Abstract
V(D)J recombination substrate choice is regulated to ensure that the appropriate gene segments are rearranged during lymphocyte development. It has been proposed that regulation of substrate usage is determined by changes in accessibility of the DNA targets. We show that Rag-mediated recombination of an episomal substrate in cells is affected by its packaging into chromatin. Chromatinized substrates were inefficiently rearranged, and methylation further reduced recombination. Disruption of nucleosomes by using butyrate on methylated substrates was sufficient to activate recombination, and dexamethasone could activate recombination in the absence of detectable transcription. Therefore, chromatin structure, and its manipulation by altering nucleosome positioning, can directly affect recombination efficiencies.
Antigen receptor genes are assembled during lymphoid development from gene segments flanked by recombination signal sequences (RSSs) that are targets for the V(D)J recombination machinery (1). This nonhomologous site-specific recombination process must be precisely controlled because of the potentially severe consequences of aberrant chromosomal recombination events. Coexpression of the lymphoid-specific genes Rag-1 and Rag-2 occurs in developing B and T cells, ensuring that only the appropriate cell types are recombinationally active (2). A second level of regulation exists to control lineage specificity. Complete Ig gene rearrangement occurs only in B cells whereas T cell receptor genes rearrange exclusively in T cells (3). Moreover, within developing B and T cells, there are temporal controls over antigen receptor rearrangement. For example, in early B cells, the Ig heavy chain is rearranged before the light chain, and D to JH gene segments are combined before V-to-DJ assembly can commence.
This remarkable ability of the recombination machinery to target only a very small subset of substrates within a given cell, together with the fact that a single recombinase is required for this process, has led to the hypothesis that the accessibility of DNA targets to the recombination machinery is tightly controlled (4). Potential loci for recombination are thought to be inaccessible until they are modified to allow the recombinational machinery access to target sites. In support of this model, forced expression of Rag-1 and Rag-2 in nonlymphoid cells allow exogenous targets to be recombined but, the endogenous Ig and T cell receptor gene segments within the same cells remain unrecombined (4).
Other properties of the endogenous loci thought to be associated with “open” DNA also correlate with recombinational activity. These properties include transcriptional activity and hypomethylation (5, 6). For example, transcription initiates within each locus to be rearranged at the time when recombination commences within that region of the genome. Extensive studies of transgenic recombination signal sequences have demonstrated correlative relationships between transcriptional transactivation and recombination frequencies; in general, transcriptionally silent transgenic substrates were refractory to recombination (7). However, certain transgenic substrates that were transcriptionally active were recombinationally silent, suggesting that transcription itself is not sufficient to render a locus recombinationally accessible and that a mechanistic relationship, if any, between these properties is complex (8, 9). Hypermethylation has been correlated with the existence of a repressed state, refractory to V(D)J recombination (5, 10). Antigen receptor loci become demethylated as they are rendered recombinationally active (11, 12). A transgenic model system was able to recapitulate the dependence on hypomethylation for recombination, suggesting that changes in CpG methylation may be another potentially important switch for Rag-mediated recombinational accessibility (13). Because these experiments have been correlative in nature, we felt that an experiment that could directly test whether these properties affected recombination efficiencies was needed.
Specifically, we wanted to determine whether DNA packaging is sufficient to silence recombination loci and whether the modulation of transcriptional activity and methylation status can explain differences in the utilization of recombination sites in vivo. Given the size and complexity of the antigen receptor loci, it is difficult to directly manipulate these properties to determine whether they are responsible for the different recombination events observed. Therefore, we devised a strategy that uses an episomal recombination substrate in which we could alter the nucleosomal structure and positioning, methylation status, and transcriptional activity of target sites within a recombination substrate to explore whether these properties directly influence substrate targeting by the V(D)J recombinase.
MATERIALS AND METHODS
Plasmids.
pSC1 was constructed as follows. First, the RSV promoter was deleted from pREP4 (Invitrogen), and the Ebna1 cassette was inactivated by the insertion of a 50-bp fragment to generate pEP4. Then, a fragment containing the luciferase gene from pMAMneoluc (CLONTECH) was subcloned into the MCS of pEP4, to generate pEL. The 23RSS along with two flanking nucleotides 5′-TGCACAGTGGTAGTACTCCACTGTCTGGCTGTACAAAAAC C-3′ was subcloned as an oligonucleotide fragment into the 3′ untranslated region of the luciferase gene in pEL, generating pEL23 (RSS in bold face, flanking nucleotides italicized). The mouse mammary tumor virus long terminal repeat (MMTVLTR) was first subcloned into pBSKS+ (NEB, Beverly, MA) and was used as a substrate for site-directed mutagenesis by using Epicurian Coli XLmutS Kanr Competent Cells (Stratagene) to embed the 12 RSS 5′-tgCACAGTGagtggtttcctgacAAAAACA-3′ (endogenous sequence in lowercase, RSS in bold). Subsequently, the MMTVLTR12RSS was subcloned into pEL23, to generate pSC1. Expression vectors for full-length murine Rag-1 and Rag-2, murine glucocorticoid receptor, and adenovirus E1a have been described elsewhere (14–16). In vitro methylation of plasmids were performed by using SssI methylase according to the manufacturer (NEB).
