Abstract
A subclass of zinc finger proteins containing a unique protein motif called the positive regulatory (PR) domain has been described. The members include the PRDI-BF1/Blimp-1 protein, the Caenorhabditis elegans egl-43 and EVI1 gene products, and the retinoblastoma interacting protein RIZ. Here we describe a member of this family, SC-1, that exhibits several distinctive features. First, SC-1 interacts with the p75 neurotrophin receptor and is redistributed from the cytoplasm to the nucleus after nerve growth factor (NGF) treatment of transfected COS cells. The translocation of SC-1 to the nucleus was specific for p75, as NGF binding to the TrkA receptor did not lead to nuclear localization of SC-1. Thus, SC-1 provides a downstream transducer for the effects of NGF through the p75 neurotrophin receptor. Under normal growth conditions, SC-1 was found predominantly in the cytoplasm. On serum-starvation, SC-1 also translocated into the nucleus. A direct correlation between nuclear expression of SC-1 with the loss of BrdUrd incorporation was observed. These results imply that SC-1 may be involved in events associated with growth arrest.
Neurotrophins influence a wide number of functions in the nervous system, including neuronal cell survival, cell differentiation and apoptosis, synaptic plasticity, and control of axonal guidance and dendritic cell growth (1–3). These actions are mediated by neurotrophin binding to two separate receptor classes, the Trk family of tyrosine kinase receptors and the p75 neurotrophin receptor, a member of the tumor necrosis factor receptor superfamily. Nerve growth factor (NGF), brain-derived neurotrophic factor, and neurotrophin 4/5 and neurotrophin 3 bind to TrkA, TrkB, and TrkC, respectively, whereas each neurotrophin is capable of binding to p75 with a similar affinity (4). In the presence of TrkA receptor, p75 can participate in the formation of high affinity binding sites and enhanced neurotrophin responsiveness leading to increased cell survival (5). In the absence of TrkA receptors, p75 can activate NF-κB and JNK activities and can generate, in specific cell populations and conditions, a death signal (6–9). This dichotomy in responses has raised questions regarding the nature of the signaling mechanisms and how specificity for the two neurotrophin receptors is encoded.
To elucidate the function of p75, an extensive two-hybrid screen was undertaken to identify proteins that interact with the cytoplasmic domain of p75. Such candidate molecules could reveal insight into the signal transduction mechanisms mediated by neurotrophins through the p75 receptor. One of the clones identified by this screen encoded a protein called SC-1 that contains six zinc finger domains and a unique domain, the positive regulatory (PR) or PRDI-BF1 and RIZ homology domain. The PR domain was previously identified as a common motif in several transcription factors, including RIZ and PRDI-BF1 (10, 18). Here, we describe the structural features of SC-1, its association with p75 neurotrophin receptor, and its subcellular localization. We find the distribution of SC-1 is strongly regulated by NGF binding to the p75 receptor and by growth conditions. Interestingly, the localization of SC-1 is not under control of NGF binding to the TrkA receptor or by other neurotrophins, such as brain-derived neurotrophic factor and neurotrophin 3. These observations define SC-1 as a protein that is differentially controlled by neurotrophin and serum conditions.
MATERIALS AND METHODS
Yeast Expression Vectors.
The Matchmaker Two-Hybrid System 2 (CLONTECH) was used to identify p75 interacting proteins, except that the cDNA library was cloned into the pGAD424 activation plasmid. Schwann cell cDNA was subcloned into EcoRI (5′) and PstI (3′) sites of pGAD424 GAL4 activation domain vector by using synthetic linkers. This library was a generous gift from Greg Lemke (Salk Institute). The bait was constructed by subcloning the rat p75 cDNA containing amino acids 244–399 into EcoRI (5′) and BamHI (3′) sites of the pAS2–1 vector to generate an in-frame fusion protein with the GAL4 DNA binding domain. The Saccharomyces cerevisiae strain Y190 was used for transformations. It contains GAL1 upstream activating sequences in front of both HIS3 and LacZ reporter genes and is responsive to GAL4 transcriptional activation. The strain was maintained on yeast extract/peptone/dextrose full media and was grown to log phase at 30°C, and then was transformed and plated onto Leu−Trp−His− selective media to allow for the growth of double transformants, which show an interaction. Positive colonies were isolated, were replated on a master plate, and were assayed for LacZ expression by a qualitative colony β-galactosidase activity assay. The library plasmid from double positive yeast colonies was rescued by transformation in HB101 and was subjected to DNA sequencing.
Isolation, Expression, and in Vitro Translation of SC-1 cDNA.
