Abstract
Modulating transcription factors is crucial to executing sophisticated gene expression programs. The silent information regulator 2 (Sir2) family of NAD-dependent protein deacetylases influences transcription by targeting proteins such as histones, p53 and forkhead-box family transcription factors. Although apparently cytoplasmic, both mammalian SIRT2 and its yeast orthologue Hst2 have been implicated in transcriptional regulation. Here, we show that Hst2 moves between the nucleus and cytoplasm, but is largely cytoplasmic owing to efficient nuclear export. This nuclear exclusion is mediated by the exportin chromosomal region maintenance 1 (Crm1) and a putative leucine-rich nuclear export sequence in Hst2, which overlaps a unique autoregulatory helix. Disruption of Hst2 export shows that nuclear exclusion inhibits the activity of Hst2 as a transcriptional repressor. Our identification of putative nuclear export sequences in numerous vertebrate SIRT2 proteins shows that active nuclear export can be a conserved mechanism for regulating Sir2 homologues.
Keywords: chromatin, deacetylase, HDAC, Saccharomyces cerevisiae
Introduction
Several members of the silent information regulator 2 (Sir2) family (sirtuins), which are defined cytologically as cytoplasmic proteins, also influence nuclear transcription. For example, although sirtuin3 (SIRT3) in mice localizes to the inner mitochondrial membrane, it affects the nuclear expression of transcription factors crucial to mitochondrial function (Shi et al, 2005). Human SIRT2 is a predominantly cytoplasmic tubulin deacetylase; however, it can associate with the homeobox transcription factor HOXA10 in both the cytoplasm and the nucleus (Afshar & Murnane, 1999; North et al, 2003; Bae et al, 2004). In Saccharomyces cerevisiae, homologue of sir two 2 (Hst2) seems to be localized to the cytoplasm even when coupled to a nuclear localization signal (Perrod et al, 2001). This is in contrast with molecular and genetic evidence that Hst2 deacetylates histones in vivo and participates in the transcriptional repression of flocculation 10 (FLO10) through direct binding of its promoter (Halme et al, 2004; Vaquero et al, 2006). Furthermore, although no defects have been reported for hst2Δ single mutants, overexpression of HST2 influences transcriptional silencing and recombination in the ribosomal DNA array, and might contribute to the effects of caloric restriction and ageing in the absence of SIR2 (Perrod et al, 2001; Lamming et al, 2005). Indications that Hst2 and other predominantly cytoplasmic sirtuins might modulate nuclear processes prompted further investigation of the role and regulation of Hst2 in vivo.
Results And Discussion
Hst2 is actively exported from the nucleus
Hst2 is a 40 kDa globular protein that resembles in size and shape other proteins that are known to diffuse freely through the nuclear pore (Shulga et al, 2000). The nuclear exclusion of Hst2 suggests that it might be prevented from entering the nucleus as part of a large cytoplasmic complex or that it might enter the nucleus but be actively returned to the cytoplasm. Size-exclusion chromatography on yeast lysate indicates that the majority of Hst2 is monomeric in vivo (supplementary Fig 1 online); therefore, exclusion of Hst2 from the nucleus is not solely due to its participation in a multimeric complex in the cytoplasm.
To investigate whether full-length Hst2 enters the nucleus, we evaluated its localization in cells treated with the nuclear export poison leptomycin B (LMB), which specifically inhibits the conserved exportin 1 (Xpo1)/chromosomal region maintenance 1 (Crm1). To evaluate export efficiency in budding yeast, we used the ‘humanized' CRM1T539C allele, which is fully export competent and LMB sensitive (Neville & Rosbash, 1999). Using a 1-h drug treatment that maintains cell viability but allows for substantial equilibration between the nucleus and cytoplasm (Neville & Rosbash, 1999), Hst2 showed diffuse staining across the entire cell with partial enrichment in 4′,6-diamidino-2-phenylindole (DAPI) staining areas in CRM1T539C cells (Fig 1). In control experiments, Hst2 remained cytoplasmic in LMB-insensitive CRM1 cells and CRM1T539C cells exposed only to an ethanol solvent (Fig 1). Thus, Hst2 transits through the nucleus, but rather than being retained, it normally undergoes efficient, Crm1-dependent nuclear export.
