Abstract
We have identified a transcription factor specifically expressed in the developing vertebrate eye. We named this gene vax2 because of the high degree of sequence similarity to the recently described vax1. Both in the human and mouse genomes, vax2 is localized in the vicinity of the emx1 gene. This mapping assignment, together with the previously reported colocalization of Vax1 and Emx2 in mouse, indicates that the vax and the emx genes may be organized in clusters. vax2 has a remarkable expression domain confined to the ventral portion of the prospective neural retina in mouse, human, and Xenopus. The overexpression of either the frog Xvax2 or the human VAX2 in Xenopus embryos leads to an aberrant eye phenotype and, in particular, determines a ventralizing effect on the developing eye. The expression domain of the transcription factor Xpax2, normally confined to the ventral developing retina, extends to the dorsal region of the retina after overexpression of vax2. On the other hand, the expression of Xvent2, a molecular marker of the dorsal retina, is strongly reduced. Furthermore, vax2 overexpression induces a striking expansion of the optic stalk, a structure deriving from the ventralmost region of the eye vesicle. Altogether, these data indicate that vax2 plays a crucial role in eye development and, in particular, in the specification of the ventral optic vesicle.
The elucidation of many basic developmental processes in mammals has been facilitated greatly by the study of model organisms. Many of the pathways involved in neural development have been found to share similarities during evolution, and numerous genes have been found to be conserved and to play similar roles even in very distantly related species. The fruit fly Drosophila melanogaster is one of the most valuable model organisms in biological research (1). There are a considerable number of genes that are highly conserved between humans and Drosophila and that play similar biological roles in the two species (2–4). We recently started a project aimed at the identification of human cDNAs significantly similar to Drosophila mutant genes. By screening the Expressed Sequence Tag database, we identified over 200 such human cDNAs, and we termed them Drosophila-related expressed sequences (DRES; refs. 5 and 6). One of the DRES genes, DRES93, was found to bear significant homology with the homeodomain of the Drosophila gene empty spiracles (ems), which plays a major role in controlling the development of the head of the fly (7). Two vertebrate homologs for this gene have been cloned, emx1 and emx2, and have been shown to be expressed in the developing dorsal telencephalon in Xenopus (8) and in the developing cerebral cortex in the mouse (9). We decided to undertake a more detailed characterization of the DRES93 cDNA, and we isolated the human, the murine, and Xenopus full-length transcripts. Sequence analysis revealed that this gene is highly similar to the recently described vax1 (ventral anterior homeobox-containing gene 1; ref. 10), and the gene was accordingly renamed vax2. vax2 turned out to be almost exclusively expressed in the ventral portion of the retina during development. To gain insight into the putative function of vax2, we injected both the human and the Xenopus transcripts into Xenopus embryos to determine the phenotypic effects of its overexpression during development. The phenotype observed confirms that this gene may play an important role in the morphogenetic processes underlying the correct development of the eye, particularly in the establishment of a correct dorsoventral pattern.
MATERIALS AND METHODS
cDNA Identification and Sequence Analysis.
To identify the human VAX2 full-length transcript, a teratocarcinoma (Stratagene 937230) cDNA library was used. For the isolation of the murine Vax2 full-length cDNA, an 11-day-old embryo (CLONTECH ML3003a) cDNA library was used. For the isolation of the full-length Xvax2 cDNA, we used a stage (st.) 28/30 Xenopus head cDNA library (a gift from R. M. Harland, University of California, Berkeley, CA). Plating, hybridization, and washing conditions have been described (11). cDNA sequence analysis as well as nucleotide and protein database searches were performed as described (5).
Genomic Mapping in Human and Mouse.
Radiation hybrid mapping assignment was performed as described (5). Oligonucleotide primers were TO-3105 (5′-GGGAAGAGTGTGTGGCTGCAG-3′) and TO-3177 (5′-CCAGCAGCTGCAAGAAAGCTAA-3′). PCR conditions were 35 cycles of 94°C for 45 s, 52°C for 45 s, and 72°C for 45 s. Genetic mapping of Vax2 and Emx1 in the mouse was achieved by using the method of de Conciliis, et al. (12).
Embryos and Histology.
Induction of ovulation in females, in vitro fertilization, and embryo culture were carried out as described (13). Staging and histological examination were performed as reported (14, 15).
In Situ Hybridization.
Mouse embryo tissue sections were prepared, and RNA in situ hybridization experiments were performed as described (16). Preparation of human embryo tissue sections and RNA in situ experiments were carried out as described (17). All procedures were approved by the Ethical Committee of Saint-Vincent de Paul Hospital in Paris. Whole-mount in situ hybridization in Xenopus embryos was performed on staged embryos as described by Harland (18). For histological examination, stained embryos were fixed in Bouin’s solution, embedded in paraffin, dewaxed in xylene, and mounted in Eukitt. When pigmented embryos were used, they were bleached after the color reaction as described (19).