A 2.1-kilobase (kb) clone containing the 3′ portion of the SC-1 cDNA was identified by the yeast screen and was used to isolate a full length 4-kb cDNA from a rat E13 brain cDNA library (Cary Lai, Scripps Institute). The cDNA was rescued into pBS by use of a helper phage. Total RNA (20 μg) was isolated from adult rat tissues and cell lines by using Trizol reagent (GIBCO/BRL) and was separated by agarose gel electrophoresis. RNA was transferred to nylon membranes (Qiagen, Chatsworth, CA) and was hybridized with a 578-base pair EcoRV/EcoRI fragment of SC-1 cDNA or a rat p75 cDNA. A coupled in vitro transcription and translation system (Promega) was used to express the full length SC-1 cDNA. The reaction was carried out at 30°C for 2 hours, and the products were verified by SDS/PAGE. The gel then was dried, fixed, and enhanced and exposed to x-ray film overnight to visualize the translation product.
Binding Assay.
In vitro translated full length SC-1 was incubated with human glutathione S-transferase (GST)–p75 fusion proteins immobilized on glutathione-Sepharose beads in 500 μl of binding buffer (10 mM Hepes/142 mM KCl/5 mM MgCl2/1 mM EDTA/0.2% Nonidet P-40) supplemented with protease inhibitors and 1% BSA. The binding was carried out at 4°C for 4–16 hours, and the beads then were washed three times with 1 ml of binding buffer without BSA. Bound proteins were eluted from the beads by boiling the latter in SDS/PAGE loading buffer for 5 min and were separated by a 7% SDS/PAGE. The gels then were fixed and exposed to x-ray film overnight for visualization of the proteins bound to the beads. The GST-p75 constructs contained the NH-terminal part including amino acids 250–322 and the COOH-terminal containing amino acids 321–399.
Cell Culture.
COS cells were maintained in DMEM supplemented with 10% FBS. Serum starvation was carried out for indicated time periods. Transfection was carried out by using the calcium phosphate procedure. After transfection, cells were allowed to recover in DMEM supplemented with 10% FBS for 1 day before any further treatments. Recombinant NGF (Genentech) was added at a concentration of 100 ng/ml for indicated times to transfected cell cultures, grown in DMEM and 10% fetal bovine serum.
Mammalian Expression Constructs of SC-1.
The cDNA for SC-1 was subcloned into pFLAG-CMV-2 expression vector (Eastman Kodak) to allow immunological detection of the protein. Two constructs were made: (i) pSC1 contained the full-length SC-1 cDNA; and (ii) pSC2 encoded the original yeast clone, which contained amino acids 490–814. pSC2 was constructed by using PCR with the following primers: 5′-primer 5′CCCAAGCTTAGGAACCGTGAAGAACAG-3′ with a HindIII site (in italics) and 3′-primer 5′TGCTCTAGAGTGGGCTGGTTATGTTTTCCC-3′ containing an XbaI site (in italics) incorporated into the respective primers. pSC1 was generated by a two-step subcloning as follows: SC1pBS was digested with EcoRV and KpnI to release a fragment containing the coding and 3′ untranslated sequences. This fragment was subcloned into pFLAG vector digested with the same enzymes to generate a plasmid termed Fsc1-Kpn; the missing 5′ sequence of the cDNA was amplified by PCR using the following primers: 5′-GGGAAGCTTATGAATGACATGAACTTGAGCCC-3′ with a HindIII site (in italics) and an endogenous ATG in bold and 5′-GGACGCTGTCAGATATCCCATTG-3′ with an endogenous EcoRV site (in italics). This fragment was purified by glass elution (Bio 101) from an agarose gel, digested with HindIII and EcoRV, and subcloned into Fsc-Kpn construct digested with the same enzymes. The construct was verified by DNA sequencing.
Immunoprecipitation and Western Blotting Procedures.
COS cells transfected with either Flag-tagged SC-1 constructs or cotransfected with rat p75 receptor [rat p75 cDNA in pCDNA3 vector from B. Carter (Vanderbilt University)] were washed with cold PBS and were scraped and centrifuged. The cells (from a 10-cm dish) were lysed in 250 μl Nonidet P-40 lysis buffer (150 mM NaCl/0.5% Nonidet P-40/50 mM Tris, pH 7.5) supplemented with protease inhibitors. Lysates were incubated on ice for 20 min and were subjected to preclearing with protein A beads (Amersham Pharmacia). The supernatants then were collected and subjected to immunoprecipitation performed by using anti-Flag antibodies, followed by Western blotting. A polyclonal antibody against the cytoplasmic region of the p75 receptor was used at a 1:5,000 dilution for immunoblots. An anti-Flag antibody (Kodak) was used at a final concentration of 1 μg/ml. Proteins were visualized by using the enhanced chemiluminescence (ECL) procedure (Amersham Pharmacia).