Cell lines and Transfections. The human embryonic kidney epithelial cell line, 293, and its derivative stably expressing Ebna-1, 293Ebna1 (Invitrogen), were maintained in DMEM containing 10% fetal bovine serum and Pen/Strep. The human osteosarcoma cell line, U2OS, was maintained in DMEM containing 15% fetal bovine serum and Pen/Strep. Dexamethasone (Sigma) was added at 1 uM, and butyrate (Sigma) was added at 7 mM 24 hours posttransfection.
DNA was introduced into cells by calcium phosphate-mediated transfection as described (16). Recombination substrate vectors (1 μg) and other vectors (5 μg) were transfected into cells plated 16 hr earlier. The cells were fed 24 hours posttransfection. Cells were either harvested 48 hours posttransfection or were selected in hygromycin (Calbiochem) to generate stable cell lines. For analysis, the cell suspension was divided, was washed in PBS, and was lysed for recovery of episomal DNA (16) and in parallel for luciferase activity by using a Promega kit according to the manufacturers’ protocol. Background luciferase activity was subtracted from the experimental samples before fold values were calculated.
Recombination Assay.
One percent of the recovered DNA was subject to PCR amplification. These PCR primers are unable to amplify a product until the V(D)J recombination reaction has occurred, bringing the primer sites in close proximity. The following oligonucleotides were used to direct the amplification of these signal-joint products: KasI, 5′-CTCTTCGGGCGCCAGCTG CCGCAG-3′, SspI, 5′-TGTGGAACGAAG TACCGAAAGGTCTTACC-3′. A 50-μl PCR amplification reaction mixture contained 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 100 ng of each oligo, and 1 unit of Taq polymerase (Boehringer Mannheim) and was subject to the following amplification profile: 1 cycle of 94°C for 4 min; (30 cycles for stable lines, and 25 cycles for transients) of 94°C for 1 min, 72°C for 2 min; 72°C for 4 min. The products were resolved on a 1.5% TBE (45 mM Tris·Borate/1 mM EDTA, pH 8.0) gel, were transferred to Hybond N (Amersham Pharmacia), and were probed with the kinased oligonucleotide SJ1 (5′-GTGGAGTACTACGAGTGTGCACAGTGAGTGGTTTCCTG-3′) that spans the newly created signal-joint. To determine relative activities, a dilution curve for the PCR amplification was generated for each experiment: The sample representing pSC1 plus RAG1/2 was serially diluted into the negative control, and the dilution series was amplified in parallel with the control and experimental samples. Values for band intensities of the radiolabeled PCR products were determined by digitizing and quantitating using macbas software (Fuji) for Fig. 1B and by PhosphorImaging (Molecular Dynamics) for Fig. 1D. pSC1 plus RAG1/2 was set as 1.0, and pSC1 without RAG1/2 set as 0. Within the range of our assay, band intensities decreased proportionally to the dilution of the pSC1 plus RAG1/2 sample.
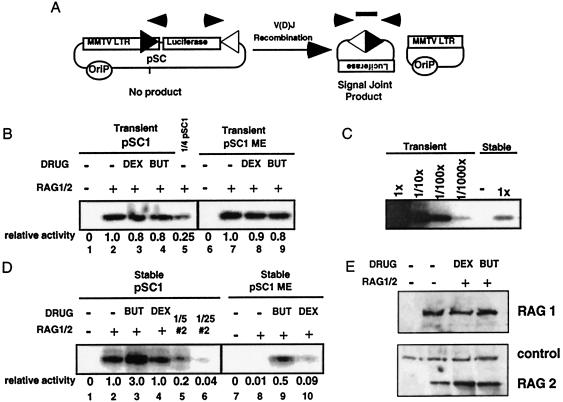
Figure 1.