Figure 1.
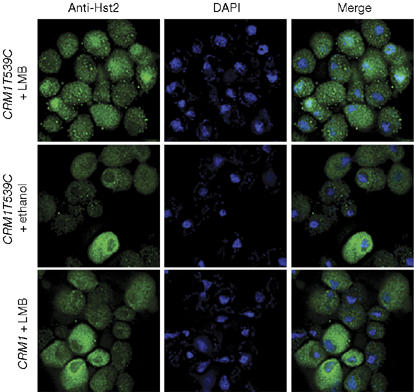
Hst2 is excluded from the nucleus by active nuclear export. Hst2 undergoes chromosomal region maintenance 1 (Crm1)-dependent nuclear export. Strains CRM1T539C and CRM1 expressing a 2μ HST2 construct were exposed to 100 ng/ml leptomycin B (LMB) or ethanol solvent for 1 h. Hst2 became diffuse throughout the cell and partially enriched in nuclear areas in the presence of the CRM1T539C allele and LMB. Hst2 remained predominantly cytoplasmic under control conditions. Hst2 is shown in green, as detected with anti-Hst2 polyclonal serum and fluorescein-conjugated secondary antibodies. DNA is counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and shown in blue. Colocalization of Hst2 and DNA when merged is indicated in cyan.
Nuclear export signal overlaps an autoregulatory helix
Crm1 recognizes a leucine (L)-rich nuclear export sequence (NES) of the form LX2−3LX2−3LXL (Bogerd et al, 1996). Analysis using the NetNES 1.1 prediction program (la Cour et al, 2004) identified a putative Hst2 NES from amino acids 306 to 317 (Fig 2A). This predicted export signal overlaps the α13 helix and may compete for the NAD binding site (Zhao et al, 2003).
Figure 2.
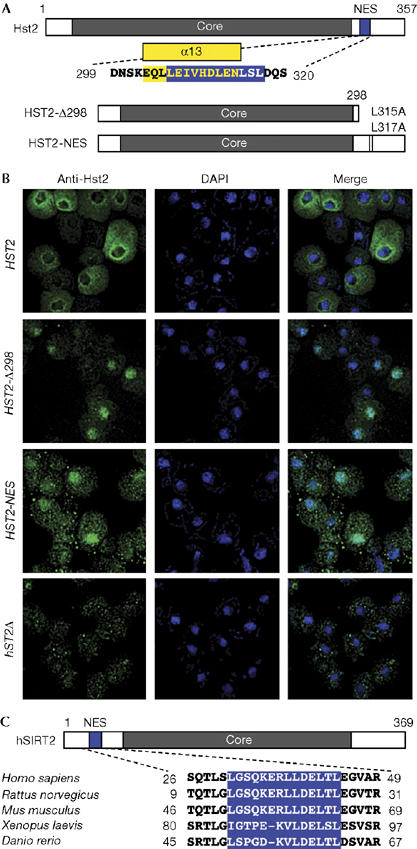
Hst2 localization is controlled by a leucine-rich nuclear export sequence. (A) A nuclear export sequence (NES; blue) was identified using the NetNES 1.1 prediction algorithms. The Hst2 α13 helix is shown in yellow. (B) Mutations in the NES region caused Hst2 to accumulate in the nucleus. Hst2 localized primarily to the cytoplasm; HST2-Δ298 and HST2-NES localized primarily to the nucleus. The hst2Δ cells illustrate low background staining. All strains are hst2Δ, bearing 2μ constructs. Immunofluorescence is as in Fig 1. DAPI, 4′,6-diamidino-2-phenylindole. (C) Alignment of NESs from vertebrate SIRT2 (hSIRT2) proteins. Blue boxes indicate NESs identified using NetNES 1.1. Multiple sequence alignments were created in Clustal W.
To confirm whether the predicted NES is functional, we assessed the localization of two Hst2 mutants. HST2-Δ298 contains amino acids 1–298, with the entire carboxy-terminal extension deleted, including the α13 helix. In contrast with wild type, HST2-Δ298 staining was specifically enriched in DAPI staining areas, indicating a predominantly nuclear localization for this mutant (Fig 2B). The presence of HST2-Δ298 in the nucleus confirms that the C-terminal extension of Hst2 is a crucial determinant of its subcellular localization.
