac TnT cDNA and the 5’ region complimentary to chicken fsTnT paired with a vector primer T7. The first PCR products were purified, either the chicken fsTnT-3 or the chicken fsTnT-4 fragment was mixed with the mouse cardiac TnT fragment and hybrid cDNA templates produced by 5 cycles of denaturation-annealing and chain elongation under standard PCR conditions. Recombinant full length cDNAs were then amplified by PCR using the flanking M13R and T7 primers and cloned into pBluescript KS(+) vector. Plasmid DNA was purified and sequenced to verify the construction of cDNA chimeras before subcloning the insert into the pAED4 expression vector as described above.
Two alternatively spliced cardiac TnT isoforms with the largest possible natural charge difference (the TnT-4 adult isoform and the TnT-1 embryonic isoform (31)) were chosen as a pair to study the naturally occurring NH2-terminal charge effect. These previously cloned adult and embryonic mouse cardiac TnT cDNAs (31) were subcloned into pAED4 vector as described above.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Protein samples were mixed with 3x SDS-PAGE sample buffer to a final concentration of 2% SDS, 0.3% bromphenol blue, 10% glycerol, 50 mM Tris-HCl, pH 6.8, heated to 80 °C for 5 min and clarified by centrifugation. Separation of the McTnT-ND proteins was carried out by electrophoresis on a 14% gel with an acrylamide:bisacrylamide ratio of 120:1 in the Tris-Tricine buffer system as previously described (32). All other protein bands were resolved on a 14% gel with an acrylamide:bisacrylamide ratio of 180:1 in the Laemmli buffer system. Resulting gels were stained with Coomassie Blue R250 to reveal the protein bands and duplicate gels transferred to a nitrocellulose membrane. The membrane was blocked with bovine serum albumin (BSA) and incubated with anti-cardiac TnT mAbs CT3 (25), 2C8 (36), or an anti-chicken fast skeletal TnT mAb 3E4 (26). Following washes with Tris-buffered saline (TBS, 136.9 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl, pH 7.4) containing 0.5% Triton X-100 and 0.05% SDS, the membranes were incubated with alkaline phosphatase-labeled anti-mouse IgG second antibody (Sigma Chemical Co., St. Louis, MO), washed as above and developed with 5-bromo-4-chloro-3-indolylphosphate and nitro blue tetrazolium substrate solution as previously described (26).
Bacterial expression and purification of TnT
The six TnT cDNA constructs described above were expressed in BL21(DE3)pLysS E. coli. Freshly transformed bacterial cells were cultured in liquid media (16 g/L Tryptone, 10 g/L yeast extract, 5 g/L NaCl, 1.32 g/L Na2HPO4, pH 7.3) containing 100 mg/L ampicillin and 25 mg/L chloramphenicol at 37 °C with vigorous shaking and induced with 0.4 mM isopropyl-1-thiol-β-D-glactoside at mid-log phase. After 3 additional hours of culture the bacterial cells were harvested by centrifugation at 4 °C.
The McTnT-ND72 and McTnT-ND91 proteins were purified by identical procedures. The bacterial pellet was suspended in 2.5 mM EDTA, 50 mM tris-HCl, pH 8.0 and lysed by three passes through a French Press cell. The bacterial lysate was clarified by centrifugation and precipitated with ammonium sulfate to obtain the 0–35% saturation fraction. Following dialysis against 0.1 mM EDTA containing 6 mM β-mercaptoethanol, the fraction was brought to 6 M urea, 0.1 mM EDTA, 6 mM β-mercaptoethanol, 20 mM sodium acetate, pH 6.0 and fractionated by chromatography on a CM52 cation-exchange column equilibrated in the same buffer. The column was eluted by a 0–500 mM linear KCl gradient and the protein peaks analyzed by SDS-PAGE. Fractions containing the NH2-terminal truncated cardiac TnT were dialyzed against 0.1 mM EDTA containing 6 mM β-mercaptoethanol, concentrated by lyophilization, re-dissolved in 6 M urea, 0.5 M KCl, 0.1 mM EDTA, 6 mM β-mercaptoethanol, 10 mM imidazole-HCl, pH 7.0, and fractionated by G75 gel filtration chromatography in the same buffer. Protein peaks were analyzed by SDS-PAGE and the fractions containing pure McTnT-ND72 or McTnT-ND91 protein were dialyzed against 0.1% formic acid and lyophilized. All purification steps were carried out at 4°C.
The Basic chimeric TnT was purified as above for the NH2-terminal truncated cardiac TnT with the following modifications. The bacterial lysate was fractionated by (NH4)2SO4 to collect the 30–50% saturation precipitation. After dialysis, the proteins were fractionated by chromatography on a CM52 column at pH 4.8 (20 mM sodium acetate buffer). The column was eluted by a 0–500 mM linear KCl gradient and the protein peaks analyzed by SDS-PAGE. Fractions containing the Basic chimeric TnT were further purified by G75 chromatography.
The Acidic chimeric TnT was purified similarly except that the 30–50% saturation (NH4)2SO4 fraction was fractionated by chromatography on a DE-52 anion-exchange column at pH 8.5 (20 mM Tris-HCl buiffer) with a 0–300 mM linear KCl gradient. The eluted protein peaks were analyzed by SDS-PAGE and fractions containing the Acidic chimeric TnT were further purified by G75 chromatography.
The embryonic and adult mouse cardiac TnTs were purified by identical procedures. The bacterial pellet was made into acetone powder and extracted by stirring with a buffer containing 6 M urea, 0.1 mM EDTA, 6 mM β-mercaptoethanol, 20 mM Tris-HCl, pH 8.0 at 4 °C for 2 h. The protein extracts were fractionated by ammonium sulfate precipitation between 30–50% saturation. After dialysis, the proteins were fractionated on a DE52 column at pH 8.0 (20 mM Tris-HCl buffer) followed by G75 chromatography as above.
Purification of other myofilament proteins
Bovine cardiac TnI was purified from adult ventricular muscle as previously described (27). Rabbit α-tropomyosin was purified from adult cardiac muscle as previously described (37).
mAb epitope analysis
The binding affinity between an antibody and its antigenic epitope depends on a three dimensional structural fit. Enzyme-linked immunosorbant assay (ELISA) epitope analysis (26) was employed to examine conformational differences between the TnT molecules. mAb CT3 against an epitope in the central domain of cardiac TnT (25) and mAb 3E4 against an NH2-terminal epitope on chicken fast skeletal TnT (26) (Fig. 1), were used to monitor conformational changes that alter the antibody binding affinity. The Acidic or Basic chimeric TnT, mouse embryonic or adult cardiac TnT were dissolved in Buffer A (0.1 M KCl, 3 mM MgCl2, 10 mM PIPES, pH 7.0) and the protein concentration determined by UV absorbance according to the following formula: (E280) (mm−1 cm −1) = 1.4 (# tyrosine) + 5.6 (# tryptophan). Five μg/mL protein solutions were used at 100 μL/well to coat microtitering plates by incubation at 4 °C overnight. After removing unbound TnT by washing once with Buffer A containing 0.05% Tween-20 (Buffer T) and blocking the plates with 150 μL/well Buffer T containing 1% BSA at room temperature for 1.5 h, the immobilized TnT was incubated with 100 μL/well serial dilutions of CT3 or 3E4 mAb in Buffer T containing 0.1% BSA at room temperature for 2 h. Following three washes with Buffer T in a 10 min period to remove the unbound first antibody, the plates were incubated with 100 μL/well horseradish peroxidase-conjugated anti-mouse immunoglobulin second antibody (Sigma, St. Louis, MO) in Buffer T containing 0.1% BSA at room temperature for 1 h. Unbound second antibody was removed by three washes as above. The binding of mAbs to a particular TnT construct was detected by H2O2/2,2’-azinobis-(3-ethylbenzthiazolinesulfonic acid) substrate reaction. The enzymatic reaction in each assay well was monitored at a series of time points by an automated microplate reader (BioRad Benchmark, Hercules, CA). A405nm values in the linear course of the color development were used to plot the antibody titration curves for quantifying the binding affinity of the mAbs to the TnT epitopes. All experiments were done in triplet.
Microtiter plate protein binding assays
An ELISA-based solid phase protein binding assay (26) was used to investigate the interactions of the TnT variants with TnI and Tm. Similar to that described above for the epitope analysis, purified TnT or BSA control was dissolved in Buffer A at 5 μg/mL and 100 μL/well used to coat microtiter plates by incubation at 4°C overnight. Unbound TnT was removed by a wash with Buffer T and the plate blocked with 150 μL/well with Buffer T containing 1% BSA at room temperature for 1.5 h. The plates were then washed three times with Buffer T over a 10 min period and incubated with 100 μL/well of serially dilutions of bovine cardiac TnI or rabbit α-Tm in Buffer T containing 0.1% BSA. Following incubation at room temperature for 2 h, the plates were washes three times with Buffer T over a 10 min period. The bound TnI or Tm was quantified via an anti-TnI mAb TnI-1 (38) or an anti-Tm mAb CH1 (39) (a gift from Prof. Jim J.-C. Lin, University of Iowa) followed by standard ELISA procedure as described above. The A405nm values in the linear course of the color development were used to construct the protein binding curves for the quantification of binding affinity between the TnT variants to Tm and TnI under various washing conditions. All experiments were done in triplicate.