In Vitro Transcription and Embryo Microinjections.
Capped synthetic RNAs were synthesized from linearized VAX2-CS2+ and Xvax2-CS2+ plasmids as described (20, 21). RNAs were injected by using a Drummond Scientific (Broomall, PA) “Nanoject” apparatus. During injection, embryos were cultured in 0.1× Marc’s modified Ringer’s solution/4% (vol/vol) Ficoll 400 (Amersham Pharmacia). The medium was gradually replaced with 0.1× Marc’s modified Ringer’s solution 12–24 h after injection.
RESULTS
cDNA Identification and Sequence Analysis.
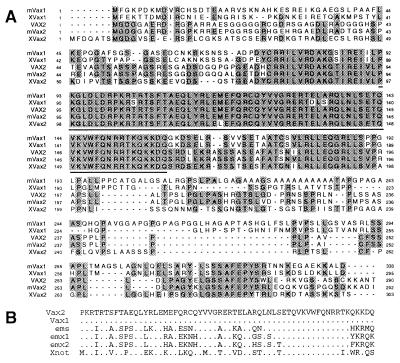
By analyzing the dres search engine (22), a collection of tblastn searches obtained by using all Drosophila protein sequences as queries, we identified a human expressed sequence tag (accession no. H92142) significantly similar to the Drosophila protein ems (accession no. P18488). This human expressed sequence tag was used as a probe on several human cDNA libraries, leading to the isolation of the full-length human VAX2 transcript (Fig. 1A). In addition, we identified the corresponding full-length murine (Vax2) and Xenopus laevis (Xvax2) cDNAs. vax2 encodes a predicted homeodomain-containing protein closely related to vax1, a gene recently identified in mouse and Xenopus laevis and likely involved in the specification and formation of the rostral and ventral forebrain in vertebrates (10). The stretch of highest sequence conservation between the murine Vax1 and Vax2 proteins was found in a region of 86 amino acids encompassing the homeodomain. Only two amino acid changes were detected in this portion, and neither of them were in the homeodomain, which is therefore identical in the two proteins. The degree of similarity is much lower in the N and C termini, with the exception of two short amino acid stretches located downstream of the homeodomain (Fig. 1B). The VAX2 homeodomain was also found to share significant sequence similarity with the homeodomains present in other proteins, as shown in Fig. 1B. Interestingly, we identified two additional forms of the human VAX2 transcript, which are predicted to encode protein products with an incomplete homeodomain almost completely lacking the third of the three helical structures that form the main structure of the homeodomain (data not shown).
Figure 1.
Sequence analysis of the vax2 cDNA. (A) Sequence alignment of the human VAX2 predicted protein with the mouse Vax2 protein (mVax2), the murine Vax1 protein (mVax1), and the Xenopus Vax1 and Vax2 proteins (XVax1 and XVax2, respectively). Amino acids that are identical in two or more proteins are shown in black. The homeodomain is underlined. (B) Amino acid sequence alignment of the VAX2 homeodomain and other related homeodomains. Residues identical to VAX2 protein are indicated by dots.
Mapping of vax2 in Human and Mouse.
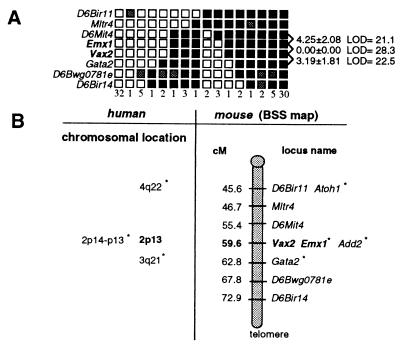
We mapped the human VAX2 gene by PCR analysis on the GeneBridge4 radiation hybrid panel (23) to chromosome 2p13, between markers D2S292 and D2S1394 (data not shown). Interestingly, the 2p13-p14 region is also known to harbor the EMX1 gene (24). We genetically mapped the murine Vax2 gene to mouse chromosome 6, between markers D6Mit4 and Gata2, in a region of synteny with human chromosome 2p13 (refs. 25 and 26; Fig. 2). Because Emx2 and Vax1 are tightly linked on mouse chromosome 19 (10), we also mapped Emx1 to see whether its chromosomal localization coincides with that of Vax2. The Emx1 gene cosegregates with Vax2, with no recombinants of 94 meioses tested. These results, together with the significant degree of sequence similarity between vax and emx genes, suggest that these genes have a common origin. They may have arisen from the tandem duplication of an ancestor gene subsequently followed by an interchromosomal duplication. It remains to be seen whether the genomic organization of vax and emx genes also has an implication in terms of spatial and temporal patterns of gene expression.
Figure 2.