In further support of the predicted NES, we characterized the HST2-NES mutant in which the two most crucial hydrophobic residues in the NES were altered without disrupting the sequence of the α13 helix (Fig 2B). Similar to HST2-Δ298, Hst2 staining in the HST2-NES mutant was significantly enriched in the nuclear DAPI staining areas when compared with wild type (Fig 2B). From these results, we conclude that the putative NES is required for efficient nuclear export.
Export negatively regulates Hst2 function
To evaluate the physiological significance of the nuclear–cytoplasmic shuttling of Hst2, hst2Δ and NES-deficient alleles of HST2 were characterized functionally. Because single hst2Δ mutants have no reported defects under standard growth conditions, we carried out an extensive phenotypic survey (Hampsey, 1997). This analysis showed that cells lacking HST2 had at least 25-fold greater resistance to the translational poison cycloheximide (CHX) compared with wild-type cells (Fig 3A). In marked contrast with the resistance of the null mutant, expression of either HST2-Δ298 or HST2-NES led to increased sensitivity to CHX (Fig 3B). When compared with cells bearing low-copy centromeric (CEN) plasmids, CHX sensitivity was further enhanced by overexpression of the mutant constructs from high-copy 2μ vectors.
Figure 3.
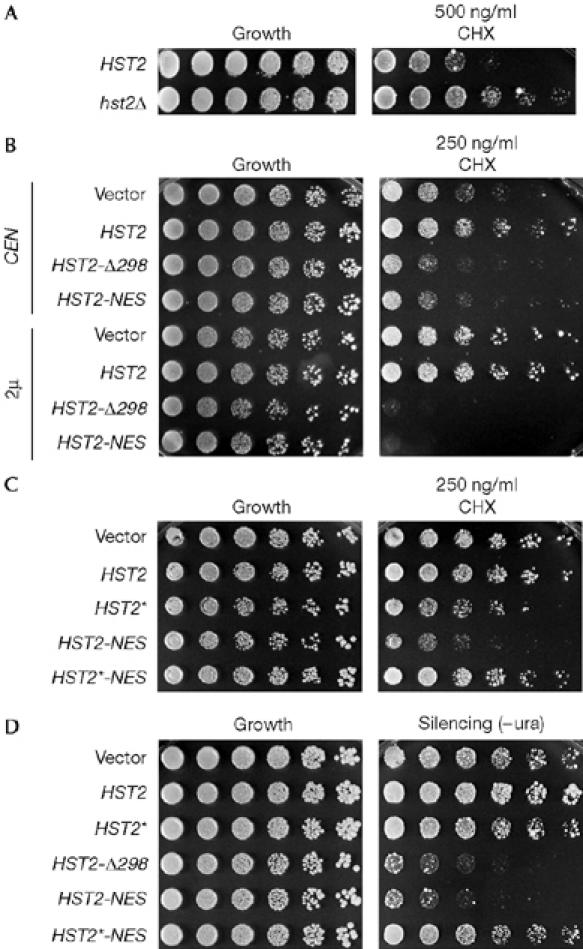
HST2 alleles lacking nuclear export sequence increase transcriptional silencing. Cultures were plated as fivefold serial dilutions. (A) The hst2Δ strain showed at least 25-fold greater growth on cycloheximide (CHX) than the HST2 strain at 37°C. (B) The HST2-Δ298 and HST2-NES mutants caused dominant, dosage-dependent sensitivity to low levels of CHX when expressed in an HST2 strain. Strains were grown on synthetic selective media at 37°C. (C) The mutation of a crucial catalytic residue in HST2*-NES restored normal growth on CHX. Growth conditions were as in (B). (D) The HST2-Δ298 and HST2-NES mutants caused marked increases in the silencing of a URA3 reporter gene integrated in the ribosomal DNA array when compared with cells carrying empty vectors. HST2*-NES did not increase silencing. CEN, centromeric; NES, nuclear export sequence; –ura, medium lacking uracil.
