Abstract
Surfactant protein D (SP-D) is an oligomeric C type lectin that promotes phagocytosis by binding to microbial surface carbohydrates. A 340-kDa glycoprotein (gp-340) has been shown to bind SP-D in the presence of calcium but does so independently of carbohydrate recognition. This protein exists both in a soluble form and in association with the membranes of alveolar macrophages. The primary structure of gp-340 has been established by molecular cloning, which yielded a 7,686-bp cDNA sequence encoding a polypeptide chain of 2,413 amino acids. The domain organization features 13 scavenger receptor cysteine-rich (SRCR) domains, each separated by an SRCR-interspersed domain, except for SRCRs 4 and 5, which are contiguous. The 13 SRCR domains are followed by two C1r/C1s Uegf Bmp1 domains separated by a 14th SRCR domain and a zona pellucida domain. gp-340 seems to be an alternative spliced form of DMBT1. Reverse transcription–PCR analysis showed that the main sites of synthesis of gp-340 are lung, trachea, salivary gland, small intestine, and stomach. Immunohistochemistry revealed strong staining for gp-340 in alveolar and other tissue macrophages. Immunostaining of the macrophage membrane was either uniform or focal in a way that suggested capping, whereas other macrophages showed strong intracellular staining within the phagosome/phagolysosome compartments. In some macrophages, SP-D and gp-340 were located in the same cellular compartment. Immunoreactive gp-340 was also found in epithelial cells of the small intestine and in the ducts of salivary glands. The distribution of gp-340 in macrophages is compatible with a role as an opsonin receptor for SP-D.
Collectins are oligomeric proteins composed of C type lectin domains connected to collagen regions (1). Three collectins are known in humans: mannan-binding lectin (MBL), a serum protein, and the lung surfactant proteins A and D (SP-A and SP-D), which are produced by epithelial cells mainly in the lung. The collectins play an important role in innate immunity by binding to specific carbohydrate structures found on the surfaces of pathogenic microorganisms, including bacteria, viruses, yeasts, and parasitic protozoa. The binding promotes effector mechanisms such as aggregation, hindrance of infection, activation of phagocytes, and initiation of phagocytosis. After binding to microbial surfaces, MBL activates the complement system through a complement activation pathway, the MBL pathway, by using two recently identified serine proteases, MASP-1 and MASP-2, to activate C4 and C2 (2).
SP-D is mainly produced by alveolar type II cells but is also present in sweat, salivary, tear, and mammary glands and in epithelial cells throughout the gastrointestinal tract. In this respect, SP-D is colocalized with IgA of the adaptive immune system (J.M. and U.H., unpublished work). Like the other collectins, SP-D binds to repeating carbohydrate moieties on pathogenic microorganisms and is considered to be a pattern-recognition molecule of the innate immune system (3). This binding enhances phagocytosis and killing of microorganisms by neutrophils and alveolar macrophages (4, 5). The binding also results in aggregation of the microorganisms and hindrance of infection. SP-D also binds directly to alveolar macrophages in the absence of microbial ligands, thereby mediating the generation of oxygen radicals (6), and acts as a potent chemotactic agent for phagocytes (7, 8). SP-D has been localized in endocytic vesicles and in lysosomal granules of alveolar macrophages (9, 10), suggesting that it is involved in receptor-mediated uptake by these cells. SP-D has also been shown to inhibit T lymphocyte proliferation and IL-2 production (11).
SP-D interacts with surfactant-associated phospholipids in vitro (12, 13), and mice made SP-D-deficient by gene targeting showed a progressive accumulation of surfactant lipids in the alveolar space. The SP-D null mice also showed an accumulation of alveolar macrophages, many of which were multinucleated and foamy in appearance. The alveolar type II cells in these animals were hyperplastic and contained giant lamellar bodies. These findings suggest that SP-D also plays a role in surfactant homeostasis (14, 15).
No receptor responsible for the effector mechanisms of SP-D has yet been characterized. A molecule, designated gp-340, which copurifies with SP-D on carbohydrate affinity columns, has been isolated and shown to bind SP-D in a calcium-dependent manner (16). It has also been established that the binding of SP-D to gp-340 is a protein–protein interaction rather than a lectin–carbohydrate interaction. Recently, it was shown that gp-340 also may bind to SP-A (17). Protein sequencing showed that gp-340 was a member of the scavenger receptor cysteine-rich (SRCR) superfamily (16).
The gp-340 molecules exist both in a soluble form and in a form associated with the alveolar macrophage membrane. In the present paper, we describe the primary structure of gp-340 and its tissue distribution. It seems probable that the molecular interaction between SP-D and gp-340 plays an important role in innate immunity on mucosal surfaces.