Immunostaining Procedures. COS cells were transiently transfected and then were fixed in cold methanol/acetone and stained with the primary anti-Flag antibody at a 1 μg/ml concentration in PBS supplemented with 3% fetal calf serum for 1 hour. Secondary anti-mouse biotin coupled antibody (Vector Laboratories) then was added at a 1:500 dilution for 1 hour, followed by 1 hour with FITC-avidin complex (Vector Laboratories; 1:250). When two different antibodies were used to detect cotransfected cells, the following procedure was used. After anti-Flag staining, the cells were blocked for 1 hour in PBS with 10% goat serum, and primary antibodies were added at 1:500 for anti-p75 polyclonal antibody 9651 (11) and 1:500 for anti-TrkA polyclonal antibody (12) for 1 hour. Anti-rabbit biotin coupled secondary antibodies (Vector Laboratories) were added at 1:500 dilution for another hour, and a rhodamine-avidin complex (Vector Laboratories) then was added at a 1:250 dilution to the cells for an additional hour.
For BrdUrd labeling, COS cells cotransfected with SC-1 and p75 cDNAs were treated with NGF for 1 hour and 10 μM BrdUrd for an additional 3 hours. Cells were fixed in cold methanol/acetone at room temperature for 5 min and were stained with anti-Flag antibody as described above. Subsequently, stained cells were treated with 2 M HCl for 10 min at room temperature and were neutralized with freshly made 0.1 M Borax (pH 8.6) for 10 min. Monoclonal anti-BrdUrd IgG1 (Dako) was applied at a 1:100 dilution in PGBA buffer (0.1 M phosphate buffer/0.1% gelatin/1% BSA/0.002% sodium azide) for 3 hours at room temperature or overnight at 4°C. Anti-mouse IgG1-Texas Red-conjugated antibody (South Biotech, Birmingham, AL) was added at a 1:100 dilution in PGBA for 1 hour at room temperature. The slides were mounted by using PBS and glycerol.
RESULTS
Isolation of p75 Interacting Protein, SC-1. A yeast expression library made from rat Schwann cell cDNA was screened for p75 interacting proteins. The entire cytoplasmic domain of the rat p75 receptor was used as a bait. One cDNA clone, called SC-1, activated both HIS3 and lacZ transcription when grown with p75 but not with laminin or a truncated Gal4 protein. SC-1 was of interest because of the presence of multiple zinc finger motifs, which also are found in tumor necrosis factor receptor-associated factor proteins that are associated with the tumor necrosis factor receptor (13) and with the p75 receptor (14).
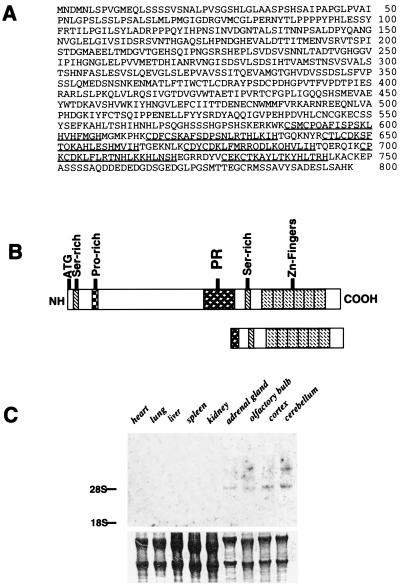
A full length cDNA of 4 kb was isolated from a rat E13 whole brain library. The predicted amino acid sequence of SC-1 indicates an ORF of 798 amino acids corresponding to a protein of 89 kDa, assuming that the first of three contiguous methionines is used as the start codon. Six zinc-fingers motifs are located at the carboxyl terminus of SC-1. The zinc fingers are of the Kruppel type, with two canonical histidine and cysteine residues (underlined in Fig. 1A). The region downstream of the zinc fingers between amino acids 749 and 791 has a highly acidic domain, containing a predominance of glutamic and aspartic acids (32%). At the N terminus are regions extremely rich in serine residues and proline sequences. This sequence represented a potential PEST sequence, which is associated with an increased sensitivity to intracellular degradation.
Figure 1.
Sequence and RNA analysis of SC-1. (A) Full-length amino acid sequence of the rat SC-1. The PR domain is indicated in italics. The zinc fingers are underlined. (B) Schematic representation of the SC-1 amino acid sequence. Six zinc fingers are located at the COOH terminus. Two serine-rich regions are indicated by steepled boxes, a proline-rich region is indicated as the squared box, and the PR domain is indicated. Below, a schematic representation of the original yeast clone coding sequence is shown for comparison containing amino acids 490–814 (truncated SC1). (C) Northern blot of SC-1 mRNA in adult rat tissues (20 μg total RNA). The blot was stained with methylene blue to visualize the amount of RNA loaded onto each lane (Lower). (D) SC-1 and p75 receptor mRNA expression in cultured Schwann cells and PC12 cells (10 μg total RNA each lane). Lanes: 1, proliferating Schwann cells; 2, quiescent Schwann cells; 3, PC12 cells. (E) An alignment of the PR domains from SC-1, Blimp, RIZ, evi1, and egl-43. The identical amino acids are boxed and shaded. Dashes indicate sequence gaps.
