Abstract
The Escherichia coli proteins H-NS is recognized as an important component among the major nucleoid-associated proteins. In studies of E. coli strains with defects in H-NS, we discovered a mutant that phenotypically restored stationary-phase viability (Rsv) of such strains. The Rsv phenotype was the result of a mutation that led to severalfold higher levels of the functionally and structurally related StpA protein. This mutation was a base pair change in the stpA structural gene, and the amino acid substitution in the StpA protein altered its turnover properties, suggesting a role for this residue in a cleavage site for proteolysis. We determined the stability of the StpA and the H-NS proteins and found that the StpA protein was degraded relatively rapidly in strains lacking functional H-NS, whereas H-NS remained stable irrespective of the presence/absence of StpA. Using protease-deficient mutants, we obtained evidence that the Lon protease was responsible for the degradation of StpA. The differential turnover of the nucleoid-associated proteins is suggested to contribute to the regulation of their stoichiometry and ratio in terms of homo- and heteromer formation. We conclude that StpA, in contrast to H-NS, is present mainly in heteromeric form in E. coli.
Keywords: nucleoid-associated protein, Lon-protease target, degradation signal, protein–protein interaction
Enterobacteria have several nucleoid-associated proteins that are important for chromosomal organization, and the H-NS protein is among the most abundant of such proteins in Escherichia coli. It affects expression of genes under certain conditions, and mutations in the genes encoding H-NS and its paralogue, StpA, affect the expression of several distinct operons both positively and negatively (reviewed in refs. 1 and 2). Expression of the hns gene is stimulated by the cold shock protein CspA (3) and the regulatory protein FIS (4, 5), but expression has also been shown to be autoregulated (4, 6, 7). Recently, expression of H-NS was shown to be under control of the DsrA RNA, although the exact mechanism remains to be elucidated (8, 9). The H-NS protein has also been suggested to affect gene expression at the posttranscriptional level—e.g., by affecting the translation of the gene encoding σS (10) and by affecting expression of the MalT protein (11).
The StpA protein has RNA chaperone activity (12), and overexpression of StpA can functionally suppress certain hns mutant features (13). Expression of stpA is negatively regulated by H-NS but stimulated by the leucine-responsive protein Lrp. Expression of stpA is also temperature dependent, with a lowered expression at 26°C as compared with 37°C (13). Overexpression of the StpA protein causes lowered expression of the hns gene (13). The H-NS and StpA proteins are able to form heterodimers in vitro (14), and evidence for StpA and H-NS coordinate activity at certain genes has also been found (13, 15).
In this report, we show evidence that the StpA protein is rapidly degraded in the absence of H-NS and that the degradation requires functional Lon protease. We isolated a mutant showing restored viability in stationary phase and found that it contained a mutation in the stpA gene encoding a more stable form of StpA.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
Strains are listed in Table 1. Plasmids used were pBSN157, a transcriptional stpA-lacZ fusion plasmid (13), and its vector pFZY1 (16).
Table 1.
Bacterial strains
| Strain | Description | Source/ref. |
|---|---|---|
| MC4100 | rpoS+, Δ(argF–lac) | 30 |
| BSNI | hns∷Cmr, | 21 |
| BSN5 | stpA60∷Kmr | 13 |
| BEU603 | trp∷Tn10, Δhns | 13 |
| RH90 | MC4100, rpoS∷Tn10 | 31 |
| JGJ150 | MC4100, hns∷Cmr | This work |
| JGJ151 | RH90, hns∷Cmr | This work |
| JGJ152 | JGJ150, stpA60∷Kmr | This work |
| JGJ201 | RH90, stpA60∷Kmr | This work |
| JGJ202 | JGJ151, stpA60∷Kmr | This work |
| JGJ210 | Rsv derivative of JGJ151 | This work |
| CAG18562 | zfi-3143∷Tn10 Kmr | 22 |
| JGJ211 | JGJ210, zfi-3143∷Tn10Kmr | This work |
| JGJ212 | MC4100, zfi-3143∷Tn10 Kmr StpAwt | This work |
| JGJ213 | MC4100, zfi-3143∷Tn10 Kmr StpAF21C | This work |
| JGJ214 | JGJ150, zfi-3143∷Tn10 Kmr StpAwt | This work |
| JGJ215 | JGJ150, zfi-3143∷Tn10 Kmr StpAF21C | This work |
| SG20250 | MC4100, lon+ | 23 |
| SG1041 | MC4100, lon-100 | 23 |
| JGJ220 | SG20250, trp∷Tn10 | This work |
| JGJ221 | SG20250, trp∷Tn10, Δhns | This work |
| JGJ222 | SG1041, trp∷Tn10 | This work |
| JGJ223 | SG1041, trp∷Tn10, Δhns | This work |
| BSN26 | MC4100, trp∷Tn10 | 11 |
| BSN28 | MC4100, trp∷Tn10, stpA60∷Kmr | 11 |
Cmr, chloramphenicol-resistant; Kmr, kanamycin-resistant.
