
The Optical Analysis of Biomolecular Machines meeting was held between 13 and 16 July 2006 at the Max Delbrück Center for Molecular Medicine in Berlin, Germany, and was organized by C. Cardoso and C. Cremer
Introduction
This meeting was attended by 160 cell biologists, biophysicists and imaging experts. The goal was to assess the current status of spatial resolution in optical microscopy in both fixed and living cells, with the idea that this cast of experts might not only accurately describe, and therefore define, the state of the art, but might also put their heads together and devise a quantum leap in the technology. It was an excellent meeting on the first score, and, although not as successful in achieving the broader second goal, a potentially historic agenda for a new era of optical microscopy had come into view by the time the meeting drew to a close.
The plenary lecture by R. Kornberg (Stanford, CA, USA) on the atomic resolution of the transcriptome set a high standard for what was to follow. X-rays do not have the same energy as the light that cell biologists use, but the implication was there nonetheless: this resolution is where one needs to be. Specifically, Kornberg discussed eukaryotic RNA polymerase II (pol II) and its numerous accessory factors, which are assembled into a machine with a mass of around 3 MDa. His biochemical results revealed details of nucleosome displacement on the upstream activator sequence of most pol-II-transcribed genes. Furthermore, his structural analysis indicates how a clamp-like portion of the machine interacts with the DNA transcription bubble and an 8–9 base-pair DNA–nascent RNA hybrid region. Other key findings from the surface representation of the structure are that: the DNA helix is energetically enforced onto the polymerase surface by the TATA-box-binding factor; transcription factor TFIIB directs the DNA path and stabilizes the early transcription complex; TFIIF captures the non-template DNA strand; and TFIIH induces negative superhelicity. What remains to be solved to complete the picture of this machine is the structure of the ‘mediator'—a 1-MDa complex that facilitates transcription once it has been initiated. Kornberg's closing remarks left little doubt that this goal will soon be attained. His talk was an inspiring launch for the meeting.
The sessions were organized so as to juxtapose presentations on advances in microscopy itself (for example, new modes of illumination and image processing) with talks on specific topics, such as fluorescent-tagging advances or single-molecule detection, together with numerous presentations on cell-biological applications. The presentations covered in this report were selected on no basis other than with the hope of providing an accurate taste of the conference's content in the epistemological sense.
Fiat lux
Recent advances have broken the theoretical diffraction-limited spatial resolution of light microscopy (around 0.2 μm) given by Ernst Abbe's famous equation relating resolution to the wavelength of incident light and the numerical aperture of the objective (Fig 1). A major contribution has been the development, by S. Hell (Göttingen, Germany), of stimulated emission depletion (STED) microscopy (reviewed by Weiss, 2000). In this technique, reversible and saturable optical transitions between two states of a fluorescent object are exploited so that the saturation level defines the size of the ultra-sharp focal spot and/or the expanded optical bandwidth. The consequence is to convert Abbe's equation to a special case, in which the effective numerical aperture is defined by the ratio of the applied intensity to the saturation intensity. Hell reported a recent application of STED microscopy to an investigation of soluble NSF attachment protein (SNAP)-receptor (SNARE) complexes in neuronal cells, resolving SNAP25 and syntaxin 1A as around 60 and 75 nm patches, respectively (see also Donnert et al (2006), in which resolution approaching 20 nm is reported). It would not be an exaggeration to say that in far-field fluorescence microscopy, a diffraction-based limit of resolution no longer exists.
Figure 1.
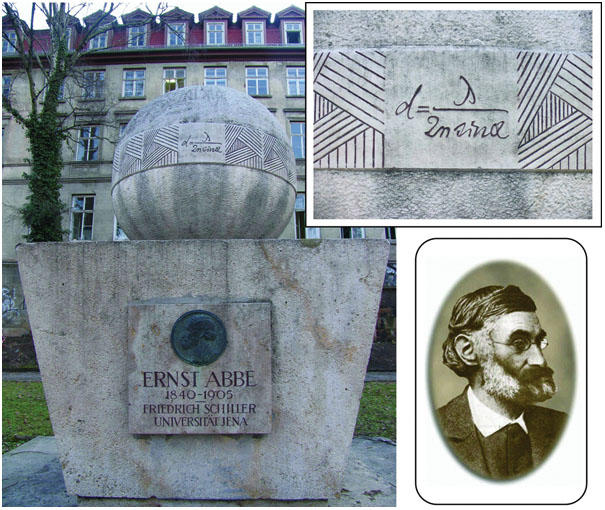
Celebrating Abbe and his equation. This monument is located in front of the Physiologisches Institut, University of Jena, Germany. The lower right inset depicts Ernst Abbe. The Carl Zeiss Company in Jena hired Abbe as a consultant, which was a key factor in the evolution of light microscopy and its commercial products.