A motif present in SC-1 between amino acids 421 and 528, shown in Fig. 1B, displayed homology to the PR domain (18). This domain was identified in several other zinc finger-containing proteins, PRDI-BF-1 (15) or Blimp-1 (16, 17), RIZ (18), the C. elegans egl-43 gene product (19), and Evi1 (20). An alignment of these domains is presented in Fig. 1E. The PR domain most closely related to SC-1 is that of Blimp, with 32.2% similarity, followed by RIZ, with 26.7% similarity. The PR sequence and the absence of a tumor necrosis factor receptor-associated factor domain makes SC-1 structurally different from the zinc finger proteins of the tumor necrosis factor receptor-associated factor subfamily (21).
To investigate the tissue distribution of SC-1, a Northern blot analysis was performed. SC-1 mRNA was found mainly in neuronal tissues, with one transcript migrating at 4.8 kb and an additional higher transcript of 6 kb found in the cerebellum (Fig. 1C). Expression of SC-1 mRNA also was detected in growing Schwann cells and PC12 cells, along with p75 mRNA (Fig. 1D).
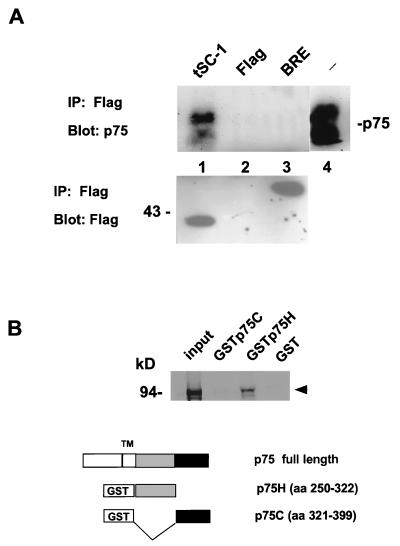
Association of SC-1 with p75. To verify the interaction between the cytoplasmic domain of p75 and SC-1, the original yeast clone was introduced into a mammalian vector with a Flag-epitope to generate pSC2. This construct was cotransfected with pcDNA3p75 in 293 cells and was subjected to immoprecipitation with anti-Flag antibodies. The p75 receptor was detected in a complex with the C-terminal SC-1 protein after immunoprecipitation (Fig. 2A). As controls, expression of the Flag vector alone and an unrelated Flag-tagged protein, Flag-Bre (22), did not result in coprecipitation of p75. Expression of Flag-tagged proteins was monitored by anti-Flag antibodies (Fig. 2B). The results of these experiments indicate that the C-terminal half of SC-1 was sufficient for binding p75.
Figure 2.
Interaction of p75 and SC-1. (A) Transfected 293 cells expressing the rat p75 cDNA; an unrelated protein, Flag-tagged BRE, and a Flag-tagged truncated SC-1 cDNA (pSC2; amino acids 490–814; Fig. 1B) were subjected to immunoprecipitation using anti-Flag antibodies, followed by blotting with the anti-p75 antibody, 9992. The arrowheads demonstrate the position of p75 on the blot, which migrated as a doublet, representing products of differential glycosylation of p75. Lanes: 1, p75 and pSC2; 2, p75 and Flag vector; 3, p75 and an unrelated protein Flag-Bre cDNA (22); 4, 293 cells transfected with p75 cDNA alone, probed with anti-p75, 9992 antibody. In the lower panel, the transfected proteins were detected after immunoprecipitation and Western blotting with anti-Flag antibodies. (B) SC-1 interacts with the juxtamembrane domain of human p75. GSTp75 fusion proteins were used for assessing the binding of in vitro translated SC-1 to p75 intracellular sequences. The binding reaction was carried out by using 3 μg of the GSTp75 fusion proteins on glutathione beads. The left-most lane represents input in vitro translated SC-1 protein. The proteins were resolved on a 7% SDS/PAGE gel. The arrowhead indicates the migration of the major in vitro translated product of SC-1.
As another means of confirming the association of SC-1 with the p75 receptor, we used GST-p75 fusion protein constructs to examine the binding of SC-1. Full length SC-1 was translated in vitro and was tested for binding to GST-p75 fusion proteins. The p75 intracellular domain contains a sequence that is related to the death domain (23) and also the death effector domain (24). Binding of SC-1 was found with a GST fusion construct containing the juxtamembrane region of the receptor but not the C-terminal portion containing the death domain of the receptor (Fig. 2B). Binding to GST protein was not observed under these conditions. It is noteworthy that the juxtamembrane region is the most evolutionarily conserved domain of the p75 receptor (25).