Mapping of Vax2 and Emx1 in the mouse genome. (A) Haplotype and linkage analysis of Vax2 and flanking loci on mouse chromosome 6 through the analysis of the (C57BL/6j × SPRET/Ei)F1 × SPRET/Ei (BSS) backcross (The Jackson Laboratory). Empty squares indicate the Mus spretus allele; solid squares indicate the C57BL/6J allele; shaded squares refer to a genotypes that are not determined. Numbers to the right, between rows, indicate recombination fractions ±SEM and logarithm of odds scores. Columns represent different haplotypes observed on chromosome 6. Numbers below columns define the number of individuals sharing each haplotype. (B) Position of Vax2 on chromosome 6 with respect to nearby markers independently mapped by others on the BSS backcross. Numbers on the left represent approximate genetic distances (in centimorgans, cM) from the most centromeric chromosome 6 marker in this cross. Chromosomal locations of four syntenic human genes, including VAX2 and EMX1, are also indicated.
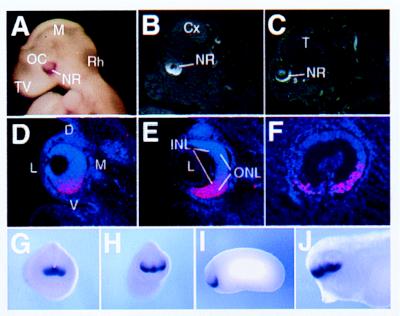
vax2 Expression in Human, Mouse, and Xenopus laevis. To determine the temporal and spatial expression of Vax2 during development, we carried out in situ hybridization experiments on mouse embryos at several developmental stages. By embryonic day 9.0 (E9.0), an asymmetric distribution of the transcript is detectable in the developing eye in mouse. Vax2 transcripts are detected at this stage by whole-mount in situ hybridization in the ventral half of the optic vesicle extending from the inferonasal to the inferotemporal retina, across the ventral furrow (Fig. 3A). This pattern of expression remains consistent throughout gestation. At E12.5, Vax2 is highly and almost exclusively expressed in the prospective inferior neural retina, whereas other parts of the eye epithelium, the prospective ciliary body, iris, and the outer layer of the optic cup, which gives rise to pigmented epithelium, are not labeled (Fig. 3B, D, and E). Within the differentiating neural retina, labeling seems to be equally strong in both the deep and superficial layers (Fig. 3D and E). Expression is also detected at a lower level from the inferior neural retina along the entire optic nerve and stalk and extends in the hypothalamus from the suprachiasmatic area to the anterior preoptic area (data not shown). Medially, the expression domain continues in the ventral telencephalon along the lamina terminalis to the septum (data not shown). At E16.5, the signal remains strong in the ventral neural retina, whereas expression is no longer detectable in the forebrain (Fig. 3C). Vax2 could not be detected in adult eye sections. The specific expression in the ventral part of the optic vesicle observed with the murine Vax2 transcript was also detected during human embryonic development by using human VAX2 cDNA as a probe (Fig. 3F).
Figure 3.
Expression pattern of vax2 in mouse (A–E), human (F), and Xenopus embryos (G–J) as determined by in situ hybridization. Vax2 expression in mouse embryos is restricted to the entire inferior neural retina, starting from E9 (A), when it is expressed in the ventral half of the optic cup, to E12.5 (B) and E16.5 (C). Eye expression in mouse at E12.5 is shown in D–E. Panels are arranged from anterior (D) to posterior (E) in coronal sections. The axis orientation of the developing eye is indicated in D: dorsal (D), ventral (V), lateral (L), and medial (M). Expression is restricted to the entire inferior neural retina, with no significant difference between inner (INL) and outer (ONL) neuroblast layers in E. (F) Sagittal section of the developing eye of a 7-week-old human embryo hybridized with the human VAX2 cDNA showing a similar pattern of expression in the ventral retina. (G–J) Whole-mount in situ hybridization showing expression of Xvax2 during Xenopus development. (G) st. 18, frontal view: the gene is expressed in the anterior neural plate. (H) st. 23, frontal view: Xvax2 is expressed in ventral regions of the forebrain vesicle and in a ventral portion of the optic vesicle. (I) st. 23, lateral view: expression as in H. (J) st. 33, lateral view: expression of Xvax2 is maintained in the ventral regions of both the telencephalon and diencephalon, as well as the retina. Other abbreviations: Cx, cerebral cortex; L, lens (except in D); M, mesencephalon (except in D); Rh, rhombencephalon; NR, neural retina; OC, optic cup; T, telencephalon; and TV, telencephalic vesicle.
The expression pattern of Xvax2 during Xenopus development, as detected by whole-mount in situ hybridization, is very similar to that observed in mouse and human. Expression begins at early neurula stage (st. 13) in the anterior neural plate (data not shown), whereas, at late neurula stage (st. 18), Xvax2 transcripts are detectable in the same area, which is fated to give rise to ventral telencephalic and diencephalic structures as well as to the eye (Fig. 3G). After neural tube closure and evagination of the optic vesicles at the early tail-bud stage (st. 23), Xvax2 expression is specifically restricted to anterior and ventral neural structures, namely the ventral part of the optic vesicles (Fig. 3 H and I) and the ventral regions of the forebrain vesicle (Fig. 3H). This expression pattern is maintained during subsequent developmental stages. In fact, in tadpole embryos (st. 33), XVax2 is specifically expressed in the ventral aspect of the retina as well as in the ventral regions of both telencephalon and diencephalon (Fig. 3J).
vax2 Overexpression Affects Eye Morphogenesis.