MATERIALS AND METHODS
cDNA Cloning and Sequencing. A pool of 29-base oligonucleotide “guessmers” with a redundancy of 384 was synthesized in accordance with the codon usage of the underlined amino acids of the gp-340 peptide SWGTV(C)DDYWDTNDANV (16). The ACG codon for threonine was omitted. The oligonucleotide sequence was 5′-TG(C/T)GA(C/T)GA(C/T)TA(C/T)TGGGA(C/T)AC(A/T/C)AA(C/T)GA(C/T)GC-3′, which was end labeled with [λ-32P]ATP and T4 kinase and used for screening a λgt11 human lung cDNA library (CLONTECH). A single clone (gp-clone 1A) was obtained from 550,000 plaques. λDNA was prepared from this clone, and the insert was subcloned into pBluescript SK(+) and sequenced. Sequencing was performed with the Cycle Sequencing Kit (Amersham Pharmacia) on double-stranded templates with M13 and T7 primers on the vector as well as gp-340 internal oligonucleotides. Sequence analysis revealed that gp-clone 1A was derived from a partially spliced mRNA. One oligonucleotide was designed from the cDNA sequence of gp-clone 1A R3OS, 5′-GATCCCTCCCTGCCCCTGCT-3′ (antisense), and used together with the λgt11 primer F11, 5′ ACTCCTGGAGCCCGTCAGTAT-3′ (sense), in a PCR with the gp-clone 1A used as template. The 600-bp PCR product was purified by electrophoresis through 0.8% (wt/vol) agarose and labeled with [α-32P]dCTP by using R3OS and F11 as primers and a Klenow large fragment as DNA polymerase. This probe was used for screening an oligo(dT)-primed λgt11 human lung cDNA library (CLONTECH) and a random- and oligo(dT)-primed λgt11 human lung cDNA library (CLONTECH). Two individual clones (gp-clones 7 and 8) were obtained from 500,000 plaques from the random- and oligo(dT)-primed library, whereas no clones were obtained from the library primed with oligo(dT) alone. λDNA was prepared from gp-clones 7 and 8. The inserts were cut with restriction enzymes and subcloned into pBluescript SK(+) and sequenced. To obtain cDNA clones from the 3′-end, oligonucleotides were designed from the expressed sequence tag clone . The zona primer-1 (5′-TTCCAACTTCCTCACAGC-3′) and 3′ primer-5 (5′-GCAGTTTCACCAAAATTC-3′) were used in a PCR with as template. The PCR product of 300-bp was purified, labeled, and used for screening an oligo(dT)-primed Uni-ZAP-XR human lung cDNA library (Stratagene). An additional six clones (gp-clones 3.1, 3.2, 5.1, 7.1, 8.1, and 8.2) were obtained, subcloned, and sequenced.
Poly(A)+ RNA from human trachea was transcribed into cDNA with the Marathon cDNA Kit (both purchased from CLONTECH) and diluted 1:250. A 5-μl portion of the dilution was used as template in a 50-μl PCR that further contained 5 μl of P3 buffer (Expand Long Template PCR System, Boehringer Mannheim), 400 μM each dNTP, 0.75 μl of a 9:1 (Taq:Pfu) polymerase mix (Taq: 5 units/μl, Perkin–Elmer; Pfu: 2 units/μl, Stratagene), and 15 pmol each primer (dmbt1.1f, 5′-ATACCACCCTAGAGGACACACC-3′, and dmbt1.2r, 5′-GCAGTTCACCAAAATTCCTT-3′). The PCR was performed as follows: an initial denaturation for 2 min at 94°C; 30 cycles of 20 s at 92°C, 30 s at 59°C, and 10 min at 68°C; and a final extension for 10 min at 72°C. The PCR products were cloned with the TOPO XL PCR Cloning Kit (Invitrogen) according to the instructions of the supplier, and insert sizes were determined by NotI–HindIII digestion and subsequent agarose gel electrophoresis. A DMBT1 clone with an insert of about 8 kilobases (kb; DMBT1/8-kb.2) was selected for sequencing by using the nested-deletion method (Nested Deletion Kit, Amersham Pharmacia). Sequencing was carried out by using dye terminator chemistry (Amersham Pharmacia) and automated DNA sequencers (Applied Biosystems model 373A/377). The cDNA clone contained frameshift mutations in codons 100 and 1,751 that were corrected by comparison with the genomic sequence of the gene (18).