Another major advance in optical microscopy has been the advent of structured illumination, which was addressed by R. Heintzmann (London, UK). This important approach is related to ‘4π microscopy', in which two opposing objectives are used to produce a standing wave superimposed on the opposing focused beams, and the constructive interference is enhanced relative to the destructive interference. As performed at present, 4π microscopy operates down to about one-seventh of the excitation wavelength and affords a z-axis resolution of around 100 nm, which is a tremendous boon for many cell biological applications. Heintzmann emphasized that there are currently two ways to circumvent the diffraction limit in optical microscopy: by saturation of stimulated emission (that is, STED) or by nonlinear structured illumination. His version of the latter approach involves patterned excitation, in which images are collected from illumination conditions created by a series of sinusoidal grids placed at different positions and orientations in the optical path, resulting in an approximately twofold (linear case) or fivefold (nonlinear case; data from M. Gustafsson, San Francisco, CA, USA) increase in resolution relative to standard wide-field microscopy.
Other new microscopy approaches were discussed, including the structured illumination-based system developed at the University of California, San Francisco, by Gustafsson and J. Sedat, which was presented at the meeting by L. Schermelleh (Planegg-Martinsreid, Germany). J. Enderlein (Jülich, Germany) gave a talk on dynamic optical saturation microscopy, which is based on the fast collection of fluorescence decay after instantaneous excitation that reduces the diffraction limit by a factor of approximately four. How these creative emerging systems will ultimately advance cell biology remains to be seen, but the first steps look extremely promising (Gustafsson, 2005).
Single-molecule detection is a longstanding goal of biological optical microscopy, particularly in living material. Of course, some molecules are larger than others, and the advances in spatial resolution mentioned above might themselves soon embrace a large molecule, such as the ribosome (diameter, around 25 nm). While awaiting further advances in spatial resolution, other innovations are opening the door to single-molecule detection. U. Kubitscheck (Bonn, Germany) described an approach based on focal-plane illumination (Fig 2A). His system creates a light sheet (height × width × thickness: 20 μm × 20 μm × 2 μm) within the focal plane, using a custom-built cylindrical lens (numerical aperture, 0.33), and the emitted fluorescence is detected using a ×60 numerical aperture 1.1 objective oriented perpendicular to the illumination path (Fig 2B). The resulting reduction in background light creates a significant increase in the signal-to-noise ratio, which is the key factor in single-molecule-detection sensitivity.
Figure 2.
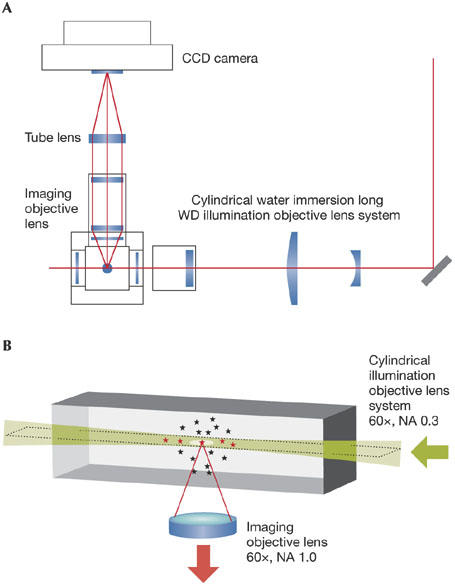
Focal-plane illumination for single-molecule detection. (A) Schematic of the illumination path and microscopy system. The resulting elimination of light from above and below the focal plane enhances single-molecule detection, typified by low signal intensities. (B) Perpendicular illumination into the focal plane.CCD, cooled charge-coupled device; NA, numerical aperture; WD, working distance.
Why fret about FRET?
Although advances in methods such as fluorescence cross-correlation microscopy and fluorescence correlation spectroscopy were not emphasized at the conference, progress in fluorescence resonance energy transfer (FRET) methods was reported by S. Rüttinger (Berlin, Germany). FRET is a non-radiative energy transfer phenomenon, in which excitation of a donor fluorophore causes transfer of resonance energy to an appropriate acceptor in close proximity (typically 1–10 nm). The magnitude of energy transferred is related inversely to the intermolecular distance (Stryer & Haugland, 1967). Rüttinger presented pulsed interleaved excitation (PIE)–FRET, which is a method that eliminates problems such as the zero-efficiency peak in the energy transfer profile. Using single-photon counting and by analysing the same data sets with fluorescence correlation spectroscopy, he was also able to quantify factors that compromise intensity ratio-based FRET measurements. It will be interesting to see the extent to which this new PIE–FRET approach can enhance the investigation of molecular interactions in living cells.