Immunoaffinity chromatographic isolation of Tn complex from mouse cardiac muscle
As described previously (40), McTnT-ND72 transgenic mice were constructed on C57BL/6 background using mouse cardiac α-myosin heavy chain (α-MHC) promoter ((41), generously provided by Dr. Jeffrey Robbins, University of Cincinnati) to direct a heart-specific expression of mouse cardiac TnT cDNA encoding cardiac TnT with the NH2-terminal variable region deleted to investigate the functional preservation in the TnT core structure. The pronucleus injection was performed at the Transgenic Core Facility at Case Western University and mouse line 14 was used in the present study. The expression of the exogenous cardiac TnT in the transgenic mouse hearts was verified by Western blots using the CT3 mAb. Another transgenic mouse line with α-MHC promoter-directed heart-specific expression of embryonic isoform of cardiac TnT in the adult cardiac muscle developed previously (28) was also used in the present study.
Troponin complex was isolated from transgenic mouse hearts by immunoaffinity chromatography similarly to that previously described (42). Troponin complex was isolated using an mAb TnI-1 against an epitope at the extreme COOH-terminus of TnI (38) that is exposed in the Tn complex providing a specific handle for the affinity purification. The TnI-1 mAb (IgG1) was purified from hybridoma ascites fluid using a Protein G column (Amersham-GE Healthcare Bio-Sciences Corp. Piscataway, NJ) and coupled to CNBr-activated Sepharose 4B (Amersham-GE Healthcare Bio-Sciences Corp. Piscataway, NJ) according to the manufacturer's protocols. The immunoaffinity isolation of Tn complex was then carried out using Sepharose 4B-TnI-1 mAb affinity column chromatography. Transgenic mouse ventricular muscle was minced into 1–2 mm3 pieces and extracted by 20 volumes (w/v) of Guba-Straub solution containing 300 mM KCl, 100 mM K2HPO4, 50 mM KH2PO4, 2.5 mM MgCl2, 1 mM EGTA, and 0.1 mM PMSF, pH 6.5 on ice for 15 m. After centrifugation at 16,000 x g at 4 °C for 20 min, the supernatant containing mainly myosin and cytosolic proteins was removed. The pellet was next extracted in 20 volume (w/v) of 1 M KCl, 10 mM Tris-HCl pH 8.0, 0.1 mM PMSF by stirring on ice for 30 min. After centrifugation as above, the extract was diluted 5-fold in TBS and loaded onto a 0.5 mL TnI-1 mAb affinity column equilibrated in TBS. The column was washed with TBS and then eluted with 50 mM glycine-HCl, pH 2.7. 0.5 mL fractions were collected into tubes containing 0.1 mL neutralizing buffer (1 M Tris-HCl, 1.5 M NaCl, 1 mM EDTA, pH 8.0) and analyzed by SDS-PAGE and Western blotting to identify the Tn peak as described above.
Incorporation of the NH2-terminal truncated cardiac TnT in the transgenic mouse cardiac Tn complex was compared with that of the intact adult and embryonic cardiac TnT by densitometric quantification of the SDS-gels and Western blots as described previously (28).
Data analysis
The DNA and protein sequence analyses were done using DNAStar computer programs. Statistical analysis of the ELISA antibody epitope analysis, protein binding assays and gel densitometry was done by Student’s t test. All values are presented as mean ± SD.
RESULTS
Preserved activity of the conserved core structure of TnT
To identify the TnT core structure required to maintain the protein-binding activity, we constructed two truncated cardiac TnT molecules with progressive NH2-terminal deletions. The deletion of the first 71 residues in the McTnT-ND72 molecule preserves the central and COOH-terminal conserved regions while the deletion of the first 90 residues in the McTnT-ND91 molecule results in a partial removal of the central conserved region (Fig. 1). The physical properties of the NH2-terminal truncated cardiac TnTs and that of the intact adult mouse cardiac TnT (TnT-4) are summarized in Table 1. The large-scale expression of the McTnT-ND72, McTnT-ND91 and adult mouse cardiac TnT in E. coli and the effective purification methods provided proteins for structural and functional characterizations (Fig. 2A). The authenticity of bacterial expressed McTnT-ND proteins was confirmed by Western blot (Fig. 2B).
Table 1.
Physical properties of the TnT proteins studied
| TnT | Number of Amino Acids | Molecular Weight | pI | NH2-terminal Charge at pH 7.0 |
|---|---|---|---|---|
| McTnT-ND72 | 221 | 26,859 | 9.87 | N/A |
| McTnT-ND91 | 201 | 24,578 | 10.00 | N/A |
| Emb McTnT (TnT-1) | 305 | 36,253 | 4.87 | −42 |
| Adult McTnT (TnT-4) | 291 | 34,546 | 5.19 | −32 |
| Acidic model TnT | 271 | 32,634 | 6.33 | −19 |
| Basic model TnT | 259 | 31,157 | 8.99 | −10 |
| Chicken Fast Sk TnT3 | 263 | 31,141 | 6.75 | −19 |
| Chicken Fast Sk TnT4 | 251 | 29,679 | 9.13 | −10 |
The physical properties of the NH2-terminal deleted mouse cardiac TnTs (McTnT-ND72 and McTnT-ND91), embryonic (Emb McTnT, TnT-1) and adult (Adult McTnT, TnT4) mouse cardiac TnTs (31), Acidic and Basic model TnT chimeras and chicken fast skeletal muscle TnT3 and TnT4 (accession number J04198) were calculated from amino acid sequences.
Figure 2. TnT protein preparations.
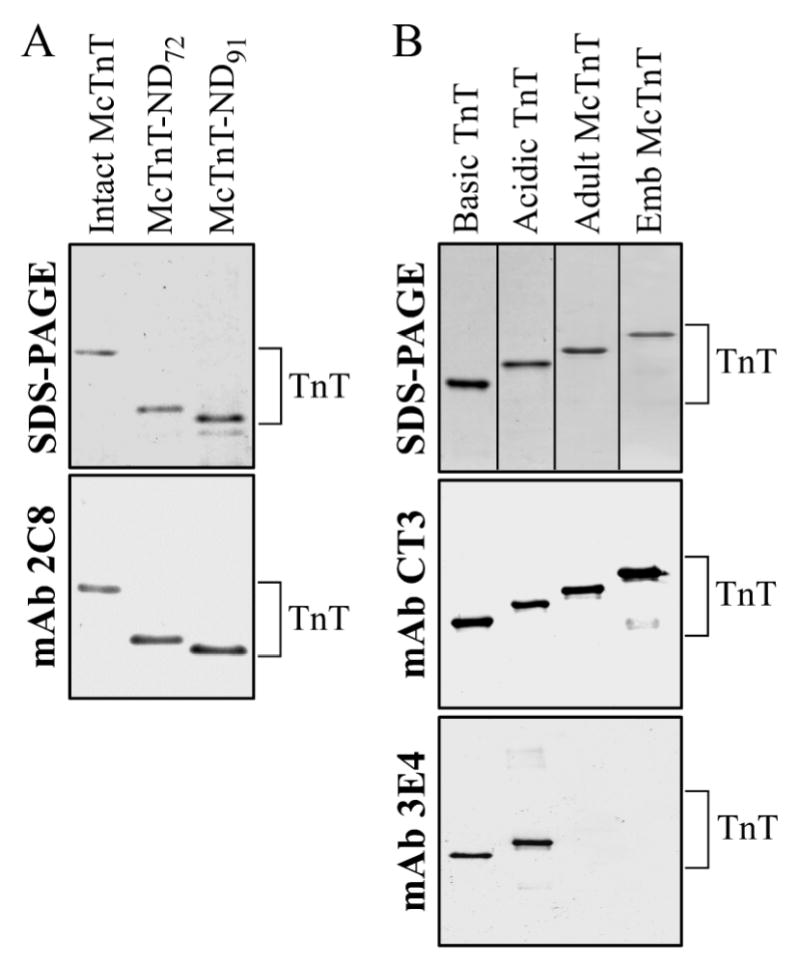
Recombinant TnT proteins were expressed in E. coli and purified as shown by the SDS-PAGE gels. (A) NH2-terminal truncated TnT (McTnT-ND72 and McTnT-ND91) were analyzed along with the intact adult mouse cardiac TnT-4 (Intact McTnT). The Western blot demonstrates that the truncated TnT proteins are positive to mAb 2C8 that recognizes an epitope in the exon 10 region (Fig. 1). (B) The Basic and Acidic chimeric TnTs were analyzed along with the adult mouse cardiac TnT-4 (Adult McTnT) and embryonic mouse cardiac TnT-1 (Emb McTnT). The Western blot demonstrates that the chimeric TnTs same as the native cardiac TnTs are positive to mAb CT3 recognizing a central cardiac TnT epitope. The chimeric TnTs are also positive to mAb 3E4 that recognizes an epitope within exon 7 of chicken fast skeletal muscle TnT. The results verified the construction of the NH2-terminal deleted and recombinant chimeric TnT molecules.
