Abstract
Polyploidy is a dominant feature of flowering plant genomes, including those of many important crop species, implying that polyploidy confers evolutionary advantages on plant species. Recent molecular studies suggest that polyploids often originate many times from the same progenitor diploids. For this to provide a broader genetic base for a polyploid species, there must be lineage recombination in the genomes of polyploids having different origins, and this has rarely been documented in recently formed wild polyploid species. Glycine tabacina, a wild relative of soybean, forms a widespread polyploid complex in Australia and the islands of the Pacific Ocean. In a sample of 40 G. tabacina plants, DNA sequence variation at one homoeologous histone H3-D locus identified three alleles, each also found in Australian diploid Glycine species. These data agree with our previous studies of chloroplast DNA variation in suggesting that this polyploid has originated several times. Both the origins of the polyploid and several independent dispersals from Australia to oceanic islands appear to have occurred within the last 30,000 years. The distributions of histone alleles, chloroplast haplotypes, and alleles at two isozyme loci were uncorrelated, and 20 multilocus genotypes were found among the 40 plants sampled. Extensive lineage recombination is thus hypothesized in the polyploid, involving migration and occasional outcrossing in this predominantly inbreeding species. The combination of multiple origins with gene exchange among lineages increases the genetic base of a polyploid and may help explain the wide colonization of polyploid G. tabacina relative to its diploid progenitors.
Polyploidy is a widespread phenomenon in flowering plants, with perhaps as many as 70% of species having experienced at least one genome duplication (1, 2). Polyploid taxa often tend to be weedy or widespread relative to congeneric or conspecific diploids, and many cultivated plant species are either recent or ancient polyploids (e.g., maize, wheat, sorghum, soybean, cotton, Brassica). The success of polyploids is often attributed to their genetic variability. This, in turn, may be due to the hybrid nature of many polyploids and to the buffering effect of their genetic system, which shelters recessive alleles from the purifying effects of selection (1).
Classic models of polyploid origin suggested the possibility of recurrent origin (3). Numerous molecular studies have confirmed the existence of far more polymorphism in polyploids than would be expected from single origins (reviewed in ref. 4), which in a widespread tetraploid species would allow the initial presence of a maximum of four alleles at a given pair of homoeologous loci. Most of these studies, however, involve only a single marker, often chloroplast DNA (cpDNA), and thus leave open the question of whether gene exchange and subsequent lineage recombination occurs among polyploids having different origins. Relatively few studies of wild plants (5, 6) have documented both multiple origins of a polyploid species and gene flow among its genetically distinct populations. Thus, it is not known whether most wild allopolyploid species exist in nature as single gene pools or, instead, as a set of isolated populations unlinked by gene exchange. Clearly, the opportunities for exploration of novel adaptive zones—a key stage in the establishment of a polyploid (7)—would be enhanced by recombination among polyploids initially having different genetic backgrounds.
Glycine tabacina is a member of Glycine subgenus Glycine, the group of wild perennial Australian relatives of the annual, northern Asian cultivated soybean. Currently, this taxon includes both diploids (2n = 40) confined to eastern Australia and tetraploids that occur not only throughout much of eastern Australia but also in the islands of the southern and west-central Pacific Ocean (8). It has been shown that there are two types of G. tabacina polyploids that differ in chloroplast DNA (cpDNA) haplotype, restriction endonuclease maps of the 18S-26S nuclear ribosomal DNA locus, and morphology. These two types share one genome but differ at the second (8, 9), with the divergent genome being derived from the egg parent (10).
One of these two polyploid taxa (designated BBB2B2 in ref. 10, but here as BBB′B′) was studied extensively for cpDNA variation, and it was shown to contain eight different chloroplast haplotypes. Six of these were identical in their restriction maps to haplotypes found in species of the B-genome group of diploids (11). Thus, the polyploid apparently originated at least six different times, and its origins in Australia and colonization of the Pacific seem to have occurred recently. Lack of nuclear ribosomal DNA restriction site variation among B-genome diploids precluded testing these hypotheses further with this nuclear locus and limited our ability to address the issue of gene exchange in these polyploids.