Growth Media and Culture Conditions.
Bacteria were grown in Luria–Bertani (LB) media (17) at 37°C with vigorous shaking (220 rpm). Growth was monitored by measuring Klett units on a Klett Summerson colorimeter, where 50 Klett units (no. KS-66, red filter) corresponds to approximately OD600 = 0.4. Where considered necessary, antibiotics (Sigma) were used at the following concentrations: carbenicillin, 25 μg⋅ml−1; chloramphenicol, 10 μg⋅ml−1; kanamycin, 25 or 50 μg⋅ml−1; spectinomycin, 100 μg⋅ml−1; and tetracycline, 7.5 μg⋅ml−1. Plates containing 40 μg⋅ml−1 of the chromogenic β-glucoside 5-bromo-4-chloro-3-indolyl β-d-glucopyranoside (Sigma) were used to monitor the Bgl phenotype expressed by hns mutant derivatives.
Stationary-Phase Survival Experiments.
Samples were taken on a daily basis from stationary-phase cultures, serially diluted in a physiological salt solution, and plated on LB plates. Viable counts were determined as colony-forming units (CFU)⋅ml−1 and plotted as log CFU⋅ml−1 versus time. The experiment was repeated two times, with similar results.
β-Galactosidase Assay.
Samples were taken from LB cultures at 50 Klett units. β-Galactosidase reactions were assayed as in ref. 18, with the exception that we used chloroform and 0.002% sodium dodecyl sulfate (SDS) to disrupt the bacteria. Data represent the mean values from assays performed in duplicate in three separate experiments, and standard errors were calculated. The numbers of plasmid-containing cells were measured as before (13).
Genetic Techniques.
Molecular genetic manipulations were performed essentially as described (19). Generalized bacteriophage P1 transduction was done as previously described (20). JGJ150 (hns∷Cmr) was derived from MC4100, which was transduced with P1 grown on BSN1 [hns∷Cmr (21)]. JGJ151 (rpoS∷Tn10, hns∷Cmr) was derived from RH90 (rpoS∷Tn10), which was transduced with P1 grown on BSN1. JGJ152 (hns∷Cmr, stpA60∷Kmr) was derived from JGJ150 that was transduced with P1 grown on BSN5 (stpA60∷Kmr). JGJ201 (rpoS∷Tn10, stpA60∷Kmr) was derived from RH90 that was transduced with P1 grown on BSN5. JGJ202 (rpoS∷Tn10, hns∷Cmr, stpA60∷Kmr) was derived from JGJ151 that was transduced with P1 grown on BSN5. JGJ210 (Rsv derivative of JGJ151) was isolated as described in Results. JGJ211 (rpoS∷Tn10, hns∷Cmr, zfi-3143∷Tn10Kmr, StpAF21C) originated from JGJ210 that was transduced with P1 grown on CAG18562 [zfi-3143∷Tn10Kmr (22)]. JGJ212 (zfi-3143∷ Tn10Kmr, StpAwt) and JGJ213 (zfi-3143∷Tn10Kmr, StpAF21C) were derived from MC4100, JGJ214 (zfi-3143∷Tn10Kmr, hns∷Cmr, StpAwt) and JGJ215 (zfi-3143∷Tn10Kmr, hns∷Cmr, StpAF21C) originated from strain JGJ150. Strains MC4100 and JGJ150 were transduced with P1 grown on JGJ211, and transductants were isolated by selection for kanamycin resistance (i.e., the zfi-3143∷Tn10Kmr marker; zfi–stpA cotransduction frequency was about 25%). To investigate whether the transductants either received the stpA allele containing the base pair substitution or whether they still possessed the wild-type (wt) allele, we used primer 40 (5′-GGGGGTACCGAAATAATCTCGCGCAGGACTG-3′) and primer 41 (5′-GGGGTCGACCTTTGTTGGTGCCGGGTTACTG-3′) to PCR amplify the stpA gene. Thereafter, we subjected the obtained fragment to EcoRI digestion. This digestion confirmed that the stpA gene of strain JGJ212 and JGJ214 contained the EcoRI site, whereas the stpA alleles in JGJ213 and JGJ215 did not. JGJ220 (trp∷Tn10) and JGJ221 (trp∷Tn10, Δhns) were derived from strain SG20250 [lon+ (23)], which had been transduced with P1 grown on BEU603 [trp∷Tn10, Δhns (13)]. Transductants were selected for growth on tetracycline plates and subsequently tested with respect to Bgl phenotype. The hns+ strain showed a negative Bgl phenotype and the hns strain showed a positive Bgl phenotype. Similarly, strains JGJ222 (trp∷Tn10) and JGJ223 (trp∷Tn10, Δhns) originated from strain SG1041 [lon (23)], which had been transduced with P1 grown on BEU603.