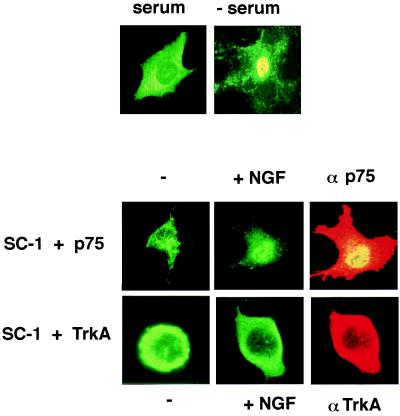
Effect of Neurotrophins on SC-1 Localization. To investigate the subcellular distribution of SC-1, we introduced the full length SC-1 into a Flag vector to produce a construct, pSC1. Expression of this protein was monitored by transfection into COS cells followed by indirect immunofluorescence using anti-Flag antibodies. SC-1 adopted a mostly cytoplasmic localization after transient transfection (Fig. 3).
Figure 3.
Subcellular localization of SC-1 is regulated by both serum and NGF. Flag-tagged SC-1 (pSC1) was either transfected alone or in combination with either p75 or TrkA into COS cells. Transfected cells were stained with anti-Flag antibodies and appropriate receptor antibodies followed by the secondary antibodies coupled to FITC for SC-1 or rhodamine for p75 or TrkA staining. (Upper) COS cells transfected with SC-1 in 10% fetal calf serum and after 1 hour of serum withdrawal. (Lower) COS cells were transfected with SC-1 and p75 cDNA or with SC-1 and TrkA cDNAs. Immunostaining for Flag-SC-1 was carried out before and after 1 hour of treatment with 100 ng/ml NGF. αp75 and αTrkA panels represent expression p75 and TrkA receptors after staining with 9651 (11) or RTA antibodies (12).
COS cells also were cotransfected with SC-1 and p75 cDNAs. The subcellular distribution of SC-1 protein was monitored with anti-Flag antibodies whereas expression of p75 receptors was detected with an anti-p75 antibody, 9651, which recognizes the extracellular domain of the receptor (11). Addition of NGF to cotransfected COS cells shifted the distribution of SC-1 toward a more nuclear localization (see Fig. 3). This effect of NGF could be observed after 1 hour of treatment. A quantitation of the subcellular distribution of cells expressing SC-1 was carried out by counting the number of cells with a cytoplasmic, nuclear, or intermediate distribution (Fig. 4). An intermediate state reflected a cell in which SC-1 protein was found both in the cytoplasm and nucleus. Also, the relocalization of SC-1 appeared to be specific for NGF as treatments with brain-derived neurotrophic factor and neurotrophin 3 did not considerably change the cytoplasmic distribution of SC-1 (data not shown). Other differences in the behavior of neurotrophins after binding to p75 have been observed (9, 26). It is not known what accounts for differential neurotrophin responsiveness, but the differences may be attributable to alternative conformational states of bound p75 (4) or differential recruitment of substrate molecules (14).
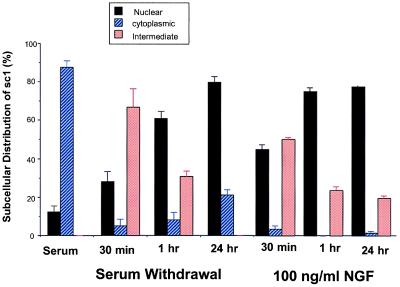
Figure 4.
Quantitation of the subcellular distribution of SC-1. Shown is quantitation of the effects of serum starvation and NGF treatment on the subcellular distribution of SC-1 in transfected COS cells. The total number of cells displaying SC-1 expression was measured and is represented as a percentage (%). Three different states were monitored: nuclear, cytoplasmic, or intermediate staining. Statistical analysis was carried out by using the χ2 test. The P values obtained varied between P < 0.001 and P < 0.01.
Effect of Serum. The distribution of SC-1 was also dramatically affected by serum conditions. After withdrawal of 10% fetal calf serum for 30 min, the distribution of SC-1 shifted to a predominately nuclear location (Fig. 3). To investigate the kinetics of SC-1 translocation to the nucleus, a time course of serum withdrawal from the transfected COS cells was carried out. We measured the localization of SC-1 after 30 min, 1 hour, and an overnight withdrawal of serum. The time course of serum withdrawal revealed that, after 30 min, >60% of the SC-1 transfected cells are found in the nucleus. By 1 hour, nearly 30% of the transfected cells SC-1 displayed immunoreactivity in the nucleus. After overnight withdrawal of serum, >80% of the transfected cells displayed SC-1 protein in the nucleus. The results summarized in Fig. 4 indicated that there is a progressive increase in the number of cells in which SC-1 protein was found in the nucleus.