Here, we report results from sequences at a single copy nuclear locus, histone H3-D, sampled from many of the same BBB′B′ accessions used previously in the cpDNA study, and from two isozyme loci. We show that these polyploids are polymorphic at the histone locus, but to a lesser extent than is true for cpDNA. Moreover, there is little correlation in the distribution of individual histone H3-D alleles, cpDNA haplotypes, and isozyme alleles, suggesting that gene exchange and recombination has been extensive among polyploid populations.
MATERIALS AND METHODS
Histone H3-D. Forty BBB′B′ polyploid accessions were sampled in this study: 39 assayed previously for cpDNA variation plus accession G1076 (from the same region as G1075) (Table 1); all were obtained from the Commonwealth Scientific and Industrial Research Organization Perennial Glycine Germplasm Collection. Accessions were selected to encompass the geographic range of the BBB′B′ polyploid and to represent all of the known cpDNA haplotypes (11). In most cases, the same DNA samples were used in this study as were used previously (11). Where new isolations were performed, usually to confirm results, a standard cetyltrimethylammonium bromide method (12) was used to isolate DNA.
Table 1.
G. tabacina polyploid accessions sampled
| Accession, G number | Provenance | cpDNA | Histone H3-D | EnpB | Mdh2B | Multilocus genotype |
|---|---|---|---|---|---|---|
| 1706 | SA, Australia | 1 | 9 | 6 | 8 | iii |
| 2709 | Vic, Australia | 1 | 9 | 6 | 5 | iv |
| 2710 | Vic, Australia | 2 | 10 | 8 | 5 | xviii |
| 1075, 1076* | ACT, Australia | 2 | 10 | 6 | 8 | xix |
| 1080 | sNSW, Australia | 2 | 9 | 8 | 8 | viii |
| 1254 | sNSW, Australia | 1 | 10 | 6 | 8 | xvi |
| 1255 | sNSW, Australia | 3 | 9 | 6 | 8 | xi |
| 1312 | sNSW, Australia | 4 | 10 | 8 | 5 | xx |
| 2511, 2762 | sNSW, Australia | 1 | 9 | 6 | 5 | iv |
| 1141 | nNSW, Australia | 4 | 9 | 8 | 8 | xii |
| 1314 | nNSW, Australia | 4 | 9 | 6 | 5 | xiii |
| 1234 | nNSW, Australia | 1 | 9 | 8 | 8 | v |
| 1329 | nNSW, Australia | 6 | 9 | 6 | 5 | xv |
| 1439 | nNSW, Australia | 2 | 9 | 6 | 5 | vii |
| 1503 | nNSW, Australia | 3 | 9 | 6 | 5 | x |
| 1262, 1862 | Qld, Australia | 1 | 10 | 6 | 5 | xvii |
| 2246, 2274 | Qld, Australia | 1 | 9 | 8 | 5 | vi |
| 2263, 2738 | Qld, Australia | 2 | 9 | 8 | 5 | ix |
| 1205, 1324, 1325 | New Caledonia | 5 | 9 | 6 | 5 | xiv |
| 1258 | New Caledonia | 1 | 5 | 6 | 5 | i |
| 1989 | New Caledonia | U | 5 | 6 | 5 | ii |
| 1863 | Fiji | 1 | 5 | 6 | 5 | i |
| 1981, 1982 | Tonga | 1 | 5 | 6 | 5 | i |
| 1984 | Vanuatu | 1 | 5 | 6 | 5 | i |
| 1986 | Vanuatu | U | 5 | 6 | 5 | ii |
| 1072 | Marianas Islands | 1 | 5 | 6 | 5 | i |
| 1226, 1306, 1338, 1382 | Ryukyu Islands, Japan | 1 | 5 | 6 | 5 | i |
| 1716 | Hikanshi, Japan | 1 | 5 | 6 | 5 | i |
| 2602 | Taiwan | 3 | 9 | 6 | 5 | x |
Accessions are grouped in states or regions of origin. The accession (G) numbers are those of Commonwealth Scientific and Industrial Research Organization Perennial Glycine Germplasm Collection. Histone H3-D allele designations are from ref. 15; cpDNA haplotype data are from ref. 11. ACT, Australian Capital Territory; sNSW and nNSW, southern and northern New South Wales; Qld, Queensland; Vic, Victoria; SA, South Australia. Accessions with the same marker genotype are grouped.