Reports of reporters
Ever since a fluorescently tagged antibody was first used to detect an intracellular target (Coons et al, 1942), the application of fluorescent probes has defined a major sector of modern cell biology, as investigated at the light microscopy level. The conference had a superb session on new approaches to the design and implementation of novel fluorescent reporters of cellular biochemistry. M. Fernandez Suárez from the A. Ting group (Cambridge, MA, USA) described fluorescent-labelling strategies based on enzyme-catalysed reactions performed in living cells. She explained how placing an acceptor peptide domain into an expressed protein of interest targets it for biotin coupling through co-expressed Escherichia coli biotin ligase (Fig 3A). This same enzyme can also conjugate ketone analogues of biotin to targets, thereby producing a desirable hydrazide functionality (Fig 3B). Other such systems in development by this group have the additional attractive feature of limiting the protein-labelling reaction to a particular intracellular organelle, on the basis of the restricted localization of the exploited enzyme to that compartment.
Figure 3.
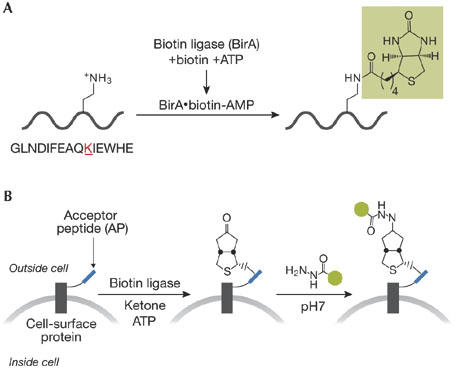
Derivatizing proteins in vivo. (A) Coupling of biotin to a protein implanted with an acceptor sequence, through biotin ligase. (B) Coupling of a ketone derivative of biotin to a target protein, again by biotin ligase, followed by hydrazide-mediated labelling with a desired tag depicted by the green circle.
D. Lorenz (Berlin, Germany) described new coumarinylmethyl esters of cyclic nucleoside monophosphates. The most promising of these compounds have high aqueous solubility, good cellular uptake, low rates of spontaneous hydrolysis of the caging group, high efficiencies of photoactivation and low cellular toxicity of the photolysed leaving groups. Lorenz has used the 8-bromoguanosine 3′,5′-cyclic monophosphate member of this reagent panel with two-photon activation to capture the instantaneous influx of Ca2+ ions into human cells expressing a cGMP-gated ion channel. Similar to the other talks in the session, this presentation coupled chemical biology with state-of-the-art microscopy and gave the audience a delightful taste of things to come. It is hoped that ‘systems biology' investigators will take heed of these empirical foundations—that is, actual experimental data.
A time to every purpose
Although applications to cell biology were evident in some of the aforementioned presentations, this was the primary focus of other sessions. D. Bazett-Jones (Toronto, Ontario, Canada) described electron spectroscopic imaging (ESI), a mode of energy-loss microscopy in which inelastic collisions of the electron beam with atoms in the specimen register information about the chemical identity of the obstacle. This principle can be used to obtain phosphorus-rich versus nitrogen-rich contour maps that, with appropriate imaging and computational steps, provide a topography of nucleic-acid-rich versus protein-rich domains in the cell. For example, this method has recently revealed that a region of the nucleolus long thought to consist solely of nascent ribosomes is in fact a landscape of ribonucleoprotein particles interspersed among RNA-deficient, protein-rich domains (Politz et al, 2005). Bazett-Jones described the application of ESI and related methods to an analysis of promyelocytic leukaemia (PML) bodies, which are dynamic intranuclear structures that have been implicated in viral replication and cancer. He reported that PML bodies undergo an unexpected degree of fusion and fission during the DNA replication period of the cell cycle, perhaps related to alterations in their normal intimacy with chromatin. The number of PML bodies also doubles during S phase, implying that their formation is linked to the dosage of certain genes.
D. Grünwald (Bonn, Germany) reported studies on the mobility and range of the inert tracer streptavidin (around 60 kDa) within the nucleus. This addresses a longstanding question in nuclear organization about the structure and rheological properties of the ground substance (if any) in the interchromatin space (Pederson, 2002). His results indicate that all regions of the nucleus are surprisingly open for molecular percolation—even heterochromatin and the apparently dense nucleoli—which is in accordance with recent findings from interference microscopy (Handwerger et al, 2005). In a companion presentation that took a prize in the poster competition, Grünwald, Kubitscheck and colleagues applied their single-molecule detection methods to follow the transit of various cargoes through nuclear-pore complexes (NPCs) at a temporal resolution of 5 ms and a localization precision of about 35 nm. Their results point to the co-existence of multiple mechanistically distinct export pathways operating in the NPCs, a welcome development that will enjoin the biochemical camp in this field.