Immunohistochemistry.
Normal human tissues and cell pellets from bronchoalveolar lavage fluid were fixed in 4% (vol/vol) formalin in PBS for 24 h and then conventionally dehydrated and embedded in paraffin. A streptavidin-biotin immunoperoxidase staining technique was used on the paraffin-embedded sections, which were treated in a microwave oven to unmask epitopes (19). A monoclonal (Hyb 213-1) or polyclonal antibody against gp-340 and a monoclonal (Hyb 245-1) antibody against SP-D were used, followed by biotin-labeled goat anti-mouse Ig (E433, Dako) or biotin-labeled goat anti-rabbit Ig (E432, Dako). After incubation with streptavidin conjugated to horse-radish peroxidase (P397, Dako), the sections were incubated with carbazole buffer [0.04% 3-amino-9-ethylcarbazole (Sigma, A-5754)/0.015% H2O2/50 mM sodium acetate buffer, pH 5.0], counterstained with Mayer’s hematoxylin, and mounted in Aquatex (Merck). The specificities of the antibodies used for immunohistochemistry are described in ref. 16. Controls were performed by replacing the primary monoclonal antibody with an unrelated monoclonal antibody of the same subclass as the gp-340 antibody or by preabsorbing the primary polyclonal antibody with highly purified gp-340.
Reverse Transcriptase–PCR (RT-PCR).
Total RNA (2 μg) isolated from various human tissues (CLONTECH) was used for first-strand synthesis by using the antisense primer DMBT1c, 5′-GAGAGGGGAACTGCGGTAG-3′. This primer corresponds to nucleotides 6,316–6,334 of gp-340 cDNA. The RNA was incubated in a volume of 20 μl with 1 unit/μl M-MuLV reverse transcriptase (Stratagene) for 1 h at 37°C. One-fourth of the resulting single-stranded cDNA was subjected to 40 cycles of PCR amplification in a volume of 30 μl (denaturing at 94°C for 30 s, annealing at 62°C for 1 min, extension at 72°C for 2 min, and a final extension at 72°C for 5 min) by means of Taq polymerase (Stratagene) with two gp-340 primers spanning the region between the SRCR 13 and the CUB 2 (a domain first found in complement component C1r/C1s; Uegf, a sea urchin epidermal growth factor-containing protein; and Bmp1, bone morphogenetic protein-1) domains. The sense primer DMBT sca9 (5′-ATTCATCCTATGGTCTA-3′) corresponded to nucleotides 5,772–5,788 of gp-340 cDNA, and the antisense primer was the same as the one used for cDNA synthesis. As a control, a single-stranded cDNA copy of β-actin was synthesized and amplified by PCR under the same conditions as gp-340, except that the PCR annealing temperature was 58°C and the primers used were 5′AKTI, 5′-GGCATGGCTTTATTTGTTTT-3′ (sense), and 3′ATKI, 5′ GTAAGCCCTGGCTGCCTC-3′ (antisense). The entire volume of each reaction mixture was subjected to electrophoresis on a 1.5% (wt/vol) agarose gel, blotted onto a nylon membrane, and hybridized with the respective PCR products labeled with [α-32P]dATP.
RESULTS AND DISCUSSION
Oligonucleotides based on the amino acid sequence of a gp-340 peptide were used to retrieve a single clone (gp-clone 1A) from a lung cDNA library. The translated sequence of this clone was found to include the sequence used to design the oligonucleotides. With this clone as template, a PCR product was generated and used for further screening of lung cDNA libraries, and a PCR product located at the 3′ end of the zona pellucida (ZP) domain was also used for screening. An additional eight cDNA clones were obtained: gp-clones 7, 8, 3.1, 3.2, 5.1, 7.1, 8.1, and 8.2. The last six included the 3′ end of the cDNA, whereas gp clones 7 and 8 were found to be internal, overlapping cDNA clones. Additional cDNA clones were found as expressed sequence tags. The sequences of these clones revealed a high homology but not identity with the 6-kb form of DMBT1, a gene recently identified as a candidate tumor suppressor gene for tumors of the central nervous system (20). Northern blotting showed that DMBT1 is expressed with transcripts of 6.0, 7.5, and 8.0 kb in fetal lung and with one transcript of 8.0 kb in adult lung. Long-range PCR with primers from the 5′ and 3′ ends of DMBT1 gave an 8.0-kb product that included the sequence data obtained from the cDNA cloning and allowed the assembly of a 7,687-bp cDNA starting with a 5′ untranslated region of 107 bp, followed by an ORF of 7,239 bp and a 3′ untranslated region of 341 bp ending with a poly(A) tail. The nucleotide sequence revealed that gp-340 and the 6.0-kb form of DMBT1 represent alternatively spliced forms of the same gene (20). The combined nucleotide sequences of gp-clones 7, 8, and DMBT1/6-kb and DMBT1/8-kb.2 have been deposited at the European Molecular Biology Laboratory nucleotide sequence database (accession nos. AF159456 and AJ243212).