Accession 1076 was not assayed for cpDNA.
PCR amplifications of histone H3-D alleles were performed by using primers described in ref. 13, which amplify an ≈600-bp region of the histone H3-D gene containing three introns (14). Use of these primers on genomic DNA amplifies both homoeologous loci (B and B′ in our genome designations); separation of these homoeologues would normally require cloning because they are nearly identical in size (13, 15). We therefore developed a strategy to sequence PCR products directly, which took advantage of the fact that B′ histone alleles all had a single EcoR-I restriction site not found in alleles of the B group. Digestion of genomic DNA with EcoR-I before amplification prohibited amplification of the B′ homoeologue, permitting direct sequencing of the B allele.
Automated dideoxy cycle sequencing was performed at the Cornell Biotechnology sequencing facility. Sequencing was performed with the 5′ amplification primer and with a reverse primer internal to the 3′ amplification primer, at the junction of the third intron and fourth exon (14). Initially, sequences were obtained for both strands of this ≈500-bp region. However, after initial results showed that only three different B alleles occurred among polyploid accessions, we screened accessions by sequencing only one strand, using the internal 3′ reverse primer.
Isozymes.
Single seeds of the 40 accessions from which histone H3-D sequences were obtained were tested for isozyme variation for endopeptidase (E.C. 3.4.-.-) and malate dehydrogenase (E.C. 1.1.1.37) by using starch gel and assay procedures of ref. 16. The zymograms for these two systems resembled those for G. tomentella in composition and complexity and were scored in similar fashion (16).
RESULTS
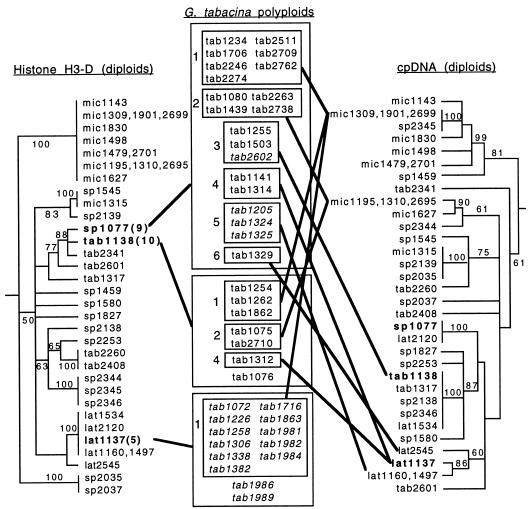
Three different histone H3-D alleles were found among the 40 accessions sampled. Each of these three alleles was identical to an allele found among accessions of the core B-genome group of diploid subgenus Glycine (alleles 5, 9, and 10 of ref. 15); the B′ homoeologue, which also occurs in these bivalent-forming allopolyploids, is not discussed here. No correlation was apparent between histone allele type and cpDNA haplotype (Table 1; Fig. 1). Each of the three histone H3-D alleles was found in different accessions having the most common haplotype (plastome 1), and the most common histone H3-D allele (9) was found in conjunction with all seven cpDNA haplotypes.
Figure 1.