Recombination efficiency is affected by the chromatin structure of the substrate. (A) Recombination substrate pSC1 contained a 12 RSS (black triangle) embedded in the MMTV LTR. The 23 RSS (white triangle) is downstream of the luciferase reporter gene. The Epstein–Barr origin of replication (Orip) is included to allow plasmid maintenance. After V(D)J recombination, signal-joint products could be amplified by PCR (arrows) and detected in a Southern blot probed with an oligonucleotide that spanned the newly formed signal joint (rectangle). The other product of the recombination reaction (the coding joint) is shown but not assayed. (B) Drug treatment does not increase the efficiency of recombination of transiently introduced substrates. pSC1 (lanes 1–4) or methylated pSC1, pSC1 ME (lanes 6–9) were cotransfected with Rag-1 and Rag-2 expression vectors (RAG1/2) into 293 cells, and the drugs butyrate (But) or dexamethasone (Dex) were added as indicated. Assays were performed as described in A. Fourfold less pSC1 DNA than in lanes 1–4 was transfected to generate lane 5. Relative activities were determined by using the titration series in which transient pSC1 plus RAG1/2 was defined as 1. (C) Stably maintained episomes are inhibited by ≈100-fold. Tenfold dilutions of transiently transfected pSC1 and RAG1/2 extract into empty extract generates a titration curve (lanes 1–4). Stably maintained pSC1 cells transfected with RAG1/2 (lane 6) or without RAG1/2 (lane 5) were amplified in parallel. (D) Stably maintained substrates are refractory to recombination, and methylation further decreases the efficiency of recombination. Cell lines stably maintaining pSC1 (lanes 1–4) or methylated pSC1, pSC1 ME (lanes 7–10) were transfected with RAG1/2 vectors and were treated with drugs as indicated. Note: the absolute amount of recombination in D, lane 2 is ≈100-fold less than in B, lane 2. Relative activities were calculated as in B except stable pSC1 plus RAG1/2 was defined as 1. (E) The levels of Rag proteins are unaffected by drug treatments. Expression vectors for Rag-1 and Rag-2 were cotransfected into 293 cells and were treated with dexamethasone or butyrate as indicated. Bands labeled control hybridized nonspecifically with the immunodetection reagents used.
Ligation-Mediated (LM) PCR Assay.
The LM-PCR assay was adapted from Zhu and Roth (17). In brief, 10% of the DNA recovered as in ref. 16 was ligated to 100 pmol preannealed BW-1/BW-2 (18) by using 4 units of T4 DNA ligase (NEB) and the supplied buffer in 100 μl at 14°C overnight. Ten micrograms of tRNA was added before phenol/chloroform extraction and ethanol precipitation. The ligations were resuspended in 10 μl of TE−1 (10 mM Tris/1 mM EDTA, pH 8.0), and 10% was used for PCR analysis. BW-1H (18) and KasI (above) were used in a 50-μl PCR reaction that contained 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 25 pmol of each oligo, and 1 unit of Taq polymerase (Boehringer Mannheim). The amplification profile was 1 cycle of 94°C for 4 min; 30 cycles of 94°C for 15 sec, 65°C for 30 sec, 72°C for 40 sec, and 72°C for 4 min. The products were resolved on a 1.5% TBE gel, were transferred to Hybond-N nylon (Amersham Pharmacia), and were hybridized to a kinased internal oligo KasII (5′-GGCCGACCTGAGGGTCGCCGGG-3′). Bands were visualized by autoradiography. Titration curves were run in parallel to determine the linearity of the assay.
Micrococcal Nuclease (MNase) Digestion.
Nuclei were isolated and digested in the presence or absence of butyrate treatment with the indicated amounts of MNase as described (19) except that the nuclei were resuspended in restriction enzyme buffer supplemented with 1 mM CaCl2 for 5 min at 30°C with the number of units indicated. Genomic DNA was purified, digested with HindIII, and run on a 1.75% TBE gel and was Southern blotted by using random primed fragment (−298 to +104) of the MMTV LTR.
Protein Analysis.
Rag proteins were detected as described (16) by using anti-Rag-1 or anti-Rag-2 (PharMingen). Other proteins were detected by isolating total cell extracts and performing Western blot analysis using anti-human glucocorticoid receptor (Affinity Bioreagents, Golden, CO) followed by horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia) and were developed by using enhanced chemiluminescence (Sigma).
RESULTS
The recombination substrate (pSC1) contained two RSSs, one with a 12-bp spacer (12 RSS) and another with a 23-bp spacer (23 RSS); these are the only DNA elements required for V(D)J recombination (ref. 20; Fig. 1A). To generate a recombination substrate on which we could control DNA access, we embedded the 12 RSS in an inducible promoter, the mouse mammary tumor virus long terminal repeat (MMTV LTR). It adopts a nucleosomal structure when stably maintained (21, 22). These nucleosomes prevent access of most transcription factors and DNA binding proteins to their cognate sites within the promoter (23). The substrate plasmid pSC1 also contained the Epstein–Barr virus origin of replication; by introducing pSC1 into 293 cells that either express EBNA1 or not, we could control whether the episome could be maintained and replicated (24). Importantly, these are low-copy nuclear episomes that are replicated only once during S phase, and, therefore, there is no increase in their copy number once stably maintained (25). These properties allow Epstein–Barr virus-based episomal vectors to adopt many of the characteristics of stably integrated DNA, such as the chromatin structure of the HIV LTR and the enhancer dependence of the myc promoter (26, 27).