Nuclear-localized HST2 mutants caused dominant, dosage-dependent sensitivity to CHX, which is in contrast with the phenotype of hst2Δ. This suggests that hyper-accumulation of Hst2 enhances its normal activity in the nucleus. To confirm that the effects of nuclear Hst2 mutants were due to increased Hst2 deacetylase activity in this region, we analysed the HST2* mutants with the crucial point mutation H135Y, analogous to the catalytically inactive sir2-H364Y mutant (Tanny et al, 1999; Imai et al, 2000). This mutation in the catalytic core of Hst2 negated sensitivity to CHX caused by point mutations in the NES (Fig 3C). This confirms that the accumulation of Hst2 in the nucleus enhances its overall activity.
Because CHX can inhibit rDNA transcription in vivo by limiting association of transcription factors with RNA pol I (Cavanaugh et al, 2002), the relative resistance of hst2Δ cells independently reinforces evidence that Hst2 is important for rDNA silencing and integrity (Perrod et al, 2001; Lamming et al, 2005). To asses directly the impact of NES-deficient HST2 alleles on rDNA silencing, we used a yeast strain with a uracil requiring 3 (URA3) reporter gene integrated into a single rDNA repeat (Smith & Boeke, 1997). HST2-Δ298 and HST2-NES alleles caused a marked increase in rDNA silencing, as indicated by their impaired growth in medium without uracil (Fig 3D). Again, the effect of the HST2-NES allele was abrogated by a crucial mutation in the catalytic core. This establishes a model in which the nuclear activities of the most abundant and enzymatically active yeast sirtuin (Smith et al, 2000; Huh et al, 2003) are restricted by efficient removal from the nucleus.
Regulating sirtuins by nuclear export
Regulation by nuclear exclusion, as shown here, has not been studied previously for Sir2 family enzymes; however, it is almost certainly not restricted to Hst2. We identified well-conserved, putative NESs in the amino-terminal extensions of numerous vertebrate SIRT2 proteins (Fig 2C; la Cour et al, 2004). This motif has been independently identified and verified as a functional NES in human SIRT2 by mutational analysis and LMB challenges (B.J. North & E. Verdin, personal communication). Furthermore, supporting the hypothesis that SIRT2 might act as a transcriptional regulator that shuttles between the nucleus and cytoplasm, a small nuclear pool of SIRT2 has been identified and the protein associates with the transcription factor HOXA10 in both cellular compartments (North et al, 2003; Bae et al, 2004). Although not functionally characterized, a potential NES has been noted in the C terminus of the cytoplasmic sirtuin of Leishmania infantum (Ouaissi, 2003) and a similar sequence exists in its orthologue in Leishmania major (data not shown).
Speculation
The presence of demonstrated or predicted nuclear export signals in virtually all ‘cytoplasmic' sirtuins supports the idea that these deacetylases have nuclear roles and that export is a conserved mechanism for their regulation. Indeed, several other chromatin-modifying enzymes, such as the class IIa histone deacetylases and the protein arginine methyltransferase 1 (PRMT1), undergo dynamic subcellular localization (Verdin et al, 2003; Herrmann et al, 2005). As knowledge of chromatin modifiers increases, we expect that other factors—the localization of which contradicts their enzymatic role—are regulated by nuclear exclusion.
Many mammalian Sir2 family members are implicated in disease. SIRT1 and SIRT3 modulate metabolic pathways linked to diabetes, but the mechanism by which the mitochondrial SIRT3 enzyme influences nuclear transcription factors is unknown (Yechoor et al, 2004; Moynihan et al, 2005; Rodgers et al, 2005; Shi et al, 2005). It has also been proposed that influences on p53, insulin levels and caloric restriction pathways might allow SIRT1, SIRT2 and SIRT3 to moderate neurodegenerative disorders such as Alzheimer's disease and Huntington's disease (Anekonda & Reddy, 2006). Indeed, SIRT2 is highly expressed in neural tissue and is downregulated in a common form of brain malignancy (Afshar & Murnane, 1999; Hiratsuka et al, 2003; Yu et al, 2005). As the findings of our study are extended to mammalian models, manipulation of the subcellular localization of Sir2 enzymes might prove to be a potent mechanism for moderating their effects on serious human illnesses.