Relationships of histone H3-D alleles and chloroplast haplotypes in G. tabacina polyploid accessions. Trees of histone H3-D alleles (15) and cpDNA haplotypes (11) are shown for the same set of diploid B-genome accessions. Diploid accessions having histone H3-D alleles identical to those of polyploid accessions are shown in bold type on both trees, and the allele number (classification from ref. 15) is given after the accession number on the histone tree. Polyploid accessions having these alleles are grouped in large boxes in the center column and are connected to corresponding diploid accessions by bold lines. Within these large boxes, polyploid accessions are grouped by cpDNA haplotype in smaller boxes, numbered to correspond to haplotype groups from ref. 11. These boxes are connected to diploid accessions in the cpDNA tree having the same haplotype. Polyploid accessions not in boxes possess plastome types for which no identical diploid accession has been sampled (1986, 1989) or have not been sampled for cpDNA (1076). Polyploid accessions from Pacific islands are italicized. lat, G. latifolia; mic, G. microphylla; tab, G. tabacina, sp., any of several unnamed B-genome species.
The geographic distribution of the histone H3-D alleles, however, was highly structured (Table 1). Despite the limited sample, the divergence in allele frequency between Pacific island accessions and Australian mainland accessions was statistically highly significant (contingency χ2 = 26.90, P < 0.001). Allele 5 was found only in island accessions and allele 10 only in the mainland sample. Allele 9, overall the most common, occurred in only four island accessions, one from Taiwan and three from New Caledonia.
Two alleles were scored at each of the two isozyme loci surveyed (EnpB and Mdh2B). Each of these two loci had one common allele, found throughout the range of the polyploid, and a second less common allele restricted to Australia. This sample of 40 tetraploid accessions were all homozygous for these two loci, but one heterozygote for endopeptidase was encountered in a sample of 97 additional BBB′B′ accessions (A.H.D.B., unpublished data). Little heterozygosity is expected in G. tabacina, because, like other perennial Glycine species, it reproduces largely by self-compatible cleistogamous flowers, and its chasmogamous flowers are self-compatible (17).
Of 84 possible multilocus genotypes (7 cpDNA × 3 histone × 2 EnpB × 2 Mdh2B), 20 were encountered (Table 1). Of these genotypes, 16 were found only among the 23 Australian accessions, and one (genotype x) was shared by one Australian and one Taiwanese accession. The other three genotypes were found only in the remaining 16 Pacific accessions. One of these genotypes (genotype i: chloroplast haplotype 1, histone allele 5, endopeptidase allele 6, MDH allele 5) was found in 11 of these accessions. A second multilocus genotype (ii) was found in two accessions, one from New Caledonia and one from Vanuatu, which were typical of Pacific island plants at their nuclear loci but had a chloroplast haplotype not identical to any known from B-genome diploids (plastome type U of ref. 11). A final genotype (xiv) was found in three New Caledonian accessions, which had the common Australian histone H3-D allele 9, chloroplast haplotype 5, and typical island alleles at both isozyme loci.
Independence of variation for the various markers, or, more technically, the absence of linkage disequilibrium, was tested for the 23 mainland accessions using contigency χ2. The two isozyme loci showed no evidence of linkage disequilibrium (χ2 = 0.01, P = 0.91). Variation at the histone-H3D locus was independent of that at EnpB (χ2 = 0.47, P = 0.49), at Mdh2B (χ2 = 0.29, P = 0.59), and at the joint two-locus isozyme haplotype (χ2 = 3.69, P = 0.30). Finally, the histone-H3D locus variation was independent of plastome type (χ2 = 1.33, P = 0.86; 22 accessions, 5 degrees of freedom). In sharp contrast, the set of 17 island accessions showed complete correlation (linkage disequilibrium) between plastome types and histone-H3D alleles (χ2 = 17, Fisher exact P = 0.00042).
The Shannon–Weaver information index (I = −∑pi ln pi) was computed for each of the set of mainland and island samples to summarize comparatively trends in genotypic diversity (18) for chloroplast and nuclear markers. Table 2 gives these estimates and the approximate standard errors. By the I-measure, there is little difference between overall plastome diversity among the mainland accessions compared with the island ones. In contrast, the nuclear diversity in the island samples in only one-third that of the mainland.
Table 2.