To address the role of vax2 in eye development, we overexpressed the gene by injection of synthetic RNA into 8-cell-stage Xenopus embryos. We observed a specific effect on eye morphogenesis in approximately 50% of the embryos examined (239 of 482) after injection of 75 pg of coding-region Xvax2 RNA either alone or in combination with 250 pg of β-galactosidase RNA into one dorsal animal blastomere (Fig. 4 and data not shown). The β-galactosidase enzymatic reaction, detected as a blue staining on the embryos, allowed us to identify the injected side. In st. 46 embryos, eye morphogenesis is normally completed (Fig. 4C); however, on the injected side, the optic fissure remains widely open, and its borders do not tend to contact each other (Fig. 4D). Moreover, in dorsal views, a giant optic stalk is seen to connect the eye and the forebrain on the injected side (Fig. 4B). This connection is in contrast with the normal situation at this stage, when the eye and the brain are connected by only the thin optic nerve (Fig. 4A). Similar effects were observed in 44% of the embryos examined (207 of 469) after injection of either 150 pg of full-length or 75 pg of coding-region human VAX2 RNA (Fig. 4E). Transverse sections of the injected embryos (Fig. 4 F–M) show the presence of a wide optic stalk-like structure abnormally connecting the injected eye and the brain (Fig. 4 F, G, J, and K). Moreover, in the most anterior sections, we observed the presence of a continuous lumen connecting the forebrain ventricle and the eye (Fig. 4 F and J). Furthermore, morphogenesis of the optic cup is strongly affected on the injected side, because the ventral retina apparently does not encircle the lens correctly. Instead, the ventral retina extends dorsally toward the brain (Fig. 4 G, H, K, and L). However, this abnormally folded retina seems to undergo neural differentiation, because it shows both a plexiform layer (Fig. 4I) and a photoreceptor layer (Fig. 4M). All of these morphological abnormalities consistently colocalized with the β-galactosidase staining. As a control, 310 embryos injected with 250–350 pg of β-galactosidase RNA alone did not show any effect on eye morphogenesis. Also, injection of transcripts from other genes involved in anterior development (e.g., Xrx1 and Xotx1) never produced the phenotype described above.
Figure 4.
Effects of vax2 overexpression on swimming tadpole embryos after injection of 75 pg of Xenopus coding-region RNA and 250 pg of β-galactosidase RNA (B, D, F–H, and J–L) or 150 pg of full-length human RNA (E, I, and M) into the left dorsal animal blastomere at the 8 cell stage, as compared to the injection of 250 pg of β-galactosidase RNA alone (A and C). (A) A st. 46 embryo injected with β-galactosidase RNA, dorsal view. On the injected side (left), the optic cup and the forebrain are connected by the thin optic nerve only (arrowhead). (B) A st. 46 embryo coinjected with Xvax2 and β-galactosidase RNAs, dorsal view. The optic cup and the forebrain are abnormally connected by a giant optic stalk (arrow). (C) The same embryo shown in A, laterally viewed from the β-galactosidase-injected side. Eye morphogenesis is complete. (D) The same embryo shown in B, laterally viewed from the vax2-injected side. The optic fissure remains wide open. (E) Ventral view of the injected side of a st. 46 embryo microinjected with VAX2 RNA. The optic fissure is open. (F–H) Transversal sections of st. 46 embryos injected with Xvax2 RNA. (J–L) Higher magnifications of F–H, respectively. On the injected side, the right side in F–H, a wide optic stalk-like structure abnormally joins the optic cup to the brain (arrows in J and K). At anterior levels (F and J), a continuous lumen can be detected connecting the brain ventricle and the eye (arrowhead in J). The defective invagination of the ventral optic cup is also evident (G, H, K, and L). (I and M) Transversal sections of a st. 46 embryo injected with VAX2 RNA showing that the aberrantly folded neural retina presents a plexiform layer (pl) and is dorsally covered by a photoreceptor layer (ph).
vax2 Is Able to Ventralize the Optic Vesicle.