When stably maintained and replicated, the episome becomes chromatinized and transcription is inhibited by the nucleosomal structure of the MMTV LTR. This nucleosomal repression can be alleviated by treatment with the glucocorticoid dexamethasone. Glucocorticoid receptor binding to the promoter disrupts the nucleosomal organization, thereby altering the chromatin structure and allowing additional factors (e.g., p300) to assemble on the promoter and thus promoting transcription (23, 28, 29). p300 is a transcriptional coactivator required for steroid receptor-dependent transcriptional transactivation (30). However, whether the histone acetylase activity of p300 is required for this transactivation is unknown. The nucleosome remodeling event mediated by nuclear hormone receptors can be uncoupled from transcriptional transactivation if transcription factor assembly on the promoter is prevented (30–32). E1a is a factor known to inactivate p300 and is present in the 293 cells we are using for our studies (33–35). Therefore, by preventing p300 access to the promoter we should be able to uncouple nucleosome remodeling by glucocorticoid receptors from transcriptional transactivation. This should allow us to examine whether alteration of nucleosomes alone is sufficient to activate recombination.
We also used the histone deacetylase inhibitor butyrate to disrupt nucleosomes more globally through an increase in the acetylation status of nucleosomes. Butyrate treatment also has been shown to activate the MMTV LTR and other steroid-responsive promoters (36, 37). As a monitor of transcription, the MMTV LTR of pSC1 drives the expression of a reporter gene, luciferase. The 23 RSS is downstream of this luciferase gene.
V(D)J recombination was induced by either transfecting cells containing the recombination substrate with expression vectors for Rag-1 and Rag-2 or by cotransfecting both the substrate and the Rag expression vectors. Rearrangement was monitored by using an assay that specifically amplifies the signal joint product of the recombination reaction (Fig. 1A).
To determine whether recombination is sensitive to differences in chromatin packaging of target sites, we first compared the efficiency of recombination of transiently introduced substrates to stably maintained substrates. We also treated the cells with either dexamethasone or butyrate to examine whether these treatments affected the efficiency of recombination. Because transiently transfected pSC1 introduced into cells lacking EBNA1 does not replicate, these DNA templates do not adopt phased nucleosomes or other repressive chromatin structures (ref. 21; also see below). We would expect that the recombination efficiency of these substrates should be high and unaffected by treatments that alter nucleosomal structure. As predicted, recombination of transiently introduced substrates was unaffected by treatment with dexamethasone or butyrate (Fig. 1B, compare lanes 2–4). The efficiency of recombination measured by PCR on the transiently introduced pSC1 was comparable to that of nonreplicating polyoma-based vectors—such as pJH200 deleted for the SV40 origin of replication—which were used in previous studies (data not shown).
In contrast to the transiently introduced substrates, cell lines expressing EBNA1 stably maintained pSC1, and the level of recombination per substrate was reduced >100-fold (Fig. 1C). Quantitation was based on titration of the transiently transfected samples diluted into untransfected cell lysate and amplified in parallel with samples generated from stable cell lines. Additionally, Southern blot analysis of total DNA isolated from purified nuclei showed a similar total episomal content in cells that either stably or transiently maintained pSC1 (see Fig. 3, compare lanes 1 and 7). Moreover, given that the efficiency of transfection of 293 cells is >80% (data not shown), the majority of cells under either condition receive the Rag genes and the substrate vector, allowing us to compare these conditions. Butyrate increased recombination of stable pSC1 3-fold (Fig. 1D, compare lanes 2 and 3), a subtle but reproducible increase. This activation of recombination was not seen with dexamethasone, which produced a <2-fold effect. Southern blot analysis of total DNA recovered showed <2-fold variability between samples (data not shown). Therefore, we conclude that replication of this recombination substrate leads to a recombinationally inhibited state that is only marginally affected by drug treatments.
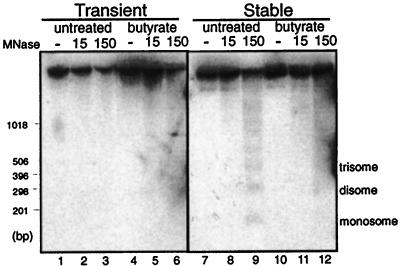
Figure 3.
Stably maintained pSC1 adopts a nucleosomal structure that can be disrupted by butyrate treatment. Nucleosome positioning on transiently (lanes 1–6) or stably maintained (lanes 7–12) pSC1, isolated either from untreated or butyrate-treated cells, was determined by treating purified nuclei with micrococcal nuclease (MNase, as indicated), and digested DNA was detected by Southern blot. Molecular weight markers are indicated on the left, and the nucleosomal ladder is indicated on the right.