In Vivo Protein Stability Experiment.
To determine the intracellular stability of StpA and H-NS we used a technique described in ref. 24. Protein stability was monitored after the protein synthesis had been inhibited by the addition of spectinomycin (100 μg⋅ml−1) to bacterial cultures grown to 50 Klett units in LB medium at 37°C. Samples to be analyzed by Western blotting were removed at indicated times.
Gel Electrophoresis and Western Blotting.
Bacteria were pelleted and resuspended in sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis (PAGE) sample buffer. The samples (sample loading was normalized according to determination of cell mass) were subjected to SDS/PAGE (15% polyacrylamide concentration) and thereafter blotted onto a 0.2-μm-pore-size poly(vinylidene difluoride) transfer membrane (Bio-Rad) by use of a semidry blotting apparatus. H-NS and StpA were purified essentially as described previously (6, 12). Polyclonal rabbit antisera directed against purified StpA or H-NS were used as the primary antibodies in Western blot analysis. To further increase the specificity of the StpA antiserum, it was adsorbed against a lysate of a stpA mutant strain. Further visualization followed the method described in ref. 11, where the membrane was developed with AttoPhos substrate (JBL Scientific, Northridge, CA) and scanned with the STORM system (Molecular Dynamics). Measurement of the StpA protein and its half-life were performed with the ImageQuant program (Molecular Dynamics).
RESULTS
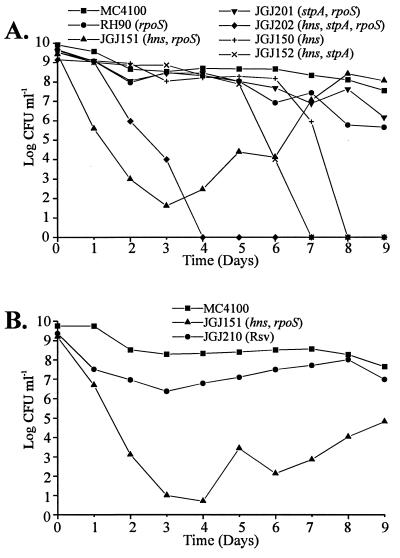
Isolation of Mutants Restoring Stationary-Phase Viability to hns Strains. Our earlier studies with an E. coli strain carrying mutations in the hns and stpA loci indicated that there might be effects on growth and viability (13). To further examine the role of H-NS and StpA on bacterial growth and viability we tested a number of different strains with such mutations in prolonged growth experiments using LB liquid medium at 37°C. Relevant antibiotics were included to ensure that the original mutations were retained. Viability was monitored by viable counts on solid medium without antibiotics (Fig. 1A). While hns+ strains showed a decrease in viability of about two orders of magnitude during extended incubation at stationary phase, the different hns mutants showed a strikingly different behavior: The strain containing mutations in hns and rpoS (JGJ151) and the strain carrying mutations in hns, stpA, and rpoS (JGJ202) displayed reduced viability immediately after they had reached the stationary phase. Within 2–3 days the viability was reduced by 6–8 orders of magnitude in such cultures (Fig. 1A). A similar reduction in viability was exhibited by rpoS+, hns mutant strains (JGJ150, JGJ152) after about 7–8 days. Cultures of strains lacking both H-NS and StpA appeared to completely lose viability upon prolonged incubation. Interestingly, after the initial large reduction in colony-forming units, cultures of the strain JGJ151 showed resumed viability after another 2–3 days, when a subpopulation appeared and reached a cell density as high as the wt strain (Fig. 1A). At day 8, we isolated single-cell colonies from JGJ151 cultures, and a selected isolate used in further studies was denoted JGJ210. Isolates showing restored viability will hereby be referred to as restored stationary-phase viability (Rsv). The Rsv phenotype was not observed with strains also carrying a mutation in the stpA gene (e.g., JGJ152, JGJ202).