TnT contains two Tm binding sites, one is located in the COOH-terminal region and the other in the central region. To investigate the binding of the NH2-terminal truncated TnT constructs to Tm, we carried out ELISA solid phase protein binding experiments. The results in Fig. 3 demonstrate the binding affinity of McTnT-ND72 to Tm was not significantly affected compared to that of the intact mouse cardiac TnT, but McTnT-ND91 showed a decreased Tm-binding affinity (the Tm concentration required for 50% maximum binding was: Intact mouse cardiac = 19.4 ± 2.2 nM, McTnT-ND72 = 20.7 ± 3.1 nM and McTnT-ND91 = 39.4 ± 8.3 nM, P < 0.001). At concentrations of Tm that render saturated binding to comparable amounts of TnT coated on the plate, there was no significant difference in the maximal binding of Tm to the three constructs (Fig. 3 inset). The results showed that deletion of the entire variable region in McTnT-ND72 retained both Tm binding sites intact while further deleting 19 residues from the central conserved region in McTnT-ND91 resulted in decreased Tm binding, indicating a destruction of the TnT core structure.
Figure 3. Selective deletion of the NH2-terminal variable region from TnT does not abolish binding to Tm.
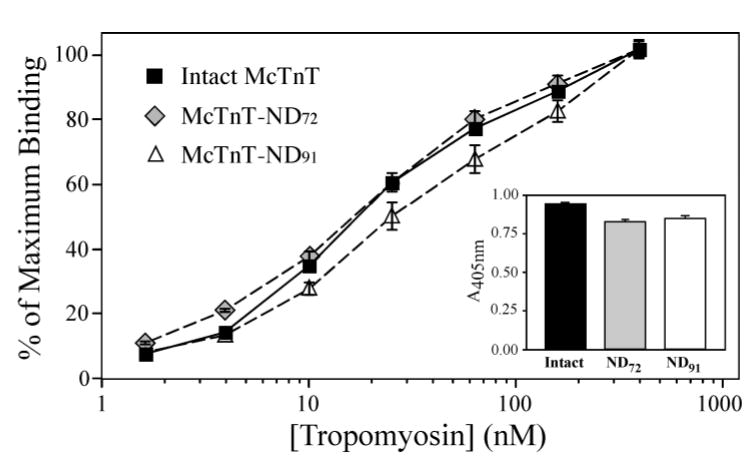
Solid-phase protein binding curves of intact mouse cardiac TnT, McTnT-ND72 and McTnT-ND91 to Tm demonstrate the effect of TnT NH2-terminal truncations. The binding affinity of McTnT-ND72 to Tm was similar to that of the intact McTnT while the Tm binding affinity of McTnT-ND91 was decreased (P<0.001). There was no significant change in the maximal binding of McTnT-ND72 and McTnT-ND91 to Tm (inset). In contrast to the preserved activity of McTnT-ND72 with only the NH2-terminal variable region deleted, the decreased binding of McTnT-ND91 to Tm indicates that deletion into the conserved region damages the TnT core structure by disrupting the central Tm binding site (Fig. 1).
Physical properties of the acidic and basic chimeric TnT model molecules
To evaluate the effect of NH2-terminal charge on the molecular conformation and protein binding activity of TnT, we constructed two chimeric model TnT molecules consisting of the NH2-terminal variable region of naturally occurring acidic (Acidic TnT) or basic (Basic TnT) chicken fast skeletal muscle TnT and the conserved central and COOH-terminal core regions of mouse cardiac TnT (Fig 1). The anticipated sequence combinations of the chimeric TnT were confirmed by Western blotting using mAb 3E4, specific for an epitope in the NH2-terminal domain of chicken fast TnT, and mAb CT3, against an epitope in the central region of cardiac TnT (Fig 2B). The physical properties of the chimeric model TnT molecules along with that of the chicken fast skeletal muscle TnT-3, TnT-4 and adult mouse cardiac TnT-4 are summarized in Table 1. In the presence of an identical central and COOH-terminal core structure, the NH2-terminal charge difference of 9 between the Acidic and Basic chimera produced an overall pI difference of 2.66, reproducing the NH2-terminal charge effect seen in the original chicken fast TnT isoforms (Table 1). These designs are consistent with the proposed universal role of the NH2-terminal variable region in regulating the overall charge of TnT.
NH2-terminal charge alters the core conformation in the TnT chimeras
The results of ELISA epitope analysis demonstrated the effect of NH2-terminal charge variation in the Basic and Acidic chimeric TnT on molecular conformation. Fig. 4A shows that the Basic TnT exhibited a higher binding affinity to mAb 3E4 than that observed for the Acidic TnT chimera. The mAb 3E4 recognizes an epitope encoded by exon 7 of chicken skeletal muscle TnT (Fig 1A). The different binding affinity of mAb 3E4 to the Basic and Acidic chimeric TnTs demonstrates the inclusion or exclusion of exon 4 and 8 segments affects local molecular conformation in the NH2-terminal domain. The Acidic chimeric TnT further exhibited a lower binding affinity to mAb CT3 when compared to that of the Basic chimeric TnT (Fig. 4B). The CT3 mAb recognizes an epitope in the central region of the mouse cardiac TnT core removed from the altered NH2-terminal region (Fig 1A). This result indicates a long-range conformational effect resulting from NH2-terminal TnT charge variation. The adult mouse cardiac TnT, with an NH2-terminus slightly more negatively charged than that of the Acidic chimeric TnT, confirmed the trend of lower binding affinity to mAb CT3 (Fig. 4B). These results demonstrate that changes in the NH2-terminal charge do not only affect the local conformation but also have long-range conformational effects on the core structure of TnT.
Figure 4. Alteration of NH2-terminal charge affects nearby and distal conformation in chimeric TnT model molecules.
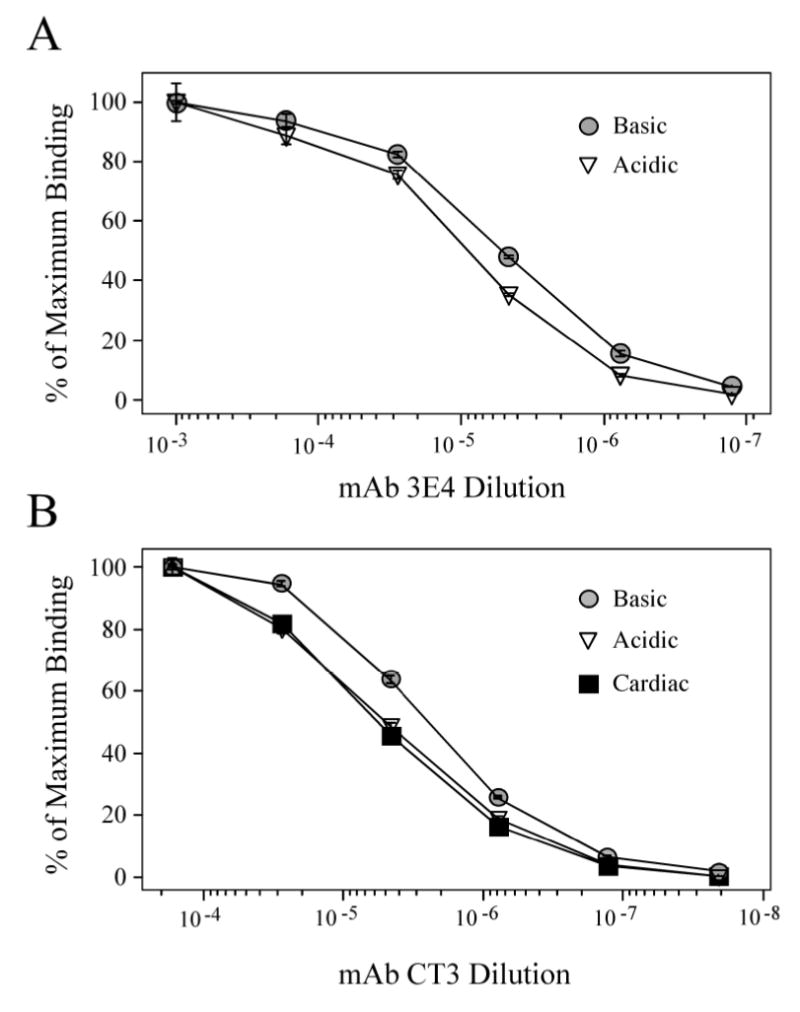
(A) ELISA epitope affinity titration curves of mAb 3E4 demonstrate that the Acidic and Basic chimeric TnTs exhibit a conformational difference at the 3E4 epitope near the NH2-terminal variation site, as defined by the difference in antibody concentrations required for 50% maximal binding. (B) Titration curves of mAb CT3 demonstrate that the Acidic and Basic chimeric TnTs and the native adult mouse cardiac TnT exhibit different conformation at the CT3 epitope in the central region (Fig. 1A), indicating remote conformational effect of the N-terminal variation.