Estimates of genotypic diversity (Shannon–Weaver information index I) and their approximate standard error (18) for 23 mainland Australian accessions of G. tabacina, compared with 17 accessions from Pacific islands
| Chloroplast genome
|
Nuclear markers
|
|||
|---|---|---|---|---|
| Mainland | Island | Mainland | Island | |
| Diversity, I | 1.34 | 1.01 | 1.65 | 0.55 |
| Standard error, I | 0.14 | 0.19 | 0.13 | 0.12 |
Estimates of plastome diversity are shown separately from those of nuclear markers (histone H3-D, EnpB and Mdh2B).
DISCUSSION
Recent Multiple Origins of the Polyploid. The G. tabacina BBB′B′ polyploid is polymorphic at one histone H3-D homoeologous locus for three alleles, each of which is identical to an allele found in accessions of diploid B-genome species. This suggests that the polyploid is of recent origin, a conclusion previously reached on the basis of similar findings for cpDNA restriction fragment variation (11). Nuclear genes in general have a much higher synonymous substitution rate than do chloroplast sequences (19). Thus, the finding of sequence identity between alleles of diploids and polyploids is even more compelling evidence of recent origin than were the earlier cpDNA results.
From divergence among the diploid Glycine B-genome species, the estimated mutation rate for the H3-D locus is approximately μ = 6 × 10−9 substitutions/site/year (15). The total length of the three H3-D introns in question is n = 278 nucleotides. Assuming the mutation process is geometric, the expected waiting time until the next mutation in two separated lineages (and therefore the expected time since the last mutation) is about (2μn)−1 = 3 × 105 years. The 95% confidence limit for the time until a single mutation is 9 × 105 years. Thus, we can be sure that any single incidence of polyploidy has occurred since that time.
The histone H3-D results support the hypothesis that the BBB′B′ polyploid originated at least as many as three times, assuming that no progenitor was heterozygous at this locus. All three events show an absence of mutation. Together they argue that the time until mutation in any one of them is of the order of 105 years. Thus, we have 95% confidence that some of the polyploid events are at most 3 × 105 years old and likely to be much more recent than that.
Determining the actual number of origins and the identities of the B-genome diploid progenitors is complicated by the possibility that combinations of histone H3-D alleles and cpDNA haplotypes unobserved so far in diploid taxa could have been present in the progenitors of the polyploid. Hybridization or lineage sorting are suggested to have produced the discordance between cpDNA haplotype and histone H3-D allele distributions observed in modern B-genome diploids (15) and presumably have occurred throughout the history of this group. This also could account for the absence from the polyploid of H3-D allele 20, which is typical of Glycine microphylla, despite the prevalence of chloroplast haplotypes (plastomes 1 and 2) that are known in diploids only from G. microphylla (11, 15).
Colonization and the Distribution of Genetic Markers. The observation of four different genotypes among Pacific Ocean accessions suggests that there have been several independent colonization events of the Pacific from a mainland gene pool. One genotype (i) predominates outside of Australia, being found both in the southern Pacific and west-central Pacific islands. None of the remaining genotypes appears to be derived from the common Pacific genotype or from one another because they possess different chloroplast haplotypes. The identity of alleles and chloroplast haplotypes between diploids and polyploids suggests that all colonization events have been recent, at most within the same time frame as the occurrence of polyploidy.
An intriguing feature of the distribution of allelic variation is that the most common Pacific histone H3-D allele is so far lacking among Australian polyploid G. tabacina accessions. This allele (allele 5) is the common allele of the Australian diploid species Glycine latifolia (15). It is also the most divergent of the three histone H3-D alleles found in the polyploid, differing by six or seven nucleotide substitutions (≈1.5%) from the other two alleles. Thus, this allele is almost certainly of Australian origin and is unlikely to have arisen in parallel from one of the other two alleles in a progenitor of Pacific accessions. It might seem reasonable to speculate that histone allele 5 is simply a rare Australian allele that occurred by chance in a colonizing polyploid. However, histone allele 5 also occurs in two New Caledonian accessions that have a different chloroplast haplotype from that found in other polyploids having this allele. These plants almost certainly represent an independent colonization event. The chance presence of this allele in two separate colonizations seems unlikely, unless the allele was once present and perhaps even common in Australian G. tabacina.