The recombinationally inhibited state of stable pSC1 led us to examine whether CpG methylation of the episome would augment the inhibition, perhaps by creating a more heterochromatin-like structure. We therefore methylated pSC1 at CpG sites in vitro to generate pSC1 ME by using SssI methylase. When transiently transfected, pSC1 ME was unaffected by drug treatment, consistent with the lack of a nucleosomal structure under these conditions (Fig. 1B, lanes 7–9), and the absolute amount of recombination was similar between the unmethylated and methylated templates, consistent with the lack of inhibition by methylation in the absence of chromatin assembly.
In contrast, stably maintained pSC1 ME was more refractory to recombination than pSC1 (100-fold) (Fig. 1D, compare lanes 8 and 2). Moreover, the addition of butyrate activated recombination 50-fold (Fig. 1D, lanes 8 and 9), almost to the basal level of the stable, unmethylated substrate. HC toxin, a more specific histone deacetylase inhibitor, also was used and generated results similar to those with butyrate treatment (data not shown). Methylation-sensitive restriction enzymes were used to verify the maintenance of the introduced methylation status of the episomes, and that drug treatment did not affect the introduced methylation status (data not shown). Dexamethasone treatment also activated recombination from this highly repressed state (9-fold) (Fig. 1D, lane 8 vs. lane 10), although to a lesser extent than butyrate. These effects were not attributable to changes in Rag protein levels as determined by Western blot analysis (Fig. 1E, compare lanes 2–4). Thus, we generated a highly repressed recombination substrate that was derepressed by drug treatments known to modify nucleosomal structures.
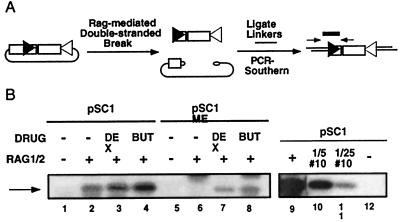
Rag proteins initiate recombination by generating RSS-dependent double-stranded breaks that are processed by the ubiquitous double-strand break repair machinery (2). To determine whether the chromatin-induced inhibition of recombination was attributable to a decrease in the accessibility of the Rag proteins to the substrates or instead to an altered efficiency of double-strand break repair, we assayed for Rag-mediated double-strand breaks (38). LM-PCR was used to quantify these double-strand breaks (Fig. 2A), and titration analysis was performed to demonstrate the linearity of the assay (Fig. 2B, lanes 9–12). The stably maintained methylated substrates were inhibited in their ability to be cleaved by the recombination machinery in the absence of drugs compared with their unmethylated counterparts (Fig. 2B, compare lane 2 to lane 6). Drug treatment could substantially activate recombination on the stably maintained methylated episomes (Fig. 2B, lanes 6–8). Because the inhibition of recombination product formation (Fig. 1D) by chromatin structure was mirrored in the LM-PCR assay, the repression of recombination is at an early step in recombination, presumably the recognition of the RSSs by the Rag proteins or, less likely, the cleavage step itself.
Figure 2.
The chromatin structure of a recombination substrate directly affects the accessibility of the recombinase for its DNA substrate. (A) Schematic diagram of the LM-PCR assay. DNA samples were ligated to blunt linkers and were subjected to PCR by using a linker primer and a second primer internal to the Rag-dependent double-strand break (arrows) at the 12 RSS (black triangle). The amplified products were subjected to Southern blot hybridization and were probed with an oligonucleotide internal to the amplified product (rectangle). (B) Measurement of double-strand break formation. Cell lines stably maintaining pSC1 or pSC1 ME were cotransfected with Rag expression vectors and were treated with drugs as indicated (lanes 1–8). Titration analysis was performed in parallel to verify the linearity of the assay (lanes 9–12). All samples were generated from the same experiment and gel. Assays were performed as described in A.
To determine directly whether butyrate treatment was having the predicted effects on the chromatin structure of pSC1, we assayed for nuclease sensitivity under different conditions. Previous studies have shown that, after replication, the MMTV LTR and other steroid-responsive promoters are packaged into a nucleosomal array that can be disrupted by butyrate treatment (22, 36, 37). We treated nuclei containing the episomes with MNase, an enzyme that digests the linker DNA between nucleosomes, allowing the position and spacing of nucleosomes on a template to be determined. The digested DNA then was probed for MMTV LTR sequences. When stably maintained, the MNase digestion pattern of the MMTV LTR showed the typical ladder of positioned nucleosomes (Fig. 3, lane 9). The ladder was not detectable when the templates were transiently transfected, consistent with these episomes not being packaged into nucleosomes (Fig. 3, lane 3). A decrease in the intensity of the full length band (≈6 kb) as the amount of MNase was increased on the transient samples (Fig. 3, lanes 1–3) indicated that the episomes were accessible to the nuclease. Additionally, the regular nucleosomal array generated on the stably maintained episomes was disrupted by butyrate treatment, as seen by the less defined ladder (Fig. 3, lane 12). The smallest DNA fragment of the nucleosome ladder in Fig. 3, lane 9 (mononucleosome) was most clearly disrupted by butyrate, as seen by the blurring of this band in lane 12. As a control, the bulk genomic DNA was visualized by ethidium bromide staining; treatment with MNase with or without butyrate treatment under transient or stable conditions led to a similar laddered digestion pattern (data not shown). These data show that the stable episomes are packaged into nucleosomes that are directly affected by butyrate treatment.