NH2-terminal charge variation in TnT model molecules affects binding to TnI and Tm
To investigate the functional significance of the long-range conformational effects of NH2-terminal charge variation on the TnT core structure, ELISA protein binding experiments showed that the Basic chimera exhibits a higher binding affinity to TnI compared to that of the Acidic chimera (Figure 5A, the TnI concentrations required for 50% maximum binding were 7.0 ± 0.9 nM and 13.7 ± 0.8 nM, respectively; P < 0.05). Consistent with this trend, the binding affinity of the more acidic adult mouse cardiac TnT to TnI was even lower than that of Acidic chimeric TnT (the TnI concentration required for 50% maximum binding was 23.3 ± 1.4 nM; P < 0.05). At TnI concentrations that render saturated binding to comparable amounts of TnT coated on the microtiter plate, the maximal binding of TnI to the Basic chimeric TnT was also significantly greater than that of the Acidic chimera and the adult mouse cardiac TnT, (Fig. 5A inset; A405nm values are 1.13 ± 0.014, 0.96 ± 0.060 and 0.955 ± 0.010, respectively; P < 0.05), indicating a stronger coupling strength that may affect the function of the Tn complex. The results demonstrate a trend that the more basic the TnT NH2-terminus the stronger the binding of the TnT molecule to TnI.
Figure 5. NH2-terminal charge affects the binding of chimeric TnTs to TnI and Tm.
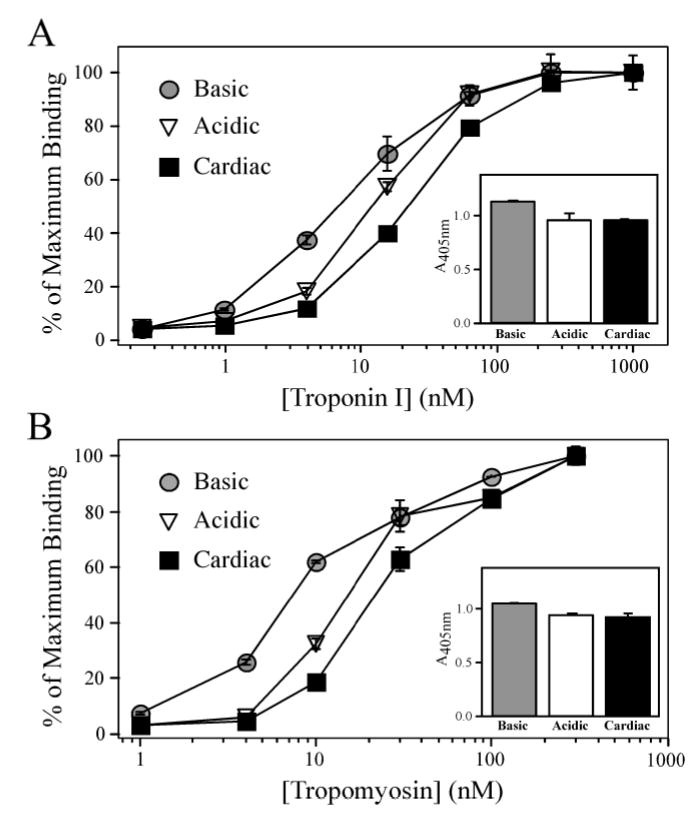
(A) Solid-phase protein binding curves demonstrate that the Basic TnT bound with higher affinity to TnI compared to that of the Acidic TnT (P < 0.05). Both Basic and Acidic chimeric TnTs exhibited higher binding affinities to TnI compared to the more acidic mouse cardiac TnT (P < 0.05). The A405nm values in the inset figure show that the Basic chimeric TnT also had a higher level of maximal binding to TnI compared to that of the Acidic chimeric TnT and mouse cardiac TnT (P < 0.05). (B) Solid-phase protein binding curves demonstrate that the Basic chimeric TnT exhibited a higher binding affinity to Tm compared to that of the Acidic chimeric TnT (P < 0.05), while the more acidic mouse cardiac TnT had the lowest binding affinity as defined by the concentration of Tm required for 50% maximum binding (P < 0.05). The A405nm values in the inset figure show that the Basic TnT exhibits a higher level of maximal binding to Tm compared to the bindings of Acidic TnT and mouse cardiac TnT (P < 0.05).
The binding of chimera TnT to Tm also exhibits a similar charge dependent trend. Figure 5B demonstrates the Tm concentrations required for 50% maximum binding to Basic and Acidic chimeric TnT were 7.7 ± 0.1 nM and 14.2 ± 1.8 nM, respectively (P < 0.05). Consistently, the binding affinity of the more acidic adult mouse cardiac TnT to Tm was even lower than that of the Acidic chimeric TnT (the Tm concentration required for 50% maximum binding was 23.5 ± 1.5 nM; P < 0.05). The maximum binding level of Basic chimeric TnT to Tm was greater than that of Acidic chimeric TnT and adult mouse cardiac TnT (Fig. 5B inset; the A405nm values are 1.04 ± 0.005, 0.93 ± 0.018 and 0.92 ± 0.031, respectively; P < 0.05), also indicating a higher coupling strength, which may affect the Tn-thin filament anchoring.
Native embryonic and adult mouse cardiac TnT isoforms exhibit conformational and functional differences
To evaluate if the long range conformational effects and altered protein interactions observed for the chimeric model TnT molecules are representative of the natural NH2-terminal charge variations occurring among TnT isoforms, we investigated embryonic and adult mouse cardiac TnT isoforms. The NH2-terminal charge of the embryonic and adult TnT isoforms differ as a result of the developmentally regulated alternative mRNA splicing of exon 5 segment (43) and the non-developmental alternative splicing of exon 4 (31) (TnT-1 and TnT-4, Fig. 1). The additive NH2-terminal charge effect of exons 4 and 5 segments imparts these two isoforms with the largest NH2-terminal charge difference (−10 in TnT-1 compared with TnT-4; Table 1) among the four cardiac TnT isoforms (44). In contrast to the chimeric TnT molecules in which 9 negative charges in the NH2-terminal region produced a pI difference of 2.66, in the two naturally occurring mouse cardiac TnT molecules the NH2-terminal charge difference of 10 only accounts for a pI difference of 0.32, due to the fact that NH2-terminal region of cardiac TnT is already highly negatively charged (Fig. 1; Table 1). ELISA epitope analysis using mAb CT3 against a central epitope shared by the two isoforms (Fig. 1) showed a higher binding affinity to the embryonic cardiac TnT than that to the adult isoform (Fig. 6). The response of CT3 epitope affinity to NH2-terminal variation here did not following the charge trend seen in the chimeric TnTs (Fig. 4) and may reflect other modulatory factors (see discussions below). Nonetheless, this result demonstrates that the naturally occurring NH2-terminal splicing alterations in TnT isoforms also have long-range conformational effects on the core structure, validating the observation from the chimeric model TnT molecules.
Figure 6. The alternative splicing-generated developmental isoforms of cardiac TnT differing in NH2-terminal charge show differences in molecular conformation.
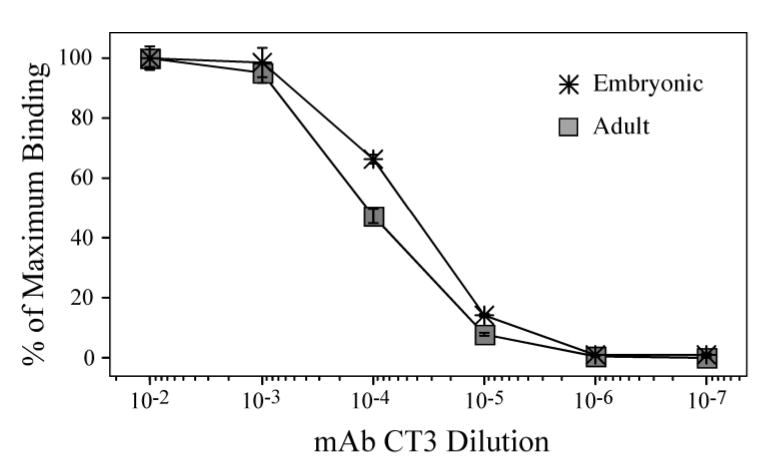
ELISA epitope affinity titration curves of mAb CT3 demonstrate that exclusion or inclusion of the exon 4 and 5 encoded segments from the adult and embryonic mouse cardiac TnT has long range conformational effects on the CT3 epitope located in the central region.
Corresponding to the conformational difference, the naturally occurring cardiac TnT isoforms also demonstrate altered interactions to TnI and Tm in the solid phase protein binding experiments (Fig. 7). The more acidic embryonic cardiac TnT exhibits a lower binding affinity to Tm compared to that of the less acidic adult cardiac TnT (Fig. 7A, the Tm concentration required for 50% maximum binding was 89.9 ± 14.1 nM and 22.3 ± 3.7 nM, respectively; P < 0.05). The embryonic cardiac TnT also exhibited a lower level of maximal binding to Tm compared to that of the adult isoform (Fig. 7A inset, A405 = 0.51 ± 0.027 and 0.81 ± 0.041, respectively; P < 0.05). These results were similar to those observed for the chimeric TnT, indicating that the more negative NH2-terminal charge produces a lower binding affinity and weaker coupling strength to Tm.