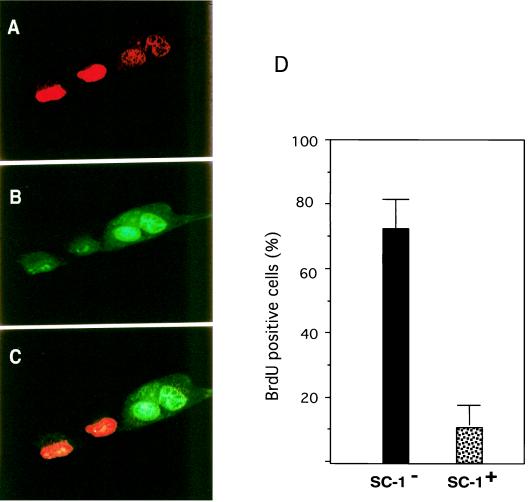
Because SC-1 belongs to a family of proteins implicated in growth arrest and cell differentiation events (10, 16, 17, 27), we assessed whether the expression of SC-1 was related to the proliferative state of COS cells. To test whether these cells had withdrawn from the cell cycle, we measured the incorporation of BrdUrd in SC-1 transfected cells. To this end, we cotransfected SC-1 and p75 into COS cells, subjected the cells to treatment with NGF for 1 hour, and then labeled with BrdUrd for another 3 hours. Staining of these cells with antibodies against BrdUrd and Flag-SC-1 revealed that those cells that were positive for BrdUrd were negative for SC-1 expression (Fig. 5) or displayed a predominately cytoplasmic distribution. The presence of SC-1 in the nucleus correlated with the lack of BrdUrd incorporation. Although speculative, in light of the function of PR proteins (17, 27), SC-1 may play a role in events leading to growth arrest.
Figure 5.
Incorporation of BrdUrd after transfection of COS-1 cells with SC-1. Expression of SC-1 was detected by using anti-Flag antibody (Kodak) and by FITC-coupled secondary antibody. Incorporation of BrdUrd in COS cells was detected by using an anti-BrdUrd primary antibody and a Texas-Red-coupled secondary antibody. (A) COS cells stained for BrdUrd. (B) COS cells stained for Flag-SC-1. (C) An overlay of A and B showing that the BrdUrd and SC-1 immunoreactivities are mutually exclusive. (D) Quantitation of BrdUrd-positive COS cells transfected with SC-1.
Effect of TrkA on SC-1 Distribution.
Because NGF was capable of inducing SC-1 translocation into the nucleus in a sustained manner, we asked whether expression of the TrkA NGF receptor also would change the distribution of SC-1 protein. We therefore cotransfected a rat TrkA cDNA with SC-1 into COS cells. SC-1 was found to be predominantly cytoplasmic after transfection (Fig. 3). No difference in its localization could be observed between untreated cells and NGF treated cells (Fig. 3). Expression of the TrkA receptor with SC-1 in COS cells prevented SC-1 from entering the nucleus under serum-poor conditions, as well as on NGF treatment. The expression of TrkA receptors was verified by using an anti-TrkA antibody (RTA; Fig. 3) that recognizes the extracellular domain of rat TrkA receptor (12). NGF binding to TrkA receptors did not have an influence on the distribution of SC-1 in COS cells, in direct contrast to the p75 receptor. Also, in cells transfected with SC-1, TrkA, and p75 cDNAs, NGF treatment did not lead to a nuclear localization for SC-1 (data not shown). The two NGF receptors therefore give divergent responses through downstream signaling components (28).
DISCUSSION
Neurotrophins promote cell differentiation and survival through a two-receptor system, involving Trk tyrosine kinase receptors and the p75 receptor, a member of the tumor necrosis factor receptor superfamily. Here, we describe the cloning of a zinc finger protein that interacts with the p75 receptor. SC-1 is a protein of 814 amino acids whose subcellular distribution is regulated by NGF and serum withdrawal. In COS cells transfected with SC-1 cDNA, SC-1 is found predominantly in the cytoplasm. However, on coexpression of SC-1 with p75 and after NGF treatment, the protein translocated to the nucleus. The kinetics of NGF-dependent translocation of SC-1 demonstrated that, after 1 hour of NGF addition, the protein was found in the nucleus. The effect on translocation of SC-1 relied on coexpression of p75 with SC-1 and was not observed in COS cells transfected with both TrkA and SC-1.