Selection on these histone sequences seems unlikely, either for the allele in Pacific accessions or against it in Australia. The three histone alleles differ only in their introns over the region sequenced, making selection on the gene product unlikely. The existence of Pacific accessions having the most common Australian histone allele argues against any functional correlation between the common Pacific allele and colonization ability. More likely there has been a selective sweep in Australia, involving a locus linked to histone H3-D, that has eliminated allele 5 there in polyploid G. tabacina accessions. In this scenario, the migration of the H3-D allele 5 must predate the selective sweep and predate recombination and selection among polyploid lineages. Migration and occasional outcrossing, evident among Australian populations from the number of multilocus genotypes present there (see below), would promote such a sweep. Populations on Pacific islands, isolated from the common mainland gene pool, would be unaffected and would retain histone allele 5.
Lineage Recombination Among Polyploids.Glycine species are predominantly selfers, although they do possess chasmogamous flowers capable of outcrossing (17), and these G. tabacina polyploids also reproduce vegetatively by means of stolons. Given these life history traits, it might be expected that there would be high correlation among the different molecular markers. It might further be predicted that the polyploid, having originated recently and several times independently, would be subdivided into several races, each distinguishable by a set of these correlated markers. The predicted number of different multilocus genotypes in this model would be seven, the number of different haplotypes for the most polymorphic of the molecular markers, cpDNA.
Instead, we observed 20 different multilocus genotypes among the 40 accessions genotyped, of a possible 84 combinations. We feel that the most likely explanation for this pattern is that recombination has occurred among an initially smaller number of genotypes. Despite the likely prevalence of inbreeding, these plants possess chasmogamous, outcrossing flowers, and hybridization appears likely among B-genome species having life histories similar to these polyploids (15). Moreover, artificial hybridization studies confirm the interfertility of polyploid accessions having different chloroplast haplotypes (8) and histone alleles, and the wide geographic distribution of G. tabacina gives ample evidence of dispersal ability. Scattered populations thus would have both the opportunity and the ability to exchange genes. Gene flow among the B-type G. tabacina was hypothesized to account for the overall morphological homogeneity of the polyploid despite the heterogeneity of chloroplast haplotypes (11). The present data from nuclear genes are in accord with this hypothesis.
Alternative hypotheses that do not involve gene exchange at the polyploid level seem less plausible. As noted above, the fact that combinations of markers found in polyploids are not observed in extant diploids could be accounted for by hybridization or sorting of ancestral polymorphisms at the diploid level. Thus, instead of gene exchange and lineage recombination among a small number of original polyploid genotypes, it is formally possible that each of the 20 multilocus genotypes observed among polyploid accessions is derived from an independent origin of the polyploid. As new multilocus genotypes are discovered in the polyploid, the number of postulated independent origins also would rise in this model. The number of independent origins required could be reduced if individuals contributing the B genome were heterozygous and could be reduced to nearly the same number required by the lineage recombination model if individuals were heterozygous at all three nuclear loci. However, heterozygosity is rare in B-genome diploids at isozyme loci (A.H.D.B., unpublished data) and for histone H3-D (15). Therefore, the likelihood that a polyploid would be formed by a heterozygous individual seems low, particularly given that outcrossing (and hence hybrid formation) itself is a relatively uncommon event in the B-genome. Finally, it is possible that alleles or haplotypes could be introgressed into polyploids by recurrent hybridization with diploids. Although also formally possible (7), the probable low success rate and the requirement for several cycles of outcrossing would seem to make this model less likely than one involving rare independent origins followed by ready migration and occasional crossing among interfertile polyploid accessions. Thus, although recognizing that alternative hypotheses exist, we favor a model in which gene exchange and lineage recombination at the polyploid level, following a smaller number of independent origins, have produced most or all of the polymorphism in the polyploid.