Figure 7. Alternative splicing-generated NH2-terminal charge difference in cardiac TnT affects binding affinity to Tm and TnI.
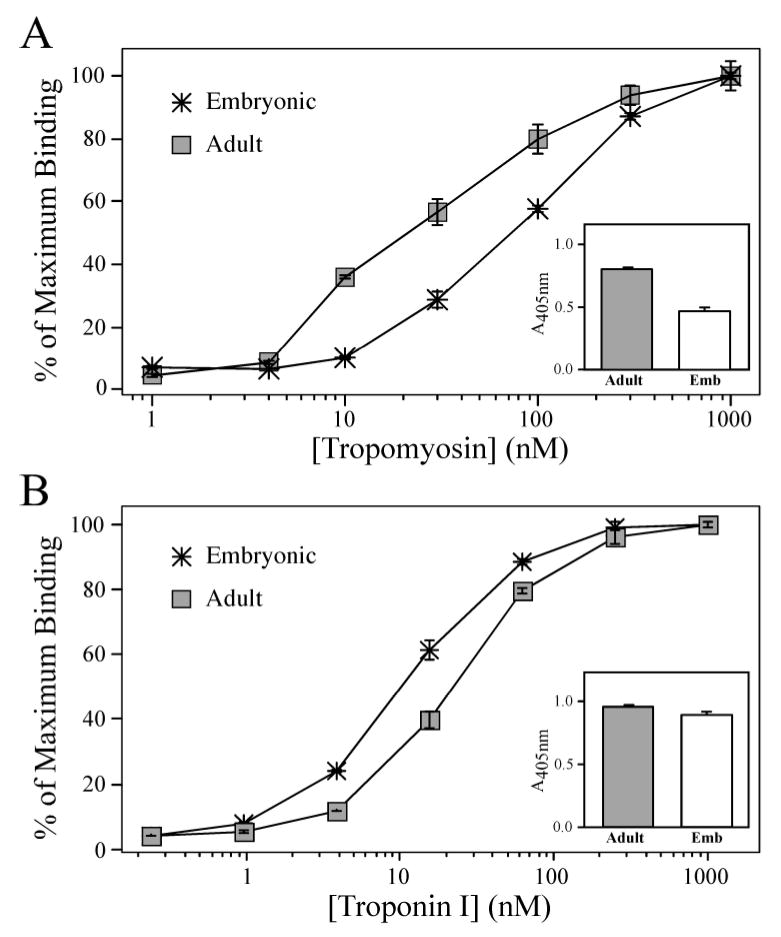
(A) Solid-phase protein binding curves demonstrate that the less acidic Adult mouse cardiac TnT immobilized to microtiter plate bound with a higher affinity to Tm than that of the more acidic Embryonic cardiac TnT, as determined by the concentration of Tm required for 50% maximum binding (P < 0.05). The Adult cardiac TnT also exhibited a higher level of maximal binding to Tm than that of the embryonic cardiac TnT (shown by the A405nm values in the inset). (B) However, the Adult cardiac TnT bound to TnI with a lower affinity compared to that of the Embryonic cardiac TnT (P < 0.05), while the A405nm values in the inset show the Adult cardiac TnT demonstrated a moderate but statistically significant higher maximal binding to TnI compared to that of the Embryonic cardiac TnT (P < 0.05).
Different from the trend of charge dependence observed in the chimeric TnTs, the more acidic embryonic mouse cardiac TnT bound to TnI with a greater affinity than that of the adult cardiac TnT (Fig. 7B, the TnI concentration required for 50% Maximum binding was; 11.4 ± 1.0 nM and 23.3 ± 1.4 nM, respectively; P < 0.05). It is worth noting that the conformational difference between mouse adult and embryonic TnTs at the CT3 epitope adjacent to the TnI-binding site (Fig. 1) also did not follow the charge trend (Fig. 6). In contrast, the maximum binding of the embryonic cardiac TnT to TnI was not higher but lower than that of the adult isoform (Fig. 7B inset, A405 values were; 0.89 ± 0.019 and 0.95 ± 0.010, respectively; P < 0.05) similar to the general trend of charge dependence. Although the data are consistent with a role for the long range effect of NH2-terminal charge on the COOH-terminal functional domain, the TnI binding behavior of the embryonic versus adult cardiac TnT isoforms suggests that the negative charge effect may have been maximized in the already highly acidic adult cardiac TnT and other factors, such as primary structure features, may further play a role in the modulatory function of TnT NH2-terminal domain. On the other hand, the binding of TnT to Tm involves an additional central site in contrast to the single COOH-terminal site binding to TnI (2). The closer proximity of the central Tm site to the NH2-terminal domain may confer a higher sensitivity of TnT-Tm interaction to TnT NH2-terminal charge variation (Fig. 7A). Nonetheless, the consistent charge effect in the cardiac TnT isoforms on the coupling strength to TnI (Fig. 7B inset) is more closely related to the function in the assembled Tn complex than the binding kinetics. Together with the Tm coupling strength (Fig. 7A), these results support the hypothesis that charge variation within the TnT NH2-terminus plays a role to fine-tune the function of the muscle thin filament.
The cardiac TnT core structure can form functional Tn complex
To verify that the proposed core structure of TnT indeed retains physiological function in vivo, we developed transgenic mouse lines that over-express the cardiac TnT core structure in the heart (Tg McTnT-ND72). In Figure 8A, SDS-PAGE and Western blots using the anti-cardiac TnT mAb CT3 demonstrate a high level of McTnT-ND72 in cardiac muscle of the transgenic mice. As shown in Figure 8B, densitometry of the Western blot demonstrates the cardiac TnT core was expressed as 49.5 ± 1.8% of the total cardiac TnT in the adult transgenic mouse ventricular muscle (Fig. 8B). All McTnT-ND72 transgenic mouse lines exhibit normal baseline life activity without apparent cardiac abnormalities. These transgenic mice expressing the cardiac TnT core in the heart muscle provide an integrated physiological system to evaluate the function of the TnT core structure in vivo.
Figure 8. Preserved myofilament incorporation and sustained in vivo function of the TnT core structure.
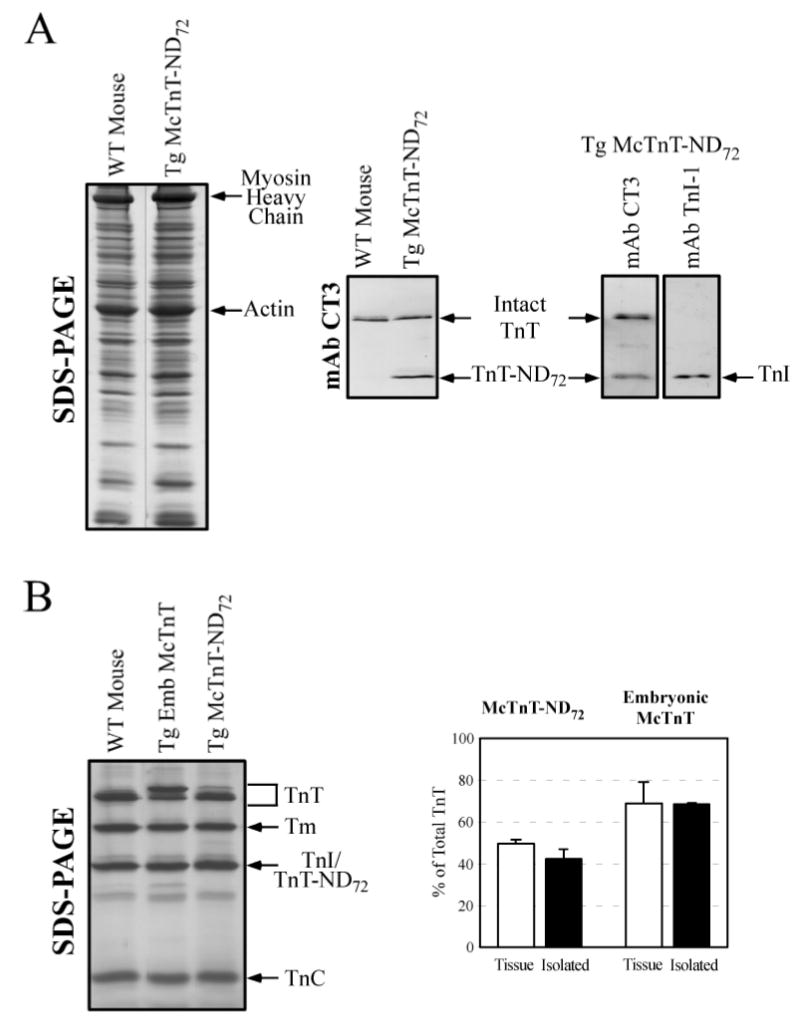
(A) SDS-PAGE and Western blot using the CT3 mAb demonstrates the high level expression of the cardiac TnT core in the heart of adult McTnT-ND72 transgenic mice. mAb TnI-1 Western blot shows that McTnT-ND72 co-migrates with the endogenous cardiac TnI. (B) Coomassie Blue staining of SDS-PAGE gels demonstrates the stoichiometric recovery of tertiary Tn complex from wild type and Tg McTnT-ND72 hearts using TnI-1 mAb immunoaffinity chromatography. As described above, the co-migration of TnT-ND72 with TnI formed a fused band in the Tn sample from the Tg McTnT-ND72 hearts. In addition to the stoichiometric recovery of Tn subunits, the TnI affinity column also recovered Tm, indicating the native biological activity of the Tn complex. High percentages of the exogenous cardiac TnT in the total cardiac tissue of transgenic mouse heart (solid bars) were recovered in the isolated Tn complex (open bars), indicating that McTnT-ND72 and Embryonic TnT both effectively incorporate into the myofilament in vivo.