Under the normal serum-rich growth conditions, SC-1 protein is found predominantly in the cytoplasm. Serum starvation of the cells induced the translocation of this protein to the nucleus. Cotransfection of the TrkA receptor with SC-1 in COS cells prevented SC-1 from entering the nucleus after NGF treatment and under serum-poor conditions. It is interesting to note that TrkA behaves in a mitogenic manner in fibroblast or non-neuronal cells (29, 30). Engagement of TrkA by NGF in fibroblastic cells might result in a proliferative state, thereby preventing SC-1 from entering the nucleus.
The effect of serum withdrawal on the subcellular localization of SC-1 led us to test whether cells with nuclear accumulation of this protein could incorporate BrdUrd as a measure of their replicative capacity. Cells expressing nuclear SC-1 did not exhibit BrdUrd incorporation. Cells incorporating BrdUrd either did not express SC-1 or expressed the protein only in the cytoplasm. This indicated a correlation between the distribution of SC-1 and the proliferative state of the cell. Thus, the subcellular distribution for SC-1 during growth conditions and its accumulation in the nucleus after serum withdrawal may be associated with changes in cell cycle progression.
Previously, increased expression of p75 was shown to increase cell cycle arrest of sympathoadrenal cells, MAH cells, that concomitantly express TrkA (31). The mechanism by which p75 exerts this effect is unclear, but the regulation of SC-1 distribution may play a role. It is interesting in this respect that, when coexpressed with TrkA, SC-1 shows exclusively cytoplasmic distribution in COS cells. Because the TrkA can act to transform cells or to promote mitogenesis, the restriction of SC-1 from a more nuclear localization may represent a mechanism to prevent growth arrest. It is possible that the integration of signals from p75 and TrkA are responsible for the coordinated induction of cell cycle arrest and eventual differentiation of the cells of neural lineage or, alternatively, the maintenance of the differentiated state of the mature cells. This hypothesis is substantiated by the observation that Blimp-1, a protein with homology to SC-1, regulates the maturation of B cells by repressing transcription of the c-myc gene (17) and other phenotypic traits (27).
A comparison of the sequence of SC-1 to other related proteins is informative. Other proteins that contain zinc finger sequences and the PR domain include the C. elegans egl-43 gene product (19), the EVI1/MDS1 fusion protein (20), and the RIZ protein (18), as well as the mouse Blimp-1 protein, which is homologous to human PRD1-BF-1 (15, 16). The comparison of the zinc finger sequences revealed that the closest relative of SC-1 is the egl-43 protein, showing 33.3% similarity. It is followed by the Blimp-1 with 25.4% and RIZ with 14% similarity. Egl-43 is required for the development of sensory neurons and the migration of the hermaphrodite-specific neurons involved in the control of egg laying (19). Blimp-1 regulates the late stage of B cell differentiation by down-regulating the expression of c-myc (17). The PRDI/Blimp-1 protein also acts as a transcriptional repressor of the β-interferon promoter (10) and exerts an apoptotic outcome after overexpression (27). The RIZ protein binds to the Rb protein (18). A role for RIZ during cell cycle arrest and apoptosis through a transcriptional repression mechanism has been recently proposed (32, 33).
The PR domain is related to the Su(var)enhancer of zeste trithorax domain, a conserved 130-amino acid sequence that is implicated in regulating chromatin function (33, 34). Comparison of the PR domains of these proteins revealed that the closest to SC-1 is Blimp with 32.2% similarity, followed by RIZ with 26.7% similarity and then by evi1 with 23.5% similarity. The PR domain of RIZ appears to be responsible for establishing protein–protein interactions. The molecular mechanism by which SC-1 is translocated into the nucleus and its roles in modulating cell cycle arrest and other neurotrophin differentiation functions are unknown. Neither is it known whether other PR domain zinc finger proteins undergo similar translocation events. It is likely that future studies of the phosphorylation status of SC-1 and its downstream target genes will reveal more fully the functions of this class of zinc finger proteins.
Acknowledgments
We thank Greg Lemke for the cDNA library; Louis Reichardt for the TrkA antisera; and Long Cao, Haeyoung Kong, Ravi Tikoo, Gus Khursigara, Margaret Tomaska, Patrizia Casaccia-Bonnfil, and Chenghua Gu for help and advise during the course of this project. The generosity of Michael Sendtner’s laboratory at the University of Wurzburg is gratefully acknowledged. We thank Stefan Wiese for his assistance with confocal microscopy. This work was supported by the National Institutes of Health.
ABBREVIATIONS
- NGF
nerve growth factor
- PR domain
positive regulatory domain
- kb
kilobase
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF176023).