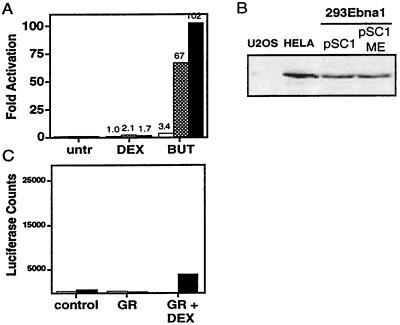
To examine whether transcription of the recombination loci correlated with recombination, we monitored the transcriptional activity of the MMTV LTR under the same conditions. Transiently transfected pSC1 should be constitutively accessible to the transcription machinery, and, in fact, the addition of butyrate had only a modest stimulatory effect (<4 fold) (Fig. 4A). In contrast, stably maintained pSC1 was highly inducible by butyrate (>50 fold) because the drug relieved the repression of chromatin on transcription. Treatment with butyrate on the methylated templates could also significantly activate transcription when they were stably maintained (>100 fold). The absolute amount of basal transcription of stably maintained pSC1 ME was 3-fold lower than its unmethylated counterpart pSC1 and <2-fold above background levels of luciferase activity (Fig. 4A, legend). Thus, for butyrate, stimulation of both transcription and recombination was coupled.
Figure 4.
Transcriptional activation does not accurately reflect accessibility of a recombination substrate. (A) Transcription of the MMTV LTR is increased by butyrate treatment when the promoter is stably maintained. Cells harboring transiently unmethylated (white), stably unmethylated (gray), or stably methylated (black) pSC1 were assayed for luciferase activity after drug treatment as indicated. The fold activation was calculated by setting the untreated samples to one, and the numerical values are listed above the bars. The average of three experiments is shown. The absolute level of luciferase activity of the untreated stable pSC1 was 1,062 ± 490; stable pSC1 ME was 323 ± 78; and that with no extract was 237 ± 76. (B) Western blot analysis reveals that glucocorticoid receptor (GR) is expressed in 293E cells as compared with U2OS cells that do not express glucocorticoid receptor and HeLa cells known to express glucocorticoid receptor (42, 43). (C) E1a can block glucocorticoid-dependent transactivation of the MMTV LTR. U2OS cells were transfected with glucocorticoid receptor in the presence (black) or absence (white) of E1a and dexamethasone (Dex) and were monitored for luciferase activity.
In contrast to butyrate, the addition of dexamethasone did not significantly activate transcription of either the transiently or stably maintained pSC1 or pSC1 ME (2-fold or less) (Fig. 4A), despite the presence of glucocorticoid receptor in these cells (Fig. 4B). Thus, for dexamethasone, transcription and recombination were uncorrelated.
This lack of dexamethasone-induced transcription was likely a consequence of adenovirus E1a in our 293 cells, as has been previously shown for other nuclear hormone receptors (31). To examine this for our substrate, we determined whether the steroid-dependent transcriptional transactivation of pSC1 in U2OS cells, which lack E1a, could be inhibited by the coexpression of E1a. We transfected U2OS cells with pSC1 and either a vector expressing E1a or a control vector. Treatment with dexamethasone activated glucocorticoid receptor-dependent transcription of the MMTV LTR that was inhibited by the coexpression of E1a (Fig. 4C). Therefore, the lack of steroid-mediated transcriptional transactivation in 293 cells can be attributed to the presence of E1a.