Figure 1.
(A) Viability test of strains in stationary phase at 37°C. (B) Viability test of the Rsv isolate JGJ210. All strains were grown in LB medium at 37°C to stationary phase (day zero). Thereafter, samples were taken each day for viable count determinations and results were plotted on a logarithmic scale versus time. CFU, colony-forming units.
To study the growth of the Rsv isolate, and to investigate whether it, unlike its parental strain JGJ151, could maintain high viability for several days in stationary phase, we re-inoculated strain JGJ210 in LB medium at 37°C and monitored its viability in a comparison with the parental strain and a wt strain (MC4100). As shown in Fig. 1B, JGJ210 did not show the same reduction in viability as the parental strain JGJ151 after the first few days, but remained stable at approximately the same level as the wt strain.
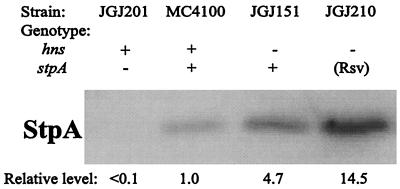
The Amount of StpA Protein Is Elevated in Rsv Isolates. One possible explanation for our results would be that the expression of StpA was increased and therefore able to complement the deficiency in H-NS. To investigate this possibility we performed Western blotting analysis with antisera directed against StpA and showing very low cross-reactivity to H-NS. As seen in Fig. 2, the level of StpA increased approximately 3-fold in JGJ210 as compared with the parental strain JGJ151.
Figure 2.
Quantitative determination of StpA protein content. Western blotting was performed with total cell extract from the following strains: MC4100 (wt), JGJ151 (hns, rpoS), JGJ210 (Rsv), and JGJ201 (rpoS, stpA) included as negative control.
The finding of an elevated level of StpA protein prompted us to study if this was caused by an increase at the transcriptional level. We therefore used a low-copy-number stpA-lacZ transcriptional fusion plasmid to measure if an increased expression could be detected from the stpA gene in trans (13), and the results are summarized in Table 2. The level of stpA-lacZ expression was lowered in the Rsv strain JGJ210 as compared with its parental strain JGJ151, and nearly as low as in the strain containing functional H-NS. This suggests that the increased level of the StpA protein in strain JGJ210 was not due to increased transcription of the stpA operon. Instead, the results indicate that the higher levels of StpA in JGJ210 led to repression of its own transcription.
Table 2.
Expression of an stpA-lacZ operon fusion
| Strain | β-Galactosidase
|
|
|---|---|---|
| pFZY1 (vector) | pBSN157 (stpA-lacZ) | |
| MC4100 (wt) | 3 ± 0.3 | 307 ± 16 |
| JGJ151 (hns, rpoS) | 3 ± 0 | 1,111 ± 130 |
| JGJ210 (Rsv) | 3 ± 0 | 442 ± 71 |
β-Galactosidase activity was monitored as described in the text. Data are given as mean ± SEM of three separate experiments.
The Restored Viability Phenotype Results from a Single Amino Acid Substitution (F21C) in the StpA Protein.
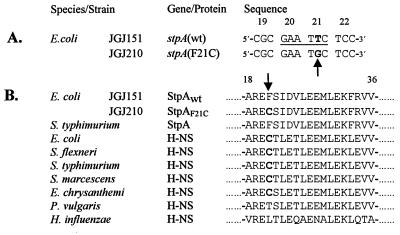
To further analyze the mutation conferring increased levels of StpA, we sequenced the stpA locus. We could not find any differences in the promoter region of JGJ210 as compared with the parental strain JGJ151 (data not shown). However, in the structural gene of stpA, we detected one base pair substitution, changing codon 21 to TGC from TTC (Fig. 3A). At the amino acid level, this substitution converted the phenylalanine at position 21 to a cysteine in strain JGJ210 (Fig. 3B). With genetic methods, we transferred the mutation to other strains. Because the substitution disrupted an EcoRI site we could easily confirm the presence of the specific mutation by EcoRI digestion of PCR-amplified chromosomal DNA (see Materials and Methods). We concluded that the Rsv phenotype of JGJ210 was due to the mutation in the stpA gene resulting in StpAF21C.