S. Fraser (Pasadena, CA, USA) presented an analysis of heart development in zebrafish embryos, which formed an unusually handsome culmination of biophysics, microscopy and developmental biology expertise. A particularly interesting finding is that, at the earliest stages of heart development, pumping is not the result of peristalsis—a fact that could have passed unnoticed without these imaging approaches. He also reported fascinating results on how blood flow influences heart development. A change in shear force of only 0.2 dyne/cm2 is sufficient to alter the gene expression pattern in the developing heart. The fact that such profound embryological insights can be obtained by these state-of-the-art imaging techniques added to the sense of wonder at this conference.
Ever since it was introduced to study the lateral mobility of proteins and lipids in the plasma membrane, fluorescence recovery after photobleaching (FRAP) has been a popular tool in cell biology, along with its variant method fluorescence loss in photobleaching (FLIP). However, similar to all methods, these have their problems and pitfalls (Sprague & McNally, 2005). S. Gorski (Bethesda, MD, USA) reported a detailed FRAP analysis of the dynamics by which RNA pol I (the enzyme that transcribes the genes for ribosomal RNA) and many of its collaborating proteins associate with the nucleolus, which is the site of ribosome formation. All these molecules constantly shuttle in and out of nucleoli; however, a major change in their average nucleolar residence time occurs during the S phase, when ribosomal gene transcription is maximal. Y. Shav-Tal (Ramat-Gan, Israel) described similar studies in which both a messenger RNA transcript and the polymerase were fluorescently tagged. The design of his experiments emphasized the average residence time of the polymerase on the gene, and revealed, among other things, an extensive degree of transcriptional pausing. R. Singer (New York, NY, USA) presented an overview of the innovative RNA-tracking methods his group has devised, which have, for example, provided new clues to how some mRNAs might be transported in a translationally repressed state to sites of activation.
Summing up
This was the best meeting on microscopy that I have ever attended and my view was shared by many others. As is typical in all meetings these days, too little time was allowed for discussion—not of the usual droll clarifications or minor points made, but unabashed devil's advocacy. On the few occasions when members of the audience rose above the usual etiquette of questions and answers, there were hints of intellectual inspiration (and speaker perspiration), although these moments were sadly infrequent and short-lived. Of course, the field of microscopy does not have provocateurs such as Francis Crick, Max Delbrück and Sydney Brenner who catalysed molecular biology by incessant talking and, in my opinion, we cell biologists are the worse for it. Perhaps Max Delbrück—for whom the host institution is named and who would have been 100 years old on 4 September 2006—was watching in the wings. I think he would have enjoyed the science, but would probably have admonished us to have taken bolder stands in the conference hall and to continue discussions into the early hours.
A few hours after the meeting ended, I walked from my hotel down to the Brandenburg Gate, its reflected light a rich gold in the setting sun. Reflection is always a good thing and, in that moment, I paused to consider whether microscopy might now stand on the edge of another Golden Age.
I believe that it does.

Thoru Pederson
Acknowledgments
I thank S. Hell for Fig 1, U. Kubitscheck for Fig 2 and M. Fernandez Suárez for Fig 3. The author's work is supported by grants from the US National Institutes of Health and the US National Science Foundation.
References
- Coons AH, Creech HJ, Jones RN, Berliner E (1942) The demonstration of pneumococcal antigen in tissues by the use of fluorescent antibody. J Immunol 45: 159–170 [Google Scholar]
- Donnert G, Keller J, Medda R, Andrei AM, Rizzoli SO, Lührmann R, Jahn R, Eggeling C, Hell SW (2006) Macromolecular-scale resolution in biological fluorescent microscopy. Proc Natl Acad Sci USA 103: 11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG (2005) Nonlinear structured illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA 102: 13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Cordero JA, Gall JG (2005) Cajal bodies, nucleoli and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell 16: 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T (2002) Dynamics and genome-centricity of interchromatin domains in the nucleus. Nat Cell Biol 4: E287–E291 [DOI] [PubMed] [Google Scholar]
- Politz JC, Polena I, Trask I, Bazett-Jones DP, Pederson T (2005) A non-ribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell 16: 3405–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, McNally JG (2005) FRAP analysis of binding: proper and fitting. Trends Cell Biol 15: 84–91 [DOI] [PubMed] [Google Scholar]
- Stryer L, Haugland RP (1967) Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci USA 57: 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S (2000) Shattering the diffraction limit of light: a revolution in fluorescence microcopy? Proc Natl Acad Sci USA 97: 8747–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]