DISCUSSION
By taking advantage of the well characterized MMTV LTR, we have demonstrated that the chromatinized state of target DNA can influence accessibility to the V(D)J recombination machinery in vivo. Comparing transiently transfected, nonreplicating pSC1 plasmid without nucleosomal organization to stably maintained, nucleosome-bound pSC1, it was evident that nucleosomes inhibited recombination >100-fold. Recent in vitro studies also have found that nucleosomal substrates are refractory to Rag-mediated cleavage (39). We then found that the repression mediated by nucleosomes was relieved by butyrate, a drug known to alter nucleosomal structures. However, the more subtle changes in chromatin introduced by dexamethasone activation of glucocorticoid receptors did not significantly affect recombination frequencies under these conditions. CpG methylation of nucleosomal templates further repressed the access of the recombinase for targets an additional 100-fold. Under these conditions, treatment with butyrate or dexamethasone, both known to alter nucleosomal packaging, increased the recombination frequency and thus, presumably, accessibility to the recombinase. By manipulating chromatin structures in vivo, we were able to decrease the accessibility of the recombination machinery for target sites by 10,000-fold. This level of inhibition may be sufficient to explain the nearly absolute specificity of recombination substrate choice in vivo. We were then able to alleviate the repression by using known modifiers of chromatin structure.
The chromatinization of substrates inhibited both transcription and recombination. However, the magnitude of inhibition on recombination was much greater (i.e., methylation of stable pSC1 decreased transcription by 3-fold and recombination by 100-fold). Whereas butyrate treatment alleviated both the repression of chromatin on transcription and recombination, dexamethasone differentially affected these processes. Steroid-mediated disruption of nucleosomes activated recombination by switching the substrate from an inactive to an active one without significant transcriptional transactivation. Although we could detect a slight increase in luciferase activity after dexamethasone treatment, the level of activation was exceedingly low and disproportionate to the recombinational activation (1.7-fold vs. 9-fold). Therefore, it is likely that the activation of the recombinase is almost exclusively attributable to the nucleosomal remodeling mediated by the glucocorticoid receptor and not transcriptional transactivation.
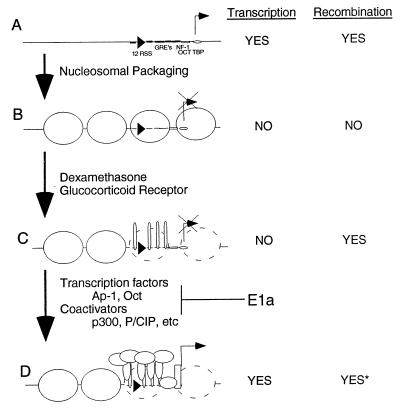
These data have led us to postulate a model for the regulation of Rag-mediated recombination by the modifications of our DNA target (Fig. 5). Transiently introduced substrates are accessible to both the transcriptional and recombinational machinery (Fig. 5A) and are thus unregulated. Activation of transcription on open substrates has no effect on recombination efficiencies (Fig. 1B; ref. 40). Nucleosomal packaging achieved through stable maintenance of targets, especially when CpG methylated, inhibits both transcription and recombination (Fig. 5B). Disruption of nucleosomes locally on the MMTV LTR by using steroid-mediated glucocorticoid receptor binding activates recombination but not necessarily transcription (Fig. 5C). Transcription requires the assembly of other transcription factors and coactivators (Fig. 5D). In our system, E1a inhibited the transition from the state in Fig. 5C to that in Fig. 5D by preventing p300 assembly on the promoter.
Figure 5.
Model of accessibility. (A) Transiently introduced recombination substrate not packaged into nucleosomes is recombinationally and transcriptionally active. (B) Stably maintained, nucleosomal targets are refractory to recombination and transcription. (C) Steroid-dependent glucocorticoid receptor binding to the MMTV LTR disrupts nucleosome activating recombination but not transcription. (D) Disruption of the nucleosomes allows assembly of transcription factors and coactivators, allowing transcriptional transactivation and presumably recombination. E1a can block the transition from C to D by preventing p300 assembly on the promoter.
That the E1a in 293 cells did not prevent the dexamethasone-induced increase in recombination implies that the activated glucocorticoid receptor is able to increase the accessibility of the recombinase to DNA even in the absence of significant transcriptional activation. How butyrate treatment activates transcription in the presence of E1a is not clear; perhaps it bypasses the requirement for specific transcription factors and coactivators. The decreased activation of recombination by steroids as compared with butyrate may be attributable to the fact that dexamethasone only disrupts the nucleosomal structure of the MMTV LTR near the 12 RSS whereas butyrate affects chromatin more globally, altering the chromatin structures at both the 12 RSS and 23 RSS (22). Experiments have demonstrated that by altering one RSS to become a better substrate for recombination, the pair of sites is used more efficiently (41). Another possibility is that modification of the chromatin structure is sufficient to activate recombination but that transcriptional transactivation potentiates the affect.
We have determined that chromatin inhibits recombination in vivo and have begun to define parameters that can alleviate this repression. It will probably require in vitro studies with reconstituted chromatin to define the specific proteins and structures that inhibit recombination and how these modifications can alleviate the repression.