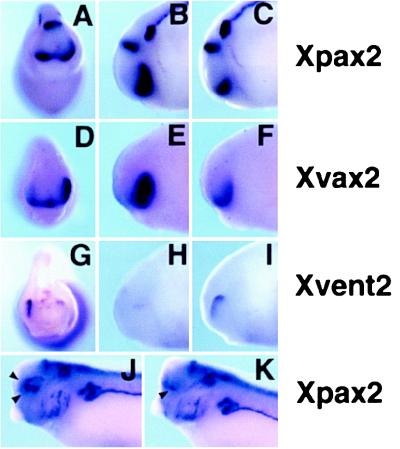
To determine whether VAX2 overexpression could alter the dorsoventral positional values within the retina, we assayed the expression pattern of the Xpax2, Xvax2, and Xvent2 genes in VAX2-injected Xenopus embryos (Fig. 5). At st. 23, Xpax2 is specifically transcribed in the ventral part of the retina (ref. 27; Fig. 5 A and C). This expression domain is strikingly expanded into the dorsal retina on the injected side (51% of the embryos; n = 41; Fig. 5 A and B). We also analyzed the expression of the endogenous Xvax2 gene in embryos unilaterally injected with the human VAX2 mRNA (Fig. 5 D–F). As for XPax2, the expression of Xvax2 was expanded dorsally on the injected side (30% of the embryos; n = 70; Fig. 5E), suggesting that vax2 is able to activate its own expression, either directly or indirectly, in the eye vesicle. On the other hand, Xvent2 expression, which is restricted to the dorsalmost region of the retina at st. 23 (ref. 28; Fig. 5 G and I), is strongly reduced or suppressed on the injected side (70% of the embryos; n = 20; Fig. 5 G and H). Similar results were obtained after injection of the Xenopus Xvax2 RNA (Xpax2 expansion: 39% of the embryos, n = 83; Xvent2 repression: 47% of the embryos, n = 80; data not shown). At the tadpole stage, Xpax2 expression is down-regulated in the ventral optic cup and is confined to the lips of the optic fissure on the control side (Fig. 5K). Remarkably, Xpax2 expression is maintained in the ventral optic cup and frequently expands into the dorsal region (Fig. 5J) on the injected side (39% of the embryos; n = 49). We conclude that vax2 overexpression is able to alter the dorsoventral patterning within the retina, changing the fate of dorsal retinal regions according to a ventral positional character.
Figure 5.
Expression of Xpax2 (A–C, J, and K), Xvax2 (D–F), and Xvent2 (G–I) genes in tail-bud (A–I) and tadpole (J and K) embryos injected with VAX2 RNA unilaterally. (A, D, and G) st. 23 injected embryos, frontal view. On the injected side (right), the Xpax2 and Xvax2 expression domains, normally confined to the ventral developing eye, are expanded dorsally (A and D), whereas the Xvent2 expression, which is dorsal in the control side, is strongly reduced (G). (B, E, and H) st. 23 injected embryos laterally viewed from the injected side. (C, F, and I) The same embryos shown in B, E, and H laterally viewed from the control side. (J and K) st. 33 injected embryos, laterally viewed from the injected (J) or the control (K) side. On the control side, Xpax2 expression in the optic cup is exclusively restricted to the lips of the optic fissure (arrowhead in K), whereas, on the injected side, Xpax2 expression expands both in ventral and anterodorsal regions of the eye cup (arrowheads in J).
vax2 Overexpression Induces an Expansion of the Optic Stalk.
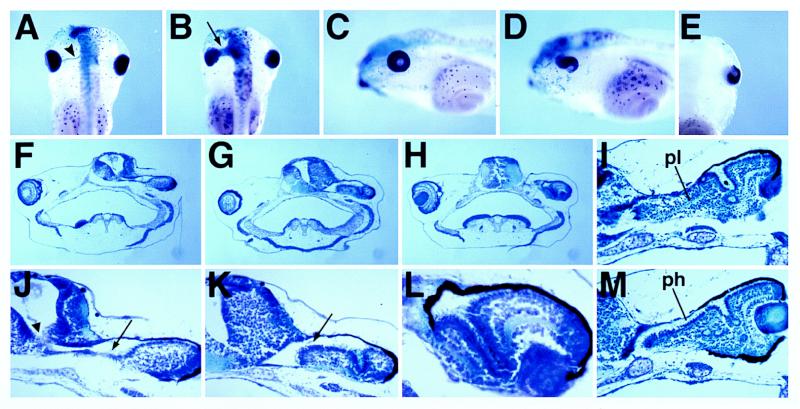
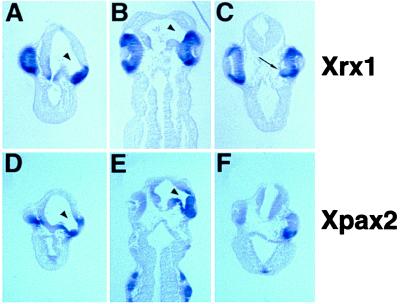
To determine whether the specification of the retinal or optic stalk territories could be affected by vax2 overexpression, we analyzed histological sections of st. 33 embryos unilaterally injected with human VAX2, both morphologically and by using Xrx1 and Xpax2 as molecular markers. Morphological analysis showed that the optic cup is clearly separated from the brain on the control side at this stage, and the two structures are connected anteriorly by only the narrow optic stalk. Conversely, on the injected side, the optic cup seems to be continuous with the anterior region of the forebrain through an enlarged optic stalk endowed with a wide central lumen (70% of the embryos; n = 37; Fig. 6 A and B and data not shown). Moreover, transverse sections at the level of the lens show that morphogenesis of the optic cup is altered at this stage on the injected side, because the ventral part of the retina seems to be incorrectly displaced toward the inside of the embryo (50% of the embryos; n = 20; Fig. 6C). At st. 33, Xrx1 expression in the eye is specifically restricted to the retina (29), whereas Xpax2 expression is confined to the optic stalk and to only the lips of the optic fissure in the optic