Abstract
Interferons (IFNs) inhibit induction by IL-4 of multiple genes in human monocytes. However, the mechanism by which IFNs mediate this inhibition has not been defined. IL-4 activates gene expression by inducing tyrosine phosphorylation, homodimerization, and nuclear translocation of the latent transcription factor, STAT6 (signal transducer and activator of transcription-6). STAT6-responsive elements are characteristically present in the promoters of IL-4-inducible genes. Because STAT6 activation is essential for IL-4-induced gene expression, we examined the ability of type I and type II IFNs to regulate activation of STAT6 by IL-4 in primary human monocytes. Pretreatment of monocytes with IFN-β or IFN-γ, but not IL-1, IL-2, macrophage colony-stimulating factor, granulocyte/macrophage colony-stimulating factor, IL-6, or transforming growth factor β suppressed activation of STAT6 by IL-4. This inhibition was associated with decreased tyrosine phosphorylation and nuclear translocation of STAT6 and was not evident unless the cells were preincubated with IFN for at least 1 hr before IL-4 stimulation. Furthermore, inhibition by IFN could be blocked by cotreatment with actinomycin D and correlated temporally with induction of the JAK/STAT inhibitory gene, SOCS-1. Forced expression of SOCS-1 in a macrophage cell line, RAW264, markedly suppressed trans-activation of an IL-4-inducible reporter as well as IL-6- and IFN-γ-induced reporter gene activity. These findings demonstrate that IFNs inhibit IL-4-induced activation of STAT6 and STAT6-dependent gene expression, at least in part, by inducing expression of SOCS-1.
The Th2-type cytokine IL-4 activates gene expression by binding to specific receptor complexes on the surface of IL-4-responsive target cells. Type I IL-