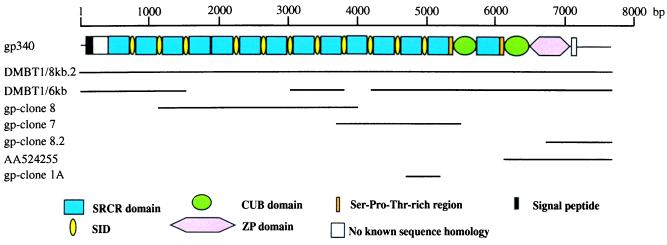
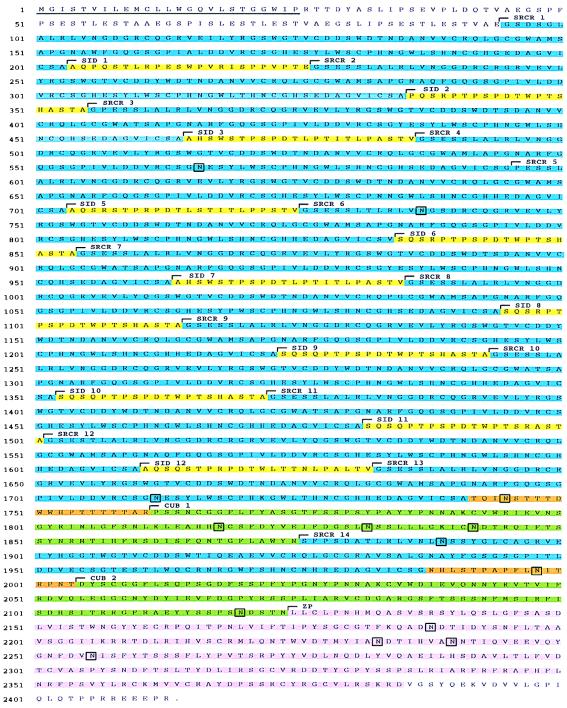
The domain organization of gp-340 is shown in Fig. 1, together with details of the cDNA clones that were characterized and the long-range PCR products. The sequence shown is the assembled sequence of DMBT1/6-kb, gp-clone 7, and gp-clone 8. The ORF encodes a polypeptide chain of 2,413 amino acids, including a possible signal peptide of 25 residues (Fig. 2). gp-340 is a glycoprotein with an apparent molecular mass of 340 kDa as determined by SDS/PAGE. When the signal peptide is omitted, the calculated molecular mass of the polypeptide chain is 258,069 kDa. The isoelectric point is calculated as 5.18, and the molar extinction coefficient at 280 nm is calculated as 605,350 M−1⋅cm−1 (corresponding to an absorbance at 280 nm of 1.0 for a solution of the unglycosylated protein chain at 0.43 mg/ml). The molecular mass of gp-340 purified from human bronchoalveolar lavage fluid is reduced to 300 kDa after deglycosylation. This reduction is consistent with the presence of the 14 putative sites for N-linked glycosylation found in the deduced amino acid sequence (Fig. 2).
Figure 1.
Structure of gp-340. The cDNA clones used to establish the sequence of gp-340 are shown together with a schematic representation of the domain structure of gp-340 drawn to scale. The positions of the cDNA clones and long-range PCR (DMBT1/6-kb and DMBT1/8-kb.2) products are indicated as solid lines. SID, SRCR-interspersed domain.
Figure 2.
The deduced amino acid sequence of gp-340. SRCR domains are shown in blue, SIDs in yellow, CUB domains in green, Ser-Pro-Thr-rich regions in orange, and the ZP domain in pink. The hydrophobic putative signal peptide is underlined. Potential glycosylation sites are shown in boxes.