cup (27). Although the extent of the Xrx1 expression domain seems to be similar in both the control and injected side (Fig. 6 A and B), the Xpax2 expression domain seems to be expanded on the injected side of the embryos showing the above described morphological abnormalities (Fig. 6 D–F). The tissue connecting the eye cup and forebrain is labeled by Xpax2, thus confirming its nature as an enlarged optic stalk (Fig. 6 D and E). Remarkably, the dorsal retina, which is contiguous with the enlarged stalk, also ectopically expresses Xpax2 (Fig. 6 D and E). Furthermore, expression of Xpax2 is maintained in the ventral retina on the injected side, as revealed by transverse sections cut at the level of the lens (Fig. 6F). Therefore, vax2 overexpression is able to concomitantly induce a widespread ectopic expression of Xpax2 within the retina, an enlargement of the optic stalk, and an aberrant folding of the ventral retina toward the brain.
Figure 6.
Expression of the Xrx1 (A–C) and Xpax2 (D–F) genes in tadpole embryos injected with VAX2 RNA unilaterally, as detected in sections after whole-mount in situ hybridization. Transversal (A and D) and frontal (B and E) sections of st. 33 injected embryos, cut at the level of the optic stalk. Both the morphology and Xpax2 expression indicate the presence of an enlarged optic stalk on the injected side (arrowhead). The extent of the Xrx1 expression domain is similar in both the injected (right) and control (left) sides. Remarkably, dorsal expansion of Xpax2 expression is detectable in the retinal tissue contiguous with the enlarged optic stalk (D). (C and D) Transversal sections of st. 33 injected embryos, cut at the level of the lens. (C) On the injected side (right), surrounding of the lens by the optic cup is abnormally ventral (arrow); the Xrx1 RNA distribution is more uniform along the ventrodorsal axis compared to the control side. (F) Ectopic Xpax2 transcription is evident in the ventral retina of the injected side (right).
DISCUSSION
In this paper, we describe the isolation and characterization of a transcription factor, vax2, strongly expressed in the developing eye in vertebrates. This gene represents the second member to be identified of a subgroup of homeobox-containing genes, the vax subfamily, another member of which, vax1, was recently reported (10). In fact, the Vax1 and Vax2 proteins share a significant sequence similarity: they are 100% identical in the homeodomain and very similar in the surrounding regions, whereas the two terminal regions, in particular the N termini, are more divergent (Fig. 1A). The Vax1 and Vax2 proteins also show significant sequence similarities (approximately 65–70% identity) with the homeodomains of the not and emx families (Fig. 1B). However, in these two cases, the sequence homology is confined to the homeodomain and does not extend to the neighboring regions, which clearly indicates that the vax genes represent an independent subfamily of homeobox genes.
Vax1 is thought to play a role in the specification and formation of the rostral and ventral forebrain in vertebrates. It is strongly expressed early in development in the rostral neural plate in the medial anterior neural ridge. At later stages, Vax1 is detected in derivatives of this territory, namely the rostromedial olfactory placode, optic nerve and disk, and anterior ventral forebrain. Vax2, on the other hand, although it shares some expression domains with Vax1 (such as the optic nerve), has a restricted pattern of expression in the developing eye, as determined in mouse, human, and Xenopus embryos. Vax2 is consistently expressed throughout gestation, starting at E9 in mouse in the inferior portion of the optic vesicle and later in the optic cup and neural retina; however, it is not detected in the dorsal regions of the above mentioned structures (Fig. 3). Therefore, the expression of Vax2 seems to define a territory: the ventral part of the developing neural retina in which fundamental processes of eye morphogenesis take place; namely, the beginning of the invagination of the optic vesicle and the formation and subsequent closure of the optic fissure.