Methods
Yeast methods. Yeast strains and plasmids are given in the supplementary information online. Standard procedures were used as described previously (Amberg et al, 2005) unless indicated.
Antiserum. Full-length recombinant Hst2 was expressed and purified as described previously (Landry et al, 2000), with the substitution of TALON affinity resin (Clontech, Palo Alto, CA, USA). Polyclonal antisera were raised in rabbits against this antigen using standard protocols (Harlow & Lane, 1998).
Immunofluorescence microscopy. hst2Δ strain LPY6623 was transformed with plasmids bearing HST2 or mutant constructs. LMB (KOSAN Biosciences Inc., Hayward, CA, USA) was added to exponentially growing cultures at a final concentration of 100 ng/ml followed by a 1 h incubation at 30°C. Immunofluorescence was carried out (Stone & Pillus, 1996; Garcia & Pillus, 2002) using a 1:200 dilution of 663-4 anti-Hst2 serum. Microscopic images, spaced at 0.2 μm increments, were collected and deconvolved as described by Rubio and Pogliano (2004).
Sequence analysis. All sequence analyses were carried out using the entire open-reading frame for each protein and processed by NetNES1.1 or Clustal W.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Fig 1
Supplemental Materials
Acknowledgments
We thank B. North and E. Verdin for communicating unpublished results; D. Urwin, S. Jacobson, P. Laurenson, J. Heilig and D. Forbes for helpful discussions and criticism of the manuscript; C. Chang and C. Cast for assistance; and J. Wang and M. Rosbash for kindly providing reagents. This work was supported by the National Institutes of Health.
References
- Afshar G, Murnane JP (1999) Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene 234: 161–168 [DOI] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Anekonda TS, Reddy PH (2006) Neuronal protection by sirtuins in Alzheimer's disease. J Neurochem 96: 305–313 [DOI] [PubMed] [Google Scholar]
- Bae NS, Swanson MJ, Vassilev A, Howard BH (2004) Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. J Biochem (Tokyo) 135: 695–700 [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol 16: 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh AH, Hirschler-Laszkiewicz I, Hu Q, Dundr M, Smink T, Misteli T, Rothblum LI (2002) Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J Biol Chem 277: 27423–27432 [DOI] [PubMed] [Google Scholar]
- Garcia SN, Pillus L (2002) A unique class of conditional sir2 mutants displays distinct silencing defects in Saccharomyces cerevisiae. Genetics 162: 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR (2004) Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415 [DOI] [PubMed] [Google Scholar]
- Hampsey M (1997) A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1998) Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory [Google Scholar]
- Herrmann F, Lee J, Bedford MT, Fackelmayer FO (2005) Dynamics of human protein arginine methyltransferase 1 (PRMT1) in vivo. J Biol Chem 280: 38005–38010 [DOI] [PubMed] [Google Scholar]
- Hiratsuka M et al. (2003) Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun 309: 558–566 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536 [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA (2005) HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 309: 1861–1864 [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA 97: 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S (2005) Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab 2: 105–117 [DOI] [PubMed] [Google Scholar]
- Neville M, Rosbash M (1999) The NES–Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18: 3746–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444 [DOI] [PubMed] [Google Scholar]
- Ouaissi A (2003) Apoptosis-like death in trypanosomatids: search for putative pathways and genes involved. Kinetoplastid Biol Dis 2: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM (2001) A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J 20: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Rubio A, Pogliano K (2004) Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J 23: 1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q (2005) SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem 280: 13560–13567 [DOI] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS (2000) Yeast nucleoporins involved in passive nuclear envelope permeability. J Cell Biol 149: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11: 241–254 [DOI] [PubMed] [Google Scholar]
- Smith JS et al. (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA 97: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Pillus L (1996) Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol 135: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D (2006) SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20: 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG (2003) Class II histone deacetylases: versatile regulators. Trends Genet 19: 286–293 [DOI] [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, Kahn CR (2004) Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci USA 101: 16525–16530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TT, McIntyre JC, Bose SC, Hardin D, Owen MC, McClintock TS (2005) Differentially expressed transcripts from phenotypically identified olfactory sensory neurons. J Comp Neurol 483: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Clements A, Marmorstein R (2003) Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol 10: 864–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig 1
Supplemental Materials