Acknowledgments
We thank T. Baker, S. Bell, and C. Roman for advice and critical reading of the manuscript. We thank T. Archer for chromatin protocols. We are grateful to K. Alexandropoulos, B. Chen, K. Collins, J. Jacob, T. Koleske, M. Porteus, I. Stancovski, and other members of the Baltimore Lab for advice and discussions. This work was supported by a National Institutes of Health grant.
ABBREVIATIONS
- RSS
recognition signal sequence
- MMTV LTR
mouse mammary tumor virus long terminal repeat
- MNase
micrococcal nuclease
- LM-PCR
ligation-mediated PCR
References
- 1.Lewis S M. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 2.Schatz D G, Oettinger M A, Schlissel M S. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 3.Willerford D M, Swat W, Alt F W. Curr Opin Genet Dev. 1996;6:603–609. doi: 10.1016/s0959-437x(96)80090-6. [DOI] [PubMed] [Google Scholar]
- 4.Schlissel M S, Stanhope B P. Semin Immunol. 1997;9:161–170. doi: 10.1006/smim.1997.0066. [DOI] [PubMed] [Google Scholar]
- 5.Mather E L, Perry R P. Proc Natl Acad Sci USA. 1983;80:4689–4693. doi: 10.1073/pnas.80.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlissel M S, Morrow T. J Immunol. 1994;153:1645–1657. [PubMed] [Google Scholar]
- 7.Sleckman B P, Gorman J R, Alt F W. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 8.Kallenbach S, Babinet C, Pournin S, Cavelier P, Goodhardt M, Rougeon F. Eur J Immunol. 1993;23:1917–1921. doi: 10.1002/eji.1830230828. [DOI] [PubMed] [Google Scholar]
- 9.Okada A, Mendelsohn M, Alt F. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh C-L, Lieber M R. EMBO J. 1992;11:315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley D E, Pollok B A, Atchison M L, Perry R P. Mol Cell Biol. 1988;8:930–937. doi: 10.1128/mcb.8.2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engler P, Haasch D, Pinkert C A, Doglio L, Glymour M, Brinster R, Storb U. Cell. 1991;65:939–947. doi: 10.1016/0092-8674(91)90546-b. [DOI] [PubMed] [Google Scholar]
- 14.Dieken E S, Meese E U, Miesfeld R L. Mol Cell Biol. 1990;10:4574–4581. doi: 10.1128/mcb.10.9.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCurrach M E, Connor T M, Knudson C M, Korsmeyer S J, Lowe S W. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roman C A, Cherry S R, Baltimore D. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhu C, Roth D B. Immunity. 1995;2:101–112. doi: 10.1016/1074-7613(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 18.Mueller P R, Wold B. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 19.Mymryk J S, Fryer C J, Jung L A, Archer T K. Methods. 1997;12:105–114. doi: 10.1006/meth.1997.0452. [DOI] [PubMed] [Google Scholar]
- 20.Hesse J E, Lieber M R, Gellert M, Mizuuchi K. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 21.Archer T K, Lefebvre P, Wolford R G, Hager G L. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 22.Richard-Foy H, Hager G L. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pina B, Bruggemeier U, Beato M. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 24.Yates J L, Warren N, Sugden B. Nature (London) 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 25.Yates J L, Guan N. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mautner J, Behrends U, Hortnagel K, Brielmeier M, Hammerschmidt W, Strobl L, Bornkamm G W, Polack A. Oncogene. 1996;12:1299–1307. [PubMed] [Google Scholar]
- 27.Stanfield O S, Griffith J D. J Mol Biol. 1996;256:503–516. doi: 10.1006/jmbi.1996.0104. [DOI] [PubMed] [Google Scholar]
- 28.Archer T, Cordingly M G, Wolford R, Hager G L. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 30.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch’ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 31.Smith C L, Onate S A, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong J, Shi Y B, Wolffe A P. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. Nature (London) 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 34.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 35.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 36.Bartsch J, Truss M, Bode J, Beato M. Proc Natl Acad Sci USA. 1996;93:10741–10746. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia V P, Jimenez L A, Castillo A I, Aranda A. DNA Cell Biol. 1997;16:421–431. doi: 10.1089/dna.1997.16.421. [DOI] [PubMed] [Google Scholar]
- 38.Roth D B, Zhu C, Gellert M. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon J, Imbalzano A N, Matthews A, Oettinger M A. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh C L, McCloskey R P, Lieber M R. J Biol Chem. 1992;267:15613–15619. [PubMed] [Google Scholar]
- 41.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 42.Nobukuni Y, Smith C L, Hager G L, Detera W S. Biochemistry. 1995;34:8207–8214. doi: 10.1021/bi00025a028. [DOI] [PubMed] [Google Scholar]
- 43.Rogatsky I, Trowbridge J M, Garabedian M J. Mol Cell Biol. 1997;17:3181–3193. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]