To investigate the function of the TnT core structure and the effect of NH2-terminal variation in forming Tn complex in vivo, we examined Tn complex isolated from cardiac muscle thin filaments of the Tg McTnT-ND72 mice, transgenic mice expressing the embryonic TnT-1 in the heart (Tg McTnT-Emb) and wild type mice by TnI-1 mAb affinity chromatography (42). As shown in Figure 8B, Tn complex isolated from Tg McTnT-ND72 hearts contained the McTnT-ND72 core as 42.2 ± 4.7% of the total TnT recovered. Tn complex isolated from Tg McTnT-Emb hearts contained embryonic TnT equal to 68.3 ± 0.4% of the total TnT recovered. These amounts of exogenous TnT incorporated in the isolated Tn complex were not statistically different from the exogenous TnT detected in the whole heart homogenates (Fig. 8B, 49.5 ± 1.8% for Tg McTnT-ND72 and 68.8 ± 10.1% for Tg McTnT-Emb). These results demonstrate that the engineered TnT core structure and the exogenous embryonic cardiac TnT effectively incorporate into the adult cardiac Tn complex in vivo.
DISCUSSION
Defining the functional core structure of TnT
The structure-function relationship of TnT has been extensively studied by characterization of the chymotryptic NH2-terminal fragment T1 (residues 1–158) and COOH-terminal fragment T2 (residues 159–259) defined two decades ago using rabbit fast skeletal muscle TnT (46, 47). The T1 region is often referred as the “tail” domain of TnT rendering the Tn complex asymmetric. Primary structures of the three TnT isoforms from many species have demonstrated that the T1 fragment consists of the highly divergent NH2-terminal region and a highly conserved portion that interacts with the Tm head-to-tail overlap (48–50). The present study investigated the hypothesis that the conserved portion of T1 (i.e., the central region of the TnT polypeptide chain) is a part of the TnT core structure while the hypervariable NH2-terminal region of T1 (i.e., the CB3 fragment corresponding to residues 1–49 in rabbit fast skeletal muscle TnT) that does not bind to any known proteins of the muscle thin filament is an auxiliary modulating structure. Investigating the TnT structure-function relationship by dissecting the molecule according to its intrinsic features rather than the exogenous protease-generated fragments will help to better understand the two-site binding between TnT and Tm as well as the functional mechanism of the NH2-terminal variable region.
By progressive NH2-terminal deletions of cardiac TnT, we investigated the proposed core structure that confers the basal protein binding activities of TnT. A selective deletion of the entire hypervariable region corresponding to the NH2-terminal 71 amino acids in mouse cardiac TnT was designed to represent the core structure of TnT (Fig. 1). This construct, i.e., McTnT-ND72, contains the whole conserved region and demonstrates preserved binding to TnI and Tm. Like the intact TnT molecule, McTnT-ND72 fully retains both Tm binding sites (Fig. 1). The increased McTnT-ND72 binding affinity to Tm demonstrates that the deletion of entire NH2-terminal variable region did not destroy the TnT core structure, although having a conformational effect on the remaining core structure with increased binding affinity to Tm.
We previously demonstrated that McTnT-ND72 exhibits preserved binding to TnI (45) through the COOH-terminal region. The present study showed that McTnT-ND72 also preserved the binding affinity to Tm. In comparison to McTnT-ND72, McTnT-ND91 lacks a portion of the conserved region and shows lower binding affinity to Tm. This difference suggests that the central Tm-binding site was disrupted by the deletion of 19 amino acids from the conserved region in McTnT-ND91, which diminishes the two-site high affinity binding between TnT and Tm. Altogether, these data suggest a TnT core structure consisting of the conserved central and COOH-terminal regions from amino acids 72 to 291 in adult mouse cardiac TnT. The functional importance of the amino acids 72-90 in the TnT core structure is supported by a previous study that a cardiac TnT fragment equivalent to McTnT-ND91 produced by caspase proteolytic truncation in myocardial ischemia reperfusion injury had impaired function (51).
The TnT core structure conveys physiological function in vivo
The preserved core activity of McTnT-ND72 shown by ELISA binding experiments using E. coli-expressed recombinant proteins was confirmed in vivo. Transgenic mice over-expressing the cardiac TnT core lacking the entire NH2-terminal variable region in the heart exhibited a normal basal life activity without apparent disruption of cardiac muscle function. Previously, we demonstrated that a COOH-terminal truncated slow skeletal muscle TnT was unable to accumulate in the muscle cells as it did not incorporate into the myofibrils (52). The large amount of McTnT-ND72 recovered in the Tn complex from the cardiac muscle thin filaments (the Guba-Straub solution extracted fraction) of the transgenic mice demonstrates that McTnT-ND72 effectively incorporates into the Tn complex and renders a functional thin filament. The McTnT-ND72 transgenic mice did not exhibit any cardiomypathy phenotype in the absence of stress stimulation, indicating the ability to sustain basal cardiac function. While full phenotypic characterization of these mice is underway, this initial observation supports the idea that deletion of the entire NH2-terminal variable region preserves the core structure and function of TnT. This is in agreement with previous findings that a restricted deletion of the NH2-terminal variable region occurs in vivo (45) and its removal does not abolish the Ca2+ activated regulation of muscle activation (20–23). These findings demonstrate that the TnT NH2-terminal variable region is nonessential to the basic regulatory function of Tn in vivo, although its deletion does alter the interaction of TnT with Tm (20, 24) and the maximal activation of the thin filament (20, 23), consistent with a modulatory function.
Regulatory function of the TnT NH2-terminal variable region
As the NH2-terminal variable region of TnT does not directly bind to other thin filament proteins, its functional significance is a standing question in understanding muscle thin filament regulation. Deletion of the 71 NH2-terminal residues from TnT did not destroy the function of the McTnT-ND72 molecule but represents a default function in the absence of NH2-terminal charge modification. The finding of decreased coupling strength of TnT to TnI and Tm resulting from the increased NH2-terminal negative charge demonstrates that the NH2-terminal variable region is modulatory to TnT function. Previous findings also indicate a functional significance of the TnT NH2-terminal variable region: Its structure is regulated by alternative RNA splicing during heart and skeletal muscle development (14); the developmentally regulated alternative splicing of the NH2-terminal region alters the production of force in skinned fibers (29, 44, 53); and its aberrant splicing is related to dilated cardiomyopathy, correlating to changes in actomyosin ATPase activation (27, 28), cardiomyocyte contractility (27, 28) and heart failure (54).
The thin filament regulatory system of muscle, including the Tn tail domain that interacts with the Tm head-to-tail overlap, is an allosteric structure in which long range conformational effects are essential to the function. Our previous studies suggest a model that the NH2-terminal variable region of TnT regulates the overall conformation and function of TnT in the Tn complex (14, 19, 25, 26). Despite the extended conformation of TnT (17–19), epitope conformational assays demonstrated the effect of NH2-terminal variation is transmitted along the TnT structure to affect the conformation of the central and COOH-terminal regions. Similarly, the Acidic and Basic chimeric TnT models result in conformational differences at both the local NH2-terminal mAb 3E4 epitope (Fig. 3A) and the mAb CT3 epitope located in the central region (Fig. 3B and 5). These findings demonstrate that the long range structural effects of the NH2-terminal variable region can be caused by variation within the NH2-terminus, which was also true in naturally occurring cardiac TnT isoforms (Fig. 6).
The effects of the NH2-terminal conformational modulation on TnI and Tm binding are also present in the model TnT molecules. The differential charge effects of the TnT NH2-terminus to regulate the binding affinity and coupling strength of TnT to TnI and Tm are worth further investigation. Altered binding affinity may effect the assembling of Tn complex, however NH2-terminal truncated (Fig. 8), aberrantly spliced cardiac TnT (28) and chicken fast skeletal muscle TnT (40) all effectively incorporate into the thin filament when over-expressed in the transgenic mouse heart. Therefore, it seems that the NH2-terminal regulation of the coupling strength between TnT and other thin filament proteins in the assembled myofibril is more critical to contractile function. Consistent with the propagation of the NH2-terminal conformational modulation along the extended TnT molecule, the TnT NH2-terminal charge variation would most effectively alter the more proximal central Tm-binding site as opposed to the distal COOH-terminal TnI binding site. This is significant to muscle regulation in that the central TnT-Tm binding site is critical to the propagation of Ca2+ activation along the thin filament (2). Stronger versus weaker coupling among the regulatory proteins may correspond to a more rigid or more flexible molecular switch to alter the activation dynamics of the myofibril. This idea is supported by findings that skinned muscle fibers containing an acidic TnT isoform exhibited decreased Ca2+-sensitivity and increased cooperativity of activation while Ca2+-sensitivity was increased and cooperativity decreased in muscle fibers containing a basic TnT isoform (40). The role of TnT NH2-terminal splicing as a mechanism to regulate the dynamics of thin filament activation is interesting and deserves further investigation.