Figure 3.
Sequence analysis of the stpA gene from the Rsv mutant. (A) DNA sequence alignment of four codons, showing the single base pair difference in boldface between the stpA sequence of strain JGJ151 and the Rsv isolate JGJ210. An arrow points out the base pair substitution, and the EcoRI site in the wt sequence is underlined. The codon positions are shown above the sequences. (B) Multiple peptide sequence alignment of StpA and H-NS peptides from several species. Boldface letters highlight the cysteine residue at position 21 (also indicated by arrow). The numbers above the sequences indicate relative position, where 1 is the N-terminal methionine. Accession numbers for the protein sequences are as follows: StpA E. coli, Sw P30017; StpA Salmonella typhimurium, GB AF009363; H-NS E. coli, Sw P08936; H-NS Shigella flexneri, Sw P09120; H-NS Salmonella typhimurium, Sw P17428; H-NS Serratia marcescens, Sw P18955; H-NS Erwinia chrysanthemi EMBL X89444; H-NS Proteus vulgaris, Sw 18818; and H-NS Haemophilus influenzae, Sw P43831. EMBL, GB, and Sw refer to the European Molecular Biology Laboratory, GenBank, and Swiss-Prot databases, respectively.
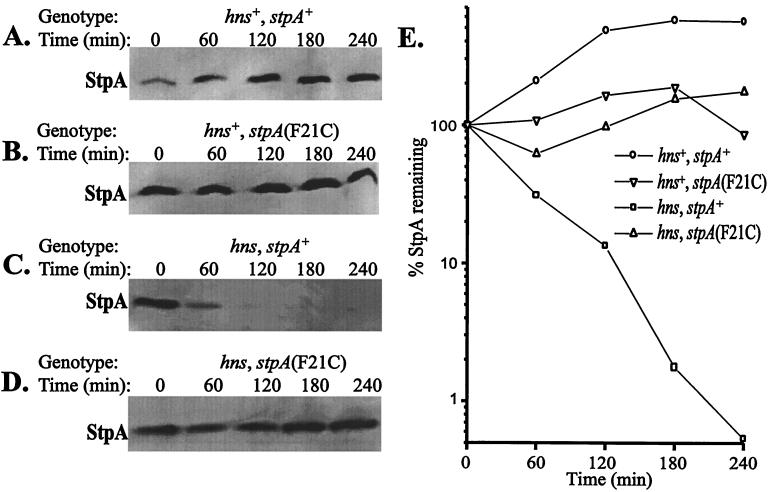
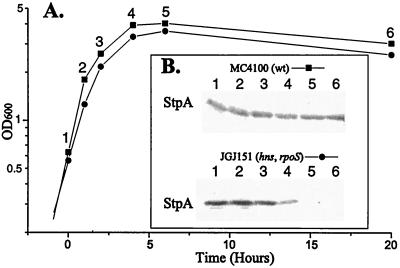
The StpA Protein Is Unstable in the Absence of H-NS. The finding that StpA levels were altered by an amino acid substitution led us to consider that it affected protein turnover. We therefore studied StpA protein stability in different genetic backgrounds. We used a method (24) in which protein synthesis is inhibited by the addition of 100 μg⋅ml−1 spectinomycin and samples are removed at different times (see Materials and Methods). Immunodetection was performed with antisera recognizing StpA but not H-NS (cf. Fig. 2). As seen in Fig. 4A and B, the StpA protein was stable throughout the duration of the experiment in a genetic background ensuring functional H-NS. Even 4 hr after the addition of spectinomycin, approximately the same level of StpA was detected. However, the level of StpAwt protein in strains lacking functional H-NS was reduced to a larger extent already 60 min after the addition of spectinomycin (Fig. 4C). Quantitative measurements of the level of StpA at the different time points were used to estimate the half-life of the protein, and StpAwt showed a half-life of about 35 min under these conditions (Fig. 4E). Interestingly, no degradation could be detected in a strain containing StpAF21C but lacking H-NS (Fig. 4D). Even after 4 hr, the level of StpAF21C was approximately the same as at the onset of spectinomycin addition. The finding that StpA was unstable in the absence of H-NS prompted us to determine whether there was a decline in StpA levels during stationary phase of the strains that lost viability. As shown in Fig. 5, the StpA levels declined in strain JGJ151 (hns, rpoS) soon after entry into stationary phase. The decline in StpA levels was independent of rpoS, since we obtained similar results (data not shown) with strain JGJ150 (hns). In the case of the Rsv isolate JGJ210, the StpA levels remained constant in the stationary phase (data not shown).