References
- 1.Levi-Montalcini R. Science. 1987;237:1154–1164. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 2.Thoenen H. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- 3.Lewin G R, Barde Y-A. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 4.Bothwell M. Annu Rev Neurosci. 1995;18:223–253. doi: 10.1146/annurev.ne.18.030195.001255. [DOI] [PubMed] [Google Scholar]
- 5.Chao M V, Hempstead B L. Trends Neurosci. 1995;19:321–326. [PubMed] [Google Scholar]
- 6.Rabizadeh S, Oh J, Zhong L T, Yang J, Bitler C M, Butcher L L, Bredesen D E. Science. 1993;261:345–348. doi: 10.1126/science.8332899. [DOI] [PubMed] [Google Scholar]
- 7.Barrett G L, Bartlett P F. Proc Natl Acad Sci USA. 1994;91:6501–6505. doi: 10.1073/pnas.91.14.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frade J M, Rodriguez-Tebar A, Barde Y-A. Nature (London) 1996;383:166–168. doi: 10.1038/383166a0. [DOI] [PubMed] [Google Scholar]
- 9.Casaccia-Bonnefil P, Carter B D, Dobrowsky R T, Chao M V. Nature (London) 1996;383:716–719. doi: 10.1038/383716a0. [DOI] [PubMed] [Google Scholar]
- 10.Ren B, Chee K J, Kim T H, Maniatis T. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber L J, Chao M V. Dev Biol. 1995;167:227–238. doi: 10.1006/dbio.1995.1019. [DOI] [PubMed] [Google Scholar]
- 12.Clary D O, Weskamp G, Austin L R, Reichardt L F. Mol Biol Cell. 1994;5:549–563. doi: 10.1091/mbc.5.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 14.Khursigara G, Orlinick J, Chao M. J Biol Chem. 1999;274:2597–2600. doi: 10.1074/jbc.274.5.2597. [DOI] [PubMed] [Google Scholar]
- 15.Keller A D, Maniatis T. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 16.Turner C A, Mack D H, Davis M M. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y, Wong K-k, Calame K. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 18.Buyse I M, Shao G, Huang S. Proc Natl Acad Sci USA. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garriga G, Guenther C, Horvitz H R. Genes Dev. 1993;7:2097–2109. doi: 10.1101/gad.7.11.2097. [DOI] [PubMed] [Google Scholar]
- 20.Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley J D, Nucifora G. Proc Natl Acad Sci USA. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arch R H, Gedrich R W, Thompson C B. Genes Dev. 1998;12:2821–2830. doi: 10.1101/gad.12.18.2821. [DOI] [PubMed] [Google Scholar]
- 22.Gu C, Castellino A, Chan J-H, Chao M V. FASEB J. 1998;12:1101–1108. doi: 10.1096/fasebj.12.12.1101. [DOI] [PubMed] [Google Scholar]
- 23.Liepinsh E, Ilag L L, Otting G, Ibanez C F. EMBO J. 1997;16:4999–5005. doi: 10.1093/emboj/16.16.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberstadt M, Huang B, Chen Z, Meadows R P, Ng S-C, Zheng L, Lenardo M J, Fesik S W. Nature (London) 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 25.Large T H, Weskamp G, Helder J C, Radeke M J, Misko T P, Shooter E M, Reichardt L F. Neuron. 1989;2:1123–1134. doi: 10.1016/0896-6273(89)90179-7. [DOI] [PubMed] [Google Scholar]
- 26.Carter B D, Kaltschmidt C, Kaltschmidt B, Offenhauser N, Bohm-Matthaei R, Baeuerle P A, Barde Y-A. Science. 1996;272:542–545. doi: 10.1126/science.272.5261.542. [DOI] [PubMed] [Google Scholar]
- 27.Messika E J, Lu P S, Sung Y-J, Yao T, Chi J-T, Chien Y-h, Davis M M. J Exp Med. 1998;188:515–525. doi: 10.1084/jem.188.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greene L A, Kaplan D R. Curr Topics Neurobiol. 1995;5:579–587. doi: 10.1016/0959-4388(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 29.Coulier F, Kumar R, Ernst M, Klein R, Martin-Zanca D, Barbacid M. Mol Cell Biol. 1990;10:4202–4210. doi: 10.1128/mcb.10.8.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein R, Jing S, Nanduri V, O’Rourke E, Barbacid M. Cell. 1991;65:189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- 31.Verdi J M, Birren S J, Ibanez C F, Persson H, Kaplan D R, Benedetti M, Chao M V, Anderson D J. Neuron. 1994;12:733–745. doi: 10.1016/0896-6273(94)90327-1. [DOI] [PubMed] [Google Scholar]
- 32.Xie M, Shao G, Buyse I M, Huang S. J Biol Chem. 1997;272:26360–26366. doi: 10.1074/jbc.272.42.26360. [DOI] [PubMed] [Google Scholar]
- 33.Huang S, Shao G, Liu L. J Biol Chem. 1998;273:15933–15939. doi: 10.1074/jbc.273.26.15933. [DOI] [PubMed] [Google Scholar]
- 34.Jenuwein T, Laible G, Dorn R, Reuter G. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]