The role of NH2-terminal charge
The hypervariable nature of the TnT NH2-terminal region imposes difficulties in understanding its role in the Ca2+-regulation of muscle contraction. Among the physical features of the NH2-terminal variable region, the charge variation demonstrates a consistent acidic to basic trend among the developmentally regulated isoforms (14, 33). Focusing on this feature, we sought to dissect out the effects of TnT NH2-terminal charge on the interaction of TnT with TnI and Tm using model TnT molecules and naturally occurring NH2-terminal splice variants. Consistent with a role for TnT NH2-terminal charge in Ca2+-regulation, all TnT molecules investigated exhibit a strong quantity-dependent effect of NH2-terminal charge on Tm binding, such that the more basic TnT coupled stronger to Tm than the more acidic TnT (Figs 4B and 6A).
Unlike the two-site (central and COOH-terminal) binding to Tm, TnT binding to TnI occurs at a single COOH-terminal site (2). In the chimeric TnT molecules, the difference in pI is large (Table 1; Basic TnT = 8.99; Acidic TnT = 6.33; adult cardiac TnT = 5.19). These chimera demonstrate significant quantity-dependent effects of NH2-terminal charge on the binding to TnI, such that the more basic TnT bound stronger (Fig 5A). On the other hand, the total NH2-terminal negative charge is strong in the cardiac TnT isofoms, diminishing the overall effect of NH2-terminal charge alterations on the pI (Table 1; adult 5.19 vs. embryonic 4.87) and TnI-binding affinity (Fig 6B) of the isoforms. This could result from the possibility that the NH2-terminal charge difference between TnT isoforms is buffered by the already large amount of negative charge in the cardiac TnT NH2-terminus to maintain the NH2-terminal charge modulation effective within the range of physiological pH.
The results reported here suggest that although the core structure of TnT confers the basal function in sustaining muscle contraction, the evolutionarily additive hypervariable NH2-terminal domain regulates TnT’s interactions within the thin filament regulatory system through charge variation-based long range conformational effects. While other mechanisms that convey the physiological function of the TnT NH2-terminal variable region remain to be investigated, the high tolerance of TnT NH2-terminal domain to structural variation or removal allows this mechanism to effectively extend the functional capacity of muscle during development and functional adaptation.
Acknowledgments
We thank Dr. Larry Smillie for the chicken fast TnT3 and TnT4 cDNA clones, Dr. Jim Lin for the CH1 mAb, and Dr. Jeffrey Robbins for the α-MHC promoter.
Footnotes
This study was supported in part by grants from the National Institutes of Health AR 048816, HL 078773 and HD 44124 (to J-PJ). BJB was supported by a Postdoctoral Fellowship from the American Heart Association Northeast Ohio Affiliate (#0325266B).
AUTHOR EMAIL ADDRESS: jpjin@northwestern.edu
Abbreviations: Acidic TnT, chimeric TnT containing an acidic NH2-terminal segment and the McTnT core; Basic TnT, chimeric TnT containing a basic NH2-terminal segment and the McTnT core; BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbant assay; McTnT, mouse cardiac TnT; ND72, deletion of the NH2-terminal 71 amino acids; ND91, deletion of the NH2-terminal 90 amino acids; pI, isoelectric point; SDS-PAGE, SDS-polyacrylamide gel electrophoresis; TBS, Tris-buffered saline; Tg, transgenic mouse; Emb, embryonic; Tm, tropomyosin; Tn, troponin; TnC, troponin C; TnI, troponin I; TnT, troponin T
References
- 1.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Perry SV. Troponin T: genetics, properties and function. J Muscle Res Cell Motil. 1998;19:575–602. doi: 10.1023/a:1005397501968. [DOI] [PubMed] [Google Scholar]
- 3.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–81. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 4.Leavis PC, Gergely J. Thin filament proteins and thin filament-linked regulation of vertebrate muscle contraction. CRC Crit Rev Biochem. 1984;16:235–305. doi: 10.3109/10409238409108717. [DOI] [PubMed] [Google Scholar]
- 5.Huang QQ, Chen A, Jin JP. Genomic sequence and structural organization of mouse slow skeletal muscle troponin T gene. Gene. 1999;229:1–10. doi: 10.1016/s0378-1119(99)00051-7. [DOI] [PubMed] [Google Scholar]
- 6.Barton PJ, Cullen ME, Townsend PJ, Brand NJ, Mullen AJ, Norman DA, Bhavsar PK, Yacoub MH. Close physical linkage of human troponin genes: organization, sequence, and expression of the locus encoding cardiac troponin I and slow skeletal troponin T. Genomics. 1999;57:102–9. doi: 10.1006/geno.1998.5702. [DOI] [PubMed] [Google Scholar]
- 7.Breitbart RE, Nadal-Ginard B. Complete nucleotide sequence of the fast skeletal troponin T gene. Alternatively spliced exons exhibit unusual interspecies divergence. J Mol Biol. 1986;188:313–24. doi: 10.1016/0022-2836(86)90157-9. [DOI] [PubMed] [Google Scholar]
- 8.Jin JP, Huang QQ, Yeh HI, Lin JJ. Complete nucleotide sequence and structural organization of rat cardiac troponin T gene. A single gene generates embryonic and adult isoforms via developmentally regulated alternative splicing. J Mol Biol. 1992;227:1269–76. doi: 10.1016/0022-2836(92)90540-z. [DOI] [PubMed] [Google Scholar]
- 9.Mak AS, Smillie LB. Structural interpretation of the two-site binding of troponin on the muscle thin filament. J Mol Biol. 1981;149:541–50. doi: 10.1016/0022-2836(81)90486-1. [DOI] [PubMed] [Google Scholar]
- 10.Pearlstone JR, Smillie LB. Binding of troponin-T fragments to several types of tropomyosin. Sensitivity to Ca2+ in the presence of troponin-C. J Biol Chem. 1982;257:10587–92. [PubMed] [Google Scholar]
- 11.Heeley DH, Smillie LB. Interaction of rabbit skeletal muscle troponin T and F-actin at physiological ionic strength. Biochemistry. 1988;27:8227–32. doi: 10.1021/bi00421a036. [DOI] [PubMed] [Google Scholar]
- 12.Jin JP, Samanez RA. Evolution of a metal-binding cluster in the NH(2)-terminal variable region of avian fast skeletal muscle troponin T: functional divergence on the basis of tolerance to structural drifting. J Mol Evol. 2001;52:103–16. doi: 10.1007/s002390010139. [DOI] [PubMed] [Google Scholar]
- 13.Jin JP, Chen A, Huang QQ. Three alternatively spliced mouse slow skeletal muscle troponin T isoforms: conserved primary structure and regulated expression during postnatal development. Gene. 1998;214:121–9. doi: 10.1016/s0378-1119(98)00214-5. [DOI] [PubMed] [Google Scholar]
- 14.Jin JP, Huang QQ, Ogut O, Chen A, Wang J. Troponin T isoform regulation and structure-function relationships. Basic Appl Myol. 2000;10:17–26. [Google Scholar]
- 15.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 16.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci U S A. 2005;102:5038–43. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabral-Lilly D, Tobacman LS, Mehegan JP, Cohen C. Molecular polarity in tropomyosin-troponin T co-crystals. Biophys J. 1997;73:1763–70. doi: 10.1016/S0006-3495(97)78206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendt T, Guenebaut V, Leonard KR. Structure of the Lethocerus troponin-tropomyosin complex as determined by electron microscopy. J Struct Biol. 1997;118:1–8. doi: 10.1006/jsbi.1996.3834. [DOI] [PubMed] [Google Scholar]
- 19.Jin JP, Root DD. Modulation of troponin T molecular conformation and flexibility by metal ion binding to the NH2-terminal variable region. Biochemistry. 2000;39:11702–13. doi: 10.1021/bi9927437. [DOI] [PubMed] [Google Scholar]
- 20.Pan BS, Gordon AM, Potter JD. Deletion of the first 45 NH2-terminal residues of rabbit skeletal troponin T strengthens binding of troponin to immobilized tropomyosin. J Biol Chem. 1991;266:12432–8. [PubMed] [Google Scholar]
- 21.Ohtsuki I, Shiraishi F, Suenaga N, Miyata T, Tanokura M. A 26K fragment of troponin T from rabbit skeletal muscle. J Biochem (Tokyo) 1984;95:1337–42. doi: 10.1093/oxfordjournals.jbchem.a134740. [DOI] [PubMed] [Google Scholar]