Figure 4.
H-NS-dependent stability and turnover of StpA. The relative stability of StpA was measured after protein synthesis was inhibited as described in the text. StpA stability was measured in the following strains: (A) JGJ212 (hns+, stpA+); (B) JGJ213 [hns+, stpA(F21C)]; (C) JGJ214 (hns, stpA+); and (D) JGJ215 [hns, stpA(F21C)]. (E) Quantitative determinations of StpA from the different experiments in A–D plotted as a function of time.
Figure 5.
Determination of StpA levels during entry into stationary phase. (A) Growth of strains MC4100 (wt) and JGJ151 (hns, rpoS) into stationary phase (the time point of 20 hr corresponds to day 0 in Fig. 1). Samples were removed at indicated times (1–6). (B) Western blotting to detect StpA was performed with total cell extracts from samples taken at times indicated in A.
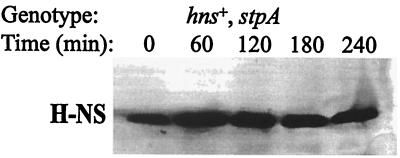
We performed similar studies of H-NS protein stability. The level of H-NS protein was unaffected both in a wt strain and in a stpA mutant strain during the 4 hr after the addition of spectinomycin (Fig. 6 and data not shown). Apart from the StpA proteins in E. coli and S. typhimurium, none of the other so-far-identified H-NS-like proteins in different bacteria has a phenylalanine at position 21 (Fig. 3B). However, as in the case of mutant StpAF21C, a cysteine at position 21 is also present in the H-NS proteins of five different bacterial species, among them the H-NS protein of E. coli (Fig. 3B).
Figure 6.
Stability of H-NS protein in the absence of StpA. The relative amount of H-NS in strain BSN28 (hns+, stpA) was measured after protein synthesis was inhibited as described in the text.
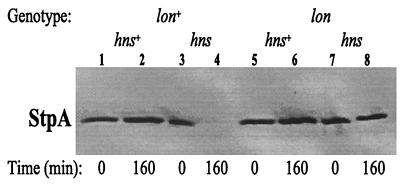
Turnover of StpA Is Lon Protease Dependent.
To test if some specific protease was responsible for the degradation of StpA, we examined the level of StpA protein in protease-deficient strains. In hns+, stpA+ strains lacking the OmpT or Lon proteases, we were not able to detect any significant difference in the steady-state levels of StpA protein according to Western blot analysis (data not shown). To examine the possible involvement of the Lon protease in the turnover of StpA, we introduced an hns allele into otherwise isogenic lon+ and lon mutant strains and tested StpA protein stability by using the same approach described above. As seen in Fig. 7, the StpA protein had completely disappeared 160 min after the addition of spectinomycin in the case of hns, lon+ cells (Fig. 7, lane 4). In contrast, strains also lacking Lon protease had a nearly unaffected level of StpA throughout the duration of the test (Fig. 7, lane 8). As detected before, the level of StpA was not affected in hns+ strains (Fig. 7, lanes 2 and 6). These results provided strong evidence that the StpA protein is a substrate for the Lon protease and that StpA in complex with H-NS is protected from proteolysis. We subsequently tested if a lon mutation per se could alter the survival of an hns mutant strain during stationary phase. The results (data not shown) indicated that the lon mutation could not restore viability to the hns mutant strain. This finding also suggested that we might not expect to find Lon-defective mutants among Rsv isolates.
Figure 7.