The present RNA injection studies in Xenopus laevis embryos suggest that vax2 is not simply a molecular marker of the prospective inferior neural retina before its morphological differentiation but plays an important role in eye morphogenesis and particularly in establishing positional information along the dorsoventral axis in the optic vesicle. The microinjection of synthetic vax2 RNA is able to alter the dorsoventral patterning within the eye, changing the fate of dorsal regions according to a ventral positional character (Fig. 6). In particular, we observed at tail-bud stages an expansion of the expression domains of Xpax2 and of the endogenous Xvax2 into the dorsal region of the optic vesicle and a concomitant inhibition of transcription of the dorsal marker Xvent2. At tadpole stages, we observed a notable enlargement of the optic stalk, a structure arising from the ventralmost region of the eye vesicle (Figs. 4 and 6). The retinal tissue contiguous with the enlarged stalk showed an ectopic dorsal expression of Xpax2, thus suggesting that the expansion of the optic stalk may be related to the alteration of dorsoventral patterning of the eye vesicle (Fig. 6). Furthermore, at swimming tadpole stages, we detected a profound alteration in the morphogenesis of the eye, including nonclosure of the optic fissure and abnormal morphogenesis of the ventral retina, which was observed protruding toward the neural tube (Figs. 4 and 6).
The pax2 gene also plays a major role in the morphogenesis of the ventral eye, (i) because it is essential in the terminal phases of the closure of the optic fissure, being required for the contact-dependent dissolution of the basal lamina between the lips of the fissure (30); and (ii) because it controls the establishment of the boundary between the optic cup and the stalk (30, 31). However, the nonclosure of the optic fissure observed in vax2-injected embryos seems to be different and more severe than that observed in cases of functional inactivation of pax2. In fact, we observed (i) that the margins of the optic fissure tend to remain very distant from each other (Fig. 4 B and D) and (ii) that the ventral retina does not invaginate correctly around the lens but instead extends medially toward the brain. Interestingly, we also observed an altered spatiotemporal regulation of Xpax2 within the retina. Normally pax2 is expressed in the ventral half of the optic vesicle early in morphogenesis (32), whereas it is down-regulated in the prospective retina shortly after its invagination when it becomes a specific marker of the optic stalk (27, 32). However, in VAX2-injected embryos, Xpax2 expression was maintained extensively in the ventral retina during morphogenesis of the optic cup. It is tempting to speculate that repression of pax2 transcription in the retina at these particular stages may be important to allow the correct invagination of the optic cup.
Remarkably, all the observed morphological abnormalities, as well as the effects on the expression of molecular markers, were fully reproduced, both qualitatively and quantitatively, after overexpression of either Xenopus or human vax2. Together with the vax2 high conservation both in protein sequence and expression pattern in the frog, mouse, and human, these data strongly suggest that vax2 acts as a conserved master regulator gene in dorsoventral patterning of the vertebrate eye. Because of the high conservation of the molecular machinery regulating eye development in Drosophila and vertebrates, it will be interesting to know whether a vax2 gene exists in the fly and whether it is involved in dorsoventral patterning of the invertebrate eye.
Previously, retinoic acid was shown to be involved in the determination of the dorsoventral patterning of the zebrafish retina (33). Interestingly, by using both morphological and molecular criteria, we observe that vax2 overexpression produces effects in the Xenopus embryo very similar to those found after retinoic acid administration in the zebrafish embryo. In fact, in both instances, the expression of ventral markers such as Pax2 is expanded in the eye vesicle, whereas that of dorsal markers is strongly reduced. In addition, both treatments induce a wide enlargement of the optic stalk. Finally, although early, postgastrulation retinoic acid treatments in zebrafish result in retinal duplications, late treatments produce an abnormal morphogenesis of the ventral retina very close to that observed after vax2 overexpression. These similarities strengthen the idea that VAX2 plays a pivotal role in the genetic control of the dorsoventral patterning within the vertebrate retina. Such a role would also point to a possible involvement of vax2 in eye anomalies caused by the alteration of morphogenetic events taking place in the ventral region of the developing eye, such as coloboma, which is caused by failure in the closure of the optic fissure.
Acknowledgments
We thank V. Marigo, G. Meroni, and R. Vignali for helpful discussion; A. Marchitiello, C. Gattuso, and the Telethon Institute of Genetics and Medicine sequencing staff for technical support; and M. Smith for help with the preparation of the manuscript. This work was supported by the Italian Telethon Foundation, by the Association Française contre les Myopathies, by European Community Grants BMH4-CT97-2341 and BIO4-CT98-0399, and by Merck Genome Research Institute Grant 37.