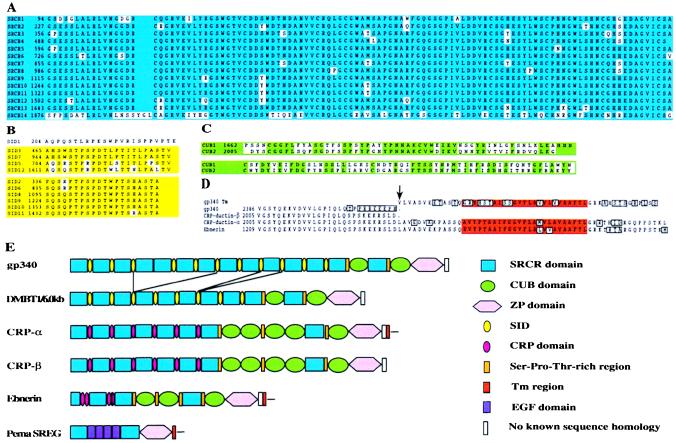
The domain organization of gp-340 features 13 SRCR domains (21) separated by SIDs, except for SRCR domains 4 and 5, which are contiguous. The SRCR domains are followed by a short Thr-rich region, a CUB domain (22, 23), a 14th SRCR domain, a Ser-Thr-Pro-rich region, a second CUB domain, and a ZP domain. Compared with the gp-340 sequence, the 6-kb form of DMBT1 lacks the C-terminal half of SIDs 3, 5, and 6, the N-terminal part of SID 7, and SRCR domains 4, 5, 6, and 7 (Fig. 3). DMBT1 also lacks the equivalent of one SID and one SRCR domain between SIDs 9 and 11 of gp-340. However, because SIDs 9 and 10 are 100% identical at the nucleotide level, as are SRCR domains 10 and 11, it is impossible to deduce exactly where the alternative splicing is located.
Figure 3.
Alignment of gp-340 domains and domain organization of gp-340 compared with other members of the SRCR superfamily. (A) Alignment of the SRCR domains of gp-340. (B) Alignment of the SIDs of gp-340. (C) Alignment of the CUB domains of gp-340. (D) Alignment of the C-terminal part of gp-340 with the same region of CRP-ductins and Ebnerin. The transmembrane (Tm) region was obtained from a genomic DMBT1 clone containing a putative exon for this region. (E) The domain organization of gp-340 is compared with the 6.0-kb form of DMBT1 and other members of group B of the SRCR superfamily that contain SRCR and ZP domains.
Identities and similarities between the different domains are shown in the sequence alignments presented in Fig. 3. A remarkable level of similarity is found among the first 13 SRCR domains (88–100% identity), and the 14th SRCR domain shows 65% identity with the other SRCR domains (Fig. 3A). All the SRCR domains have eight conserved cysteine residues and belong to group B of the SRCR family (21). The SID domains are Ser-Thr-Pro-rich (60%) and can be separated in two groups showing high within-group similarity (Fig. 3B). The two CUB domains are 52% identical (Fig. 3C). The ZP domain is followed by a stretch of 28 residues that show a high degree of similarity to equivalent regions of CRP-ductin (24) and Ebnerin (25) (Fig. 3D). Two cDNA variants of CRP-ductin have been characterized. CRP-ductin-α is an alternative spliced form of CRP ductin-β that lacks the Tm region and the short cytoplasmic tail (24). All the isolated gp-340/DMBT1 cDNA clones have a stop codon after residue 2,413, and this stop codon seems to be placed similarly to that in CRP-ductin-β (Fig. 3D). The intron/exon structure of the gp-340/DMBT1 gene has been characterized (18), and an exon encoding a putative Tm region of gp-340/DMBT1 was identified approximately 1.8 kb from the stop codon. Alignment of this exon with the same region of CRP-ductin and Ebnerin showed that it starts exactly where CRP-ductin-α continues the sequence of CRP-ductin-β (Fig. 3D). Thus far, no cDNA clones coding for the Tm region of gp-340/DMBT1 have been obtained.
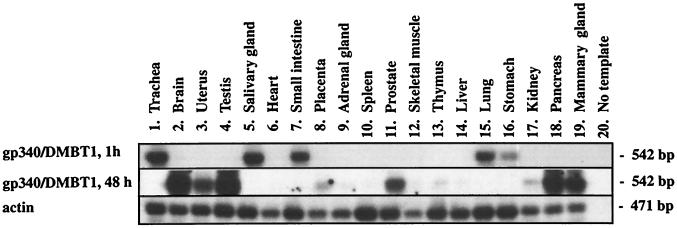
Northern blot analysis of DMBT1 revealed an 8-kb transcript in fetal and adult lung, and additional bands of 6.0 and 7.5 were visible in fetal lung and adult small intestine (20). Western blotting of bronchoalveolar lavage fluid showed two closely spaced bands corresponding to molecular masses of approximately 340 kDa, and one band corresponded to 200 kDa (16). The top band could represent the translated form of the full-length 8-kb transcript, whereas the lower bands could be alternatively spliced forms of the DMBT1 gene. By performing RT-PCR on 19 different tissues, it was shown that the main sites of synthesis of gp-340/DMBT1 are lung, trachea, salivary gland, small intestine, and stomach (Fig. 4). Minor sites of synthesis were brain, testis, uterus, prostate, pancreas, and mammary gland, and little or no synthesis was found in heart, placenta, adrenal gland, spleen, skeletal muscle, thymus, liver, or kidney (Fig. 4).