It has long been recognized that a potential advantage of polyploid species is their ability to harbor greater genetic diversity than is possible for their diploid relatives, primarily because of fixed hybridity at each homoeologous locus (1, 4). In recent years, much attention has been given to the potential for a broader genetic base in polyploid species at many homologous loci attributable to multiple origins (reviewed in ref. 4). With gene exchange and recombination, independent contributions from genetically diverse diploids enrich the gene pool of an entire polyploid species, instead of leading to a series of isolated polyploid races, each individually depauperate genetically. In G. tabacina, a large number of multilocus genotypes coexist in a morphologically coherent but widely distributed species. That G. tabacina has colonized areas where no diploid Glycine species have been found, despite similar opportunities for dispersal (20), is perhaps caused in part by the advantages of this source of genetic diversity.
Acknowledgments
The authors thank Randy Bayer, Curt Brewbaker, Joe Miller, and two anonymous reviewers for comments on drafts of the manuscript. Work was supported by U.S. National Science Foundation Grant DEB 9614984.
ABBREVIATION
- cpDNA
chloroplast DNA
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Grant V. Plant Speciation. New York: Columbia Univ. Press; 1981. [Google Scholar]
- 2.Masterson J. Science. 1994;264:421–423. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 3.Harlan J R, de Wet J M J. Bot Rev. 1975;41:361–390. [Google Scholar]
- 4.Soltis D E, Soltis P S. Crit Rev Plant Sci. 1993;12:243–273. [Google Scholar]
- 5.Brochmann C, Elven R. Evol Trends Plants. 1992;6:111–124. [Google Scholar]
- 6.Segraves K A, Thompson J N, Soltis P S, Soltis D E. Mol Ecol. 1999;8:253–262. [Google Scholar]
- 7.Ramsey J, Schemske D W. Annu Rev Ecol Syst. 1998;29:467–501. [Google Scholar]
- 8.Singh R J, Kollipara K P, Hymowitz T. Genome. 1987;29:490–497. [Google Scholar]
- 9.Hymowitz T, Singh R J, Kollipara K P. Plant Breeding Rev. 1998;16:289–317. [Google Scholar]
- 10.Doyle J J, Doyle J L, Brown A H D. Aust Syst Bot. 1990;3:125–136. [Google Scholar]
- 11.Doyle J J, Doyle J L, Brown A H D, Grace J P. Proc Natl Acad Sci USA. 1990;87:714–717. doi: 10.1073/pnas.87.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 13.Doyle J J, Kanazin V, Shoemaker R C. Mol Phylogenet Evol. 1996;6:438–447. doi: 10.1006/mpev.1996.0092. [DOI] [PubMed] [Google Scholar]
- 14.Kanazin V, Blake T, Shoemaker R C. Mol Gen Genet. 1996;250:137–147. doi: 10.1007/BF02174172. [DOI] [PubMed] [Google Scholar]
- 15.Doyle J J, Doyle J L, Brown A H D. Mol Biol Evol. 1999;16:354–362. doi: 10.1093/oxfordjournals.molbev.a026116. [DOI] [PubMed] [Google Scholar]
- 16.Grant J E, Brown A H D, Grace J P. Aust J Bot. 1984;32:665–677. [Google Scholar]
- 17.Schoen D, Brown A H D. Evolution (Lawrence, Kans) 1991;45:1651–1664. doi: 10.1111/j.1558-5646.1991.tb02670.x. [DOI] [PubMed] [Google Scholar]
- 18.Brown A H D, Weir B S. In: Isozymes in Plant Genetics and Breeding Part A. Tanksley S D, Orton T J, editors. Amsterdam: Elsevier; 1983. pp. 219–239. [Google Scholar]
- 19.Gaut B S. Evol Biol. 1998;30:93–120. [Google Scholar]
- 20.Hymowitz T, Singh R J, Larkin R P. Micronesica. 1990;23:5–13. [Google Scholar]