ABBREVIATIONS
- En
embryonic day n
- st. n
stage n
Footnotes
References
- 1.Rubin G M. Science. 1988;240:1453–1459. doi: 10.1126/science.3131880. [DOI] [PubMed] [Google Scholar]
- 2.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 3.Li Q Y, Newbury-Ecob R A, Terrett J A, Wilson D I, Curtis A R, Yi C H, Gebuhr T, Bullen P J, Robson S C, Strachan T, et al. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 4.Mounkes L C, Jones R S, Liang B-C, Gelbart W, Fuller M T. Cell. 1992;71:925–937. doi: 10.1016/0092-8674(92)90389-t. [DOI] [PubMed] [Google Scholar]
- 5.Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio S, Coluccia E, Zollo M, et al. Nat Genet. 1996;13:167–174. doi: 10.1038/ng0696-167. [DOI] [PubMed] [Google Scholar]
- 6.Banfi S, Borsani G, Bulfone A, Ballabio A. Hum Mol Genet. 1997;6:1745–1753. doi: 10.1093/hmg/6.10.1745. [DOI] [PubMed] [Google Scholar]
- 7.Dalton D, Chadwick R, McGinnis W. Genes Dev. 1989;3:1940–1956. doi: 10.1101/gad.3.12a.1940. [DOI] [PubMed] [Google Scholar]
- 8.Pannese M, Lupo G, Kablar B, Boncinelli E, Barsacchi G, Vignali R. Mech Dev. 1998;73:73–83. doi: 10.1016/s0925-4773(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 9.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 10.Hallonet M, Hollemann T, Wehr R, Jenkins N A, Copeland N G, Pieler T, Gruss P. Development (Cambridge, UK) 1998;125:2599–2610. doi: 10.1242/dev.125.14.2599. [DOI] [PubMed] [Google Scholar]
- 11.Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, et al. Nature (London) 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 12.de Conciliis L, Marchitiello A, Wapenaar M C, Borsani G, Giglio S, Mariani M, Consalez G G, Zuffardi O, Franco B, Ballabio A, et al. Genomics. 1998;51:243–250. doi: 10.1006/geno.1998.5348. [DOI] [PubMed] [Google Scholar]
- 13.Newport J, Kirschner M. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 14.Andreazzoli M, Pannese M, Boncinelli E. Development (Cambridge, UK) 1997;124:1733–1743. doi: 10.1242/dev.124.9.1733. [DOI] [PubMed] [Google Scholar]
- 15.Nieuwkoop P, Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North–Holland; 1967. [Google Scholar]
- 16.Rugarli E I, Lutz B, Kuratani S C, Wawersik S, Borsani G, Ballabio A, Eichele G. Nat Genet. 1993;4:19–25. doi: 10.1038/ng0593-19. [DOI] [PubMed] [Google Scholar]
- 17.Fougerousse F, Durand M, Suel L, Pourquie O, Delezoide A L, Romero N B, Abitbol M, Beckmann J S. Genomics. 1998;48:145–156. doi: 10.1006/geno.1997.5160. [DOI] [PubMed] [Google Scholar]
- 18.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 19.Mayor R, Morgan R, Sargent M G. Development (Cambridge, UK) 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- 20.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupp R A, Snider L, Weintraub H. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 22.Guffanti A, Banfi S, Simon G, Ballabio A, Borsani G. Trends Genet. 1997;13:79–80. doi: 10.1016/s0168-9525(97)01015-9. [DOI] [PubMed] [Google Scholar]
- 23.Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czary N, Spillett D, Muselet D, Prud’Homme J-F, Dib C, Auffray C, et al. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- 24.Kastury K, Druck T, Huebner K, Barletta C, Acampora D, Simeone A, Faiella A, Boncinelli E. Genomics. 1994;22:41–45. doi: 10.1006/geno.1994.1343. [DOI] [PubMed] [Google Scholar]
- 25.Tisminetzky S, Devescovi G, Tripodi G, Muro A, Bianchi G, Colombi M, Moro L, Barlati S, Tuteja R, Baralle F E. Gene. 1995;167:313–316. doi: 10.1016/0378-1119(95)00591-9. [DOI] [PubMed] [Google Scholar]
- 26.Ueyama H, Inazawa J, Nishino H, Han Xiang D, Ochiai Y, Ohkubo I. Genomics. 1995;25:720–723. doi: 10.1016/0888-7543(95)80016-f. [DOI] [PubMed] [Google Scholar]
- 27.Heller N, Brandli A W. Mech Dev. 1997;69:83–104. doi: 10.1016/s0925-4773(97)00158-5. [DOI] [PubMed] [Google Scholar]
- 28.Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. Development (Cambridge, UK) 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- 29.Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. Mech Dev. 1997;61:187–198. doi: 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- 30.Torres M, Gomez Pardo E, Gruss P. Development (Cambridge, UK) 1996;122:3381–3391. doi: 10.1242/dev.122.11.3381. [DOI] [PubMed] [Google Scholar]
- 31.Macdonald R, Scholes J, Strahle U, Brennan C, Holder N, Brand M, Wilson S W. Development (Cambridge, UK) 1997;124:2397–2408. doi: 10.1242/dev.124.12.2397. [DOI] [PubMed] [Google Scholar]
- 32.Nornes H O, Dressler G R, Knapik E W, Deutsch U, Gruss P. Development (Cambridge, UK) 1990;109:797–809. doi: 10.1242/dev.109.4.797. [DOI] [PubMed] [Google Scholar]
- 33.Hyatt G A, Schmitt E A, Marsh Armstrong N, McCaffery P, Drager U C, Dowling J E. Development (Cambridge, UK) 1996;122:195–204. doi: 10.1242/dev.122.1.195. [DOI] [PubMed] [Google Scholar]