StpA stability in Lon-protease-deficient E. coli. Measurement of StpA stability in JGJ220 (hns+, lon+), JGJ221 (hns, lon+), JGJ222 (hns+, lon), and JGJ223 (hns, lon) strains. The relative amount of StpA was monitored by Western blotting after protein synthesis was inhibited.
DISCUSSION
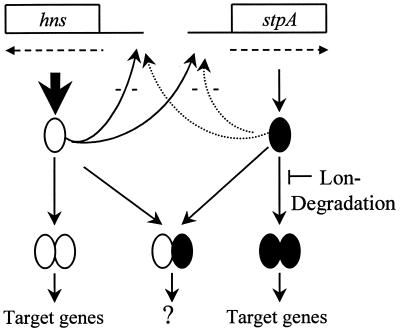
The present data show that there is differential turnover of the nucleoid-associated proteins H-NS and StpA. The StpA protein was subjected to relatively rapid degradation in vivo when H-NS protein was absent (Fig. 4C). On the other hand, no such effect was observed in the case of the H-NS protein, which appeared quite stable in the absence of StpA (Fig. 6). In the presence of H-NS, however, turnover of StpA was not detected during the 4-hr-long experiment (Fig. 4A). These results suggest that there are very low levels of StpA monomers and homodimers normally in vivo because they would be degraded rapidly. Apparently, the bacteria have different mechanisms to ensure appropriate levels of H-NS and StpA to maintain certain cellular properties. At the transcriptional level there is both autoregulatory control of their expression and cross-talk regulation between the hns and stpA genes (4, 6, 7, 13). While neither the regulationally active nor the protease-sensitive molecular form of StpA (i.e., monomers and oligomers, respectively) is determined, it is evident that the levels of StpA will be governed by both synthesis and degradation. Fig. 8 shows a schematic summary of the molecular forms and turnover of StpA and H-NS and their regulation. On the basis of the present evidence, we conclude that StpA is present mainly in heteromeric complexes with H-NS in E. coli. We postulate that such heteromers are functionally different from, and presumably have other targets than, H-NS or StpA homomers.
Figure 8.
Schematic summary of molecular forms and turnover of StpA and H-NS. Lon-protease-dependent StpA degradation is indicated in the case of StpA in accordance with the present data. Coordinate regulation at the transcriptional level has been demonstrated earlier (13, 15).
The observation that the StpA protein remained stable in a lon mutant strain during a long period of time strongly supports the conclusion that Lon is the only protease degrading StpA. There is an interesting similarity in our findings to earlier studies on the nucleoid-associated HU proteins in E. coli (25). It was shown that one of the two HU proteins, HU1 (also referred to as HUβ or HupB) is subject to Lon-dependent degradation (25). However, there are also differences between the features shown by the HU proteins and features shown by the H-NS/StpA proteins. The two HU subunits occur in homodimer and heterodimer forms with varying composition during the growth cycle, and the heterodimer formation was reported to be required for long-term survival in stationary phase (26). In the case of H-NS/StpA our present data show that formation of heteromers per se is not essential for viability, but that homomers of either protein H-NSwt and StpAF21C may ensure long-term viability (Fig. 1).