Figure 4.
RT-PCR analysis of gp-340/DMBT1 expression in human tissues. The Southern blots were exposed for 1 h for all tissues or for 48 h omitting tissues 1, 5, 7, 15, and 16 to avoid overexposure.
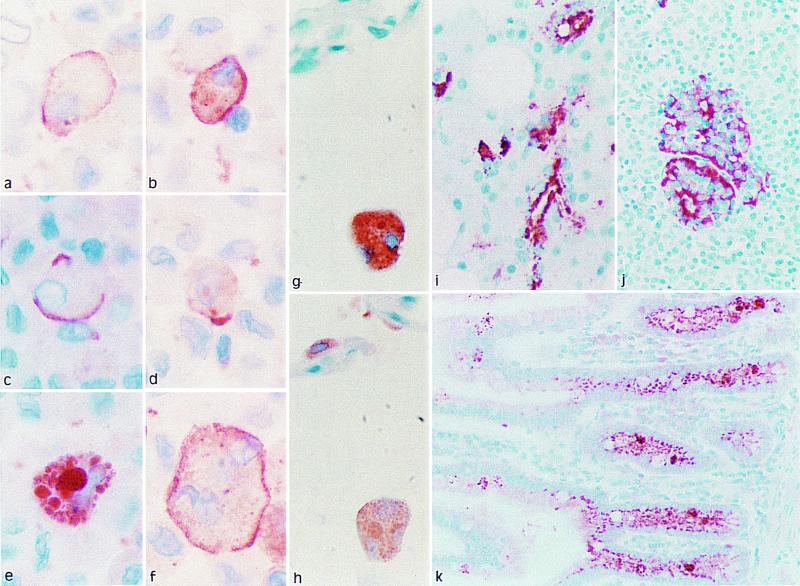
Immunohistochemistry revealed strong immunostaining for gp-340 in the alveolar macrophages of the lung (Fig. 5 a–g). In some macrophages, gp-340 was found to be uniformly distributed on the membrane. Other macrophages showed a focal membrane staining (capping), and yet other macrophages were strongly stained in the phagosome/phagolysosome compartments. In some macrophages, SP-D and gp-340 were found colocalized in the same cell compartment (Fig. 5 g–h). This distribution of gp-340 in the macrophages is compatible with the concept that gp-340 is an opsonin receptor for SP-D. Immunoreactive gp-340 was also found in the epithelial cells of the small intestine (Fig. 5k) and in the ducts of salivary glands (Fig. 5i). In the pancreas, gp-340 was located in certain cells of the islets of Langerhans (Fig. 5j). The main site of synthesis of CRP-ductin is the small intestine, but CRP-ductin is also found in the exocrine pancreatic and hepatic ducts. By using RT-PCR, we have found that CRP-ductin also is expressed in the lung (J. Madsen and U.H., unpublished work). Ebnerin is found specifically in the secretory duct epithelial cells of the lingual salivary glands of the rat and does not seem to be expressed in either lung or large intestine. Pema-SREG was cloned from a gut cDNA library from the sea lamprey Petormyzon marinus (Fig. 3E; ref. 26). This molecule features two SRCR domains that are separated by four EGF domains and a ZP domain followed by a Tm region and a cytoplasmatic tail. The biological function of CRP-ductin and Ebnerin are unknown, but the domain organization of these two molecules and gp-340 indicates that they are homologous proteins in human, mouse, and rat. However, functional studies showing that murine SP-D binds to CRP-ductin are needed to determine whether this homology is the case.
Figure 5.
Immunohistochemistry of gp-340 in normal human tissues with monoclonal (a–g, i, and j) or polyclonal (k) antibody directed against gp-340. (a–f) Sections of human alveolar macrophages, obtained by cytospin and stained by the monoclonal antibody directed against gp-340. (g and h) Serial sections of human lung stained with monoclonal antibodies directed against gp-340 (g) or SP-D (h). (i–k) Immunoreactive gp-340 was also found in salivary ducts (i), in certain cells of the pancreatic islets (j), and in epithelial cells of the small intestine (k).
Previously, DMBT1 has been proposed as a candidate tumor suppressor gene implicated in the carcinogenesis of brain tumors (20). This conclusion was reached based on the findings of homozygous deletions and lack of expression in medulloblastomas and glioblastoma multiforms. Similar results were obtained for a substantial fraction of gastrointestinal and lung tumors (27, 28). The finding that gp-340 is an alternative spliced form of the DMBT1 gene raises the possibility that DMBT1 may not be a classical tumor suppressor gene but rather play a role in the interaction of tumor cells and the immune system.
Recently, MBL has been shown to recognize and bind specifically to oligosaccharide ligands expressed on the surfaces of a human colorectal carcinoma (29). Recombinant vaccinia virus carrying the human MBL gene was shown to possess a potent growth-inhibiting activity against human colorectal carcinoma cells transplanted in nude mice when administered by intratumoral or subcutaneous injection; a significant prolongation of life span of tumor-bearing mice resulted from the treatment. This effect seems to be a consequence of local production of MBL, and it was anticipated that the killing of the tumor cell was mediated by complement activation and lysis of the tumor cell. Surprisingly, a mutant form of MBL, which had essentially no complement-activating activity, was nearly as active as wild-type MBL, and the results therefore indicate that MBL has a cytotoxic activity on its own (29).
Recently, gp-340 also has been shown to bind SP-A in a calcium-dependent manner that is independent of the latter’s lectin activity (17), but gp-340 neither affected the binding of SP-A to alveolar macrophages nor inhibited the ability of SP-A to stimulate macrophage chemotaxis. It was shown that gp-340 alone stimulates the random migration of alveolar macrophages in a manner independent of SP-A-stimulated chemotaxis.
Taken together, the present findings indicate that gp-340 may be an opsonin receptor for SP-A and SP-D located in tissue macrophages throughout the body and in epithelial cells lining the gastrointestinal tract. gp-340/DMBT1 is expressed in several different molecular forms, and the lung collectins bind to at least one of these forms. We suggest that this interaction may be involved in clearance of microorganisms; in addition, because SP-D has a carbohydrate-binding profile similar to that of MBL, we speculate that SP-D together with gp-340 may be in involved in the immune surveillance of cancer.
Acknowledgments
We thank Dr. Stefan Wiemann and Dr. Inge Krebs, Deutsche Krebsforschungszentrum. This work was supported by the Danish Medical Research Council, Michaelsen Fonden, the Novo Nordisk Foundation, Fonden til Lægevidenskabens Fremme, Nationalforeningens Fond, The Benzon Foundation, and Deutsche Krebshilfe Grant 10-1260-Po2.
ABBREVIATIONS
- EGF
epithelial growth factor
- CUB
C1r/C1s Uegf Bmp1
- SRCR
scavenger receptor cysteine-rich
- RT-PCR
reverse transcription–PCR
- kb
kilobase
- ZP
zona pellucida
- SID
SRCR-interspersed domain
- Tm
transmembrane
Footnotes
References
- 1.Weis W I, Taylor M E, Drickamer K. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 2.Thiel S, Vorup-Jensen T, Stover C M, Schwaeble W, Laursen S B, Poulsen K, Willis A C, Eggleton P, Hansen S, Holmskov U, et al. Nature (London) 1997;386:506–510. doi: 10.1038/386506a0. [DOI] [PubMed] [Google Scholar]
- 3.Janeway C A., Jr Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 4.Madan T, Eggleton P, Kishore U, Strong P, Aggrawal S S, Sarma P U, Reid K B. Infect Immun. 1997;65:3171–3179. doi: 10.1128/iai.65.8.3171-3179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartshorn K L, Crouch E, White M R, Colamussi M L, Kakkanatt A, Tauber B, Shepherd V, Sastry K N. Am J Physiol. 1998;274:L958–L969. doi: 10.1152/ajplung.1998.274.6.L958. [DOI] [PubMed] [Google Scholar]
- 6.van Iwaarden J F, Shimizu H, Van Golde P H, Voelker D R, van Golde L M. Biochem J. 1992;286:5–8. doi: 10.1042/bj2860005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crouch E C, Persson A, Griffin G L, Chang D, Senior R M. Am J Respir Cell Mol Biol. 1995;12:410–415. doi: 10.1165/ajrcmb.12.4.7695920. [DOI] [PubMed] [Google Scholar]
- 8.Cai G Z, Griffin G L, Senior R M, Longmore W J, Moxley M A. Am J Physiol. 1999;276:L131–L136. doi: 10.1152/ajplung.1999.276.1.L131. [DOI] [PubMed] [Google Scholar]
- 9.Voorhout W F, Veenendaal T, Kuroki Y, Ogasawara Y, van Golde L M, Geuze H J. J Histochem Cytochem. 1992;40:1589–1597. doi: 10.1177/40.10.1527377. [DOI] [PubMed] [Google Scholar]
- 10.Crouch E, Kuan S F, Persson A. In: Progressive Respiratory Research. Müller B, von Wichert P, editors. Vol. 27. Basel: Karger; 1994. pp. 132–140. [Google Scholar]
- 11.Borron P J, Crouch E C, Lewis J F, Wright J R, Possmayer F, Fraher L J. J Immunol. 1998;161:4599–4603. [PubMed] [Google Scholar]
- 12.Kuroki Y, Gasa S, Ogasawara Y, Shiratori M, Makita A, Akino T. Biochem Biophys Res Commun. 1992;187:963–969. doi: 10.1016/0006-291x(92)91291-w. [DOI] [PubMed] [Google Scholar]
- 13.Persson A, Gibbons B J, Shoemaker J D, Moxley M A, Longmore W J. Biochemistry. 1992;31:12183–12189. doi: 10.1021/bi00163a030. [DOI] [PubMed] [Google Scholar]
- 14.Botas C, Poulain F, Akiyama J, Brown C, Allen L, Goerke J, Clements J, Carlson E, Gillespie A M, Epstein C, et al. Proc Natl Acad Sci USA. 1998;95:11869–11874. doi: 10.1073/pnas.95.20.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korfhagen T R, Sheftelyevich V, Burhans M S, Bruno M D, Ross G F, Wert S E, Stahlman M T, Jobe A H, Ikegami M, Whitsett J A, et al. J Biol Chem. 1998;273:28438–28443. doi: 10.1074/jbc.273.43.28438. [DOI] [PubMed] [Google Scholar]
- 16.Holmskov U, Lawson P, Teisner B, Tornoe I, Willis A C, Morgan C, Koch C, Reid K B. J Biol Chem. 1997;272:13743–13749. doi: 10.1074/jbc.272.21.13743. [DOI] [PubMed] [Google Scholar]
- 17.Tino M J, Wright J R. Am J Respir Cell Mol Biol. 1999;20:759–768. doi: 10.1165/ajrcmb.20.4.3439. [DOI] [PubMed] [Google Scholar]
- 18.Mollenhauer, J., Holmskov, U., Wiemann, S., Krebs, I., Herbertz, S., Madsen, J., Kioschis, P., Coy, J. & Poustka, A. (1999) Oncogene, in press. [DOI] [PubMed]
- 19.Shi S R, Key M E, Kalra K L. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 20.Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus K K, von Deimling A, Poustka A. Nat Genet. 1997;17:32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- 21.Resnick D, Pearson A, Krieger M. Trends Biochem Sci. 1994;19:5–8. doi: 10.1016/0968-0004(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 22.Bork P, Beckmann G. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 23.Romero A, Romao M J, Varela P F, Kolln I, Dias J M, Carvalho A L, Sanz L, Topfer-Petersen E, Calvete J J. Nat Struct Biol. 1997;4:783–788. doi: 10.1038/nsb1097-783. [DOI] [PubMed] [Google Scholar]
- 24.Cheng H, Bjerknes M, Chen H. Anat Rec. 1996;244:327–343. doi: 10.1002/(SICI)1097-0185(199603)244:3<327::AID-AR5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 25.Li X J, Snyder S H. J Biol Chem. 1995;270:17674–17679. doi: 10.1074/jbc.270.30.17674. [DOI] [PubMed] [Google Scholar]
- 26.Mayer W E, Tichy H. Gene. 1995;164:267–271. doi: 10.1016/0378-1119(95)94092-z. [DOI] [PubMed] [Google Scholar]
- 27.Mori M, Shiraishi T, Tanaka S, Yamagata M, Mafune K, Tanaka Y, Ueo H, Barnard G F, Sugimachi K. Br J Cancer. 1999;79:211–213. doi: 10.1038/sj.bjc.6690035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu W, Kemp B L, Proctor M L, Gazdar A F, Minna J D, Hong W K, Mao L. Cancer Res. 1999;59:1846–1851. [PubMed] [Google Scholar]
- 29.Ma Y, Uemura K, Oka S, Kozutsumi Y, Kawasaki N, Kawasaki T. Proc Natl Acad Sci USA. 1999;96:371–375. doi: 10.1073/pnas.96.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]