- 22.Fujita S, Maeda K, Maeda Y. Expression in Escherichia coli and a functional study of a beta-troponin T 25 kDa fragment of rabbit skeletal muscle. J Biochem (Tokyo) 1992;112:306–8. doi: 10.1093/oxfordjournals.jbchem.a123896. [DOI] [PubMed] [Google Scholar]
- 23.Chandra M, Montgomery DE, Kim JJ, Solaro RJ. The N-terminal region of troponin T is essential for the maximal activation of rat cardiac myofilaments. J Mol Cell Cardiol. 1999;31:867–80. doi: 10.1006/jmcc.1999.0928. [DOI] [PubMed] [Google Scholar]
- 24.Fisher D, Wang G, Tobacman LS. NH2-terminal truncation of skeletal muscle troponin T does not alter the Ca2+ sensitivity of thin filament assembly. J Biol Chem. 1995;270:25455–60. doi: 10.1074/jbc.270.43.25455. [DOI] [PubMed] [Google Scholar]
- 25.Jin JP, Chen A, Ogut O, Huang QQ. Conformational modulation of slow skeletal muscle troponin T by an NH(2)-terminal metal-binding extension. Am J Physiol Cell Physiol. 2000;279:C1067–77. doi: 10.1152/ajpcell.2000.279.4.C1067. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Jin JP. Conformational modulation of troponin T by configuration of the NH2-terminal variable region and functional effects. Biochemistry. 1998;37:14519–28. doi: 10.1021/bi9812322. [DOI] [PubMed] [Google Scholar]
- 27.Biesiadecki BJ, Jin JP. Exon skipping in cardiac troponin T of turkeys with inherited dilated cardiomyopathy. J Biol Chem. 2002;277:18459–68. doi: 10.1074/jbc.M200788200. [DOI] [PubMed] [Google Scholar]
- 28.Biesiadecki BJ, Elder BD, Yu ZB, Jin JP. Cardiac troponin T variants produced by aberrant splicing of multiple exons in animals with high instances of dilated cardiomyopathy. J Biol Chem. 2002;277:50275–85. doi: 10.1074/jbc.M206369200. [DOI] [PubMed] [Google Scholar]
- 29.Ogut O, Granzier H, Jin JP. Acidic and basic troponin T isoforms in mature fast-twitch skeletal muscle and effect on contractility. Am J Physiol. 1999;276:C1162–70. doi: 10.1152/ajpcell.1999.276.5.C1162. [DOI] [PubMed] [Google Scholar]
- 30.Jin JP, Zhang J, Wang J. An embryonic alternative transcription initiation site and the 5'-upstream structure of mouse cardiac troponin T gene. Biochem Biophys Res Commun. 1995;214:1168–74. doi: 10.1006/bbrc.1995.2409. [DOI] [PubMed] [Google Scholar]
- 31.Jin JP, Wang J, Zhang J. Expression of cDNAs encoding mouse cardiac troponin T isoforms: characterization of a large sample of independent clones. Gene. 1996;168:217–21. doi: 10.1016/0378-1119(95)00803-9. [DOI] [PubMed] [Google Scholar]
- 32.Jin JP. Cloned rat cardiac titin class I and class II motifs. Expression, purification, characterization, and interaction with F-actin. J Biol Chem. 1995;270:6908–16. [PubMed] [Google Scholar]
- 33.Wang J, Jin JP. Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene. 1997;193:105–14. doi: 10.1016/s0378-1119(97)00100-5. [DOI] [PubMed] [Google Scholar]
- 34.Ogut O, Jin JP. Developmentally regulated, alternative RNA splicing-generated pectoral muscle-specific troponin T isoforms and role of the NH2-terminal hypervariable region in the tolerance to acidosis. J Biol Chem. 1998;273:27858–66. doi: 10.1074/jbc.273.43.27858. [DOI] [PubMed] [Google Scholar]
- 35.Smillie LB, Golosinska K, Reinach FC. Sequences of complete cDNAs encoding four variants of chicken skeletal muscle troponin T. J Biol Chem. 1988;263:18816–20. [PubMed] [Google Scholar]
- 36.Jin JP, Brotto MA, Hossain MM, Huang QQ, Brotto LS, Nosek TM, Morton DH, Crawford TO. Truncation by Glu180 nonsense mutation results in complete loss of slow skeletal muscle troponin T in a lethal nemaline myopathy. J Biol Chem. 2003;278:26159–65. doi: 10.1074/jbc.M303469200. [DOI] [PubMed] [Google Scholar]
- 37.Smillie LB. Preparation and identification of alpha- and beta-tropomyosins. Methods Enzymol. 1982;85(Pt B):234–41. doi: 10.1016/0076-6879(82)85023-4. [DOI] [PubMed] [Google Scholar]
- 38.Jin JP, Yang FW, Yu ZB, Ruse CI, Bond M, Chen A. The highly conserved COOH terminus of troponin I forms a Ca2+-modulated allosteric domain in the troponin complex. Biochemistry. 2001;40:2623–31. doi: 10.1021/bi002423j. [DOI] [PubMed] [Google Scholar]
- 39.Lin JJ, Chou CS, Lin JL. Monoclonal antibodies against chicken tropomyosin isoforms: production, characterization, and application. Hybridoma. 1985;4:223–42. doi: 10.1089/hyb.1985.4.223. [DOI] [PubMed] [Google Scholar]
- 40.Huang QQ, Brozovich FV, Jin JP. Fast skeletal muscle troponin T increases the cooperativity of transgenic mouse cardiac muscle contraction. J Physiol 520 Pt. 1999;1:231–42. doi: 10.1111/j.1469-7793.1999.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramaniam A, Gulick J, Neumann J, Knotts S, Robbins J. Transgenic analysis of the thyroid-responsive elements in the alpha-cardiac myosin heavy chain gene promoter. J Biol Chem. 1993;268:4331–6. [PubMed] [Google Scholar]
- 42.Yu ZB, Zhang LF, Jin JP. A proteolytic NH2-terminal truncation of cardiac troponin I that is up-regulated in simulated microgravity. J Biol Chem. 2001;276:15753–60. doi: 10.1074/jbc.M011048200. [DOI] [PubMed] [Google Scholar]
- 43.Jin JP, Lin JJ. Isolation and characterization of cDNA clones encoding embryonic and adult isoforms of rat cardiac troponin T. J Biol Chem. 1989;264:14471–7. [PubMed] [Google Scholar]
- 44.Gomes AV, Guzman G, Zhao J, Potter JD. Cardiac troponin T isoforms affect the Ca2+ sensitivity and inhibition of force development. Insights into the role of troponin T isoforms in the heart. J Biol Chem. 2002;277:35341–9. doi: 10.1074/jbc.M204118200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Biesiadecki BJ, Jin JP. Selective removal of the NH2-terminal variable region of cardiac troponin T in ischemia-reperfusion by myofibril-associated u-calpain clevage. Biochemistry in press. 2006 doi: 10.1021/bi060273s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohtsuki I. Molecular arrangement of troponin-T in the thin filament. J Biochem (Tokyo) 1979;86:491–7. doi: 10.1093/oxfordjournals.jbchem.a132549. [DOI] [PubMed] [Google Scholar]
- 47.Heeley DH, Watson MH, Mak AS, Dubord P, Smillie LB. Effect of phosphorylation on the interaction and functional properties of rabbit striated muscle alpha alpha-tropomyosin. J Biol Chem. 1989;264:2424–30. [PubMed] [Google Scholar]
- 48.Heeley DH, Golosinska K, Smillie LB. The effects of troponin T fragments T1 and T2 on the binding of nonpolymerizable tropomyosin to F-actin in the presence and absence of troponin I and troponin C. J Biol Chem. 1987;262:9971–8. [PubMed] [Google Scholar]
- 49.Ishii Y, Lehrer SS. Two-site attachment of troponin to pyrene-labeled tropomyosin. J Biol Chem. 1991;266:6894–903. [PubMed] [Google Scholar]
- 50.Schaertl S, Lehrer SS, Geeves MA. Separation and characterization of the two functional regions of troponin involved in muscle thin filament regulation. Biochemistry. 1995;34:15890–4. doi: 10.1021/bi00049a003. [DOI] [PubMed] [Google Scholar]
- 51.Communal C, Sumandea M, de Tombe P, Narula J, Solaro RJ, Hajjar RJ. Functional consequences of caspase activation in cardiac myocytes. Proc Natl Acad Sci U S A. 2002;99:6252–6. doi: 10.1073/pnas.092022999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Huang QQ, Breckenridge MT, Chen A, Crawford TO, Morton DH, Jin JP. Cellular fate of truncated slow skeletal muscle troponin T produced by Glu180 nonsense mutation in amish nemaline myopathy. J Biol Chem. 2005;280:13241–9. doi: 10.1074/jbc.M413696200. [DOI] [PubMed] [Google Scholar]
- 53.Tobacman LS, Lee R. Isolation and functional comparison of bovine cardiac troponin T isoforms. J Biol Chem. 1987;262:4059–64. [PubMed] [Google Scholar]
- 54.Anderson PA, Malouf NN, Oakeley AE, Pagani ED, Allen PD. Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ Res. 1991;69:1226–33. doi: 10.1161/01.res.69.5.1226. [DOI] [PubMed] [Google Scholar]