Survival of E. coli in stationary phase is known to depend on RpoS, and there are presumably many alterations in cell physiological features when bacteria adapt to the altered conditions (27). Mutants that may take over stationary-phase cultures were earlier shown to be altered in the rpoS gene (28). The very drastic reduction in viability shown here with hns, stpA mutants suggests that products from some gene(s) affected by H-NS/StpA per se cause cell death. In absence of RpoS the cells lost viability at once, whereas in the case of rpoS+ derivatives the loss of viability was delayed (Fig. 1). The reason for this difference is not known, but evidently the combination of H-NS and RpoS deficiency made the bacteria particularly unfit to the stationary phase. In the present study, we did not observe any Rsv revertants with the rpoS+, hns strain after the prolonged incubations. The reason for this failure is unclear, but perhaps progressive changes in the culture medium (e.g., nutrient depletion and pH alterations) did not allow for efficient growth of such putative mutants. The fact that StpAF21C restored viability in the absence of H-NS implied that it was a gain-of-function mutation. The results indicated that the StpAF21C protein was able to repress the transcription of stpA (Table 2) and therefore was still functional in gene regulatory systems despite the amino acid substitution. Our results furthermore suggest that the increased level of StpA in a StpAF21C strain (Fig. 2) was caused by the inability of the Lon protease to degrade StpAF21C (Fig. 4D). This could be because the mutation changes the conformation of StpA to a more compact, stable form. Another explanation would be that the mutation makes the StpA protein uncleavable by the Lon protease. This latter possibility is further supported by the comparison between the StpA sequence, and the Lon protease consensus cleavage site proposed (29): ΦX3–4LS(L,X)X5SXΦ, in which Φ represents a hydrophobic side chain. This consensus fits well with the StpA sequence: ΦX4LX8Φ where the first Φ is at position 21 (Fig. 3B). The conversion of a phenylalanine to a cysteine in StpAF21C results in a polar instead of a hydrophobic amino acid at the first residue of the consensus. This alteration could reduce the interaction between StpA and Lon. The isolation of the StpAF21C mutant protein provides an example in which loss of the Lon protease substrate phenotype is genetically selected because of its essential physiological importance. It remains to be elucidated if there is a specific growth phase or condition when StpA forms homodimers to some greater extent. Also, the question about correlation with Lon expression is of interest. Further work concerning the interaction between StpAwt and StpAF21C with the Lon protease will, we hope, shed more light on the molecular action of the Lon protease and the degradation signal(s) in such substrate proteins. Evidently, the present system offers good selective possibilities for genetic analysis of such signals and determinants.
Acknowledgments
We thank Drs. R. Hengge-Aronis, S. Gottesman, and C. Gross for bacterial strains. We are grateful to M. Persson and S. Wang for assistance with the experiments. This work was supported by grants from the Swedish Natural Science Research Council, the Swedish Medical Research Council, and the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine.
ABBREVIATION
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Atlung T, Ingmer H. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 2.Williams R M, Rimsky S. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 3.La Teana A, Brandi A, Falconi M, Spurio R, Pon C L, Gualerzi C O. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falconi M, Higgins N P, Spurio R, Pon C L, Gualerzi C O. Mol Microbiol. 1993;10:273–282. [PubMed] [Google Scholar]
- 5.Falconi M, Brandi A, La Teana A, Gualerzi C O, Pon C L. Mol Microbiol. 1996;19:965–975. doi: 10.1046/j.1365-2958.1996.436961.x. [DOI] [PubMed] [Google Scholar]
- 6.Dersch P, Schmidt K, Bremer E. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 7.Ueguchi C, Kakeda M, Mizuno T. Mol Gen Genet. 1993;236:171–178. doi: 10.1007/BF00277109. [DOI] [PubMed] [Google Scholar]
- 8.Lease R A, Cusick M E, Belfort M. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashino T, Ueguchi C, Mizuno T. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson J, Dagberg B, Richet E, Uhlin B E. J Bacteriol. 1998;180:6117–6125. doi: 10.1128/jb.180.23.6117-6125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang A, Derbyshire V, Salvo J L, Belfort M. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 13.Sondén B, Uhlin B E. EMBO J. 1996;15:4970–4980. [PMC free article] [PubMed] [Google Scholar]
- 14.Williams R M, Rimsky S, Buc H. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 16.Koop A H, Hartley M E, Bourgeois S. Gene. 1987;52:245–256. doi: 10.1016/0378-1119(87)90051-5. [DOI] [PubMed] [Google Scholar]
- 17.Bertani G. J Bacteriol. 1951;63:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Willets N S, Clark A J, Low B. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsman K, Sondén B, Göransson M, Uhlin B E. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trisler P, Gottesman S. J Bacteriol. 1984;160:184–191. doi: 10.1128/jb.160.1.184-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geuskens V, Mhammedi-Alaoui A, Desmet L, Toussaint A. EMBO J. 1992;11:5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnefoy E, Almeida A, Rouviere-Yaniv J. Proc Natl Acad Sci USA. 1989;86:7691–7695. doi: 10.1073/pnas.86.20.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claret L, Rouviere-Yaniv J. J Mol Biol. 1997;273:93–104. doi: 10.1006/jmbi.1997.1310. [DOI] [PubMed] [Google Scholar]
- 27.Hengge-Aronis R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd. Ed. Neidhardt F C, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1497–1512. [Google Scholar]
- 28.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 29.Gottesman S, Maurizi M R. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 31.Hengge-Aronis R, Fischer D. Mol Microbiol. 1992;6:1877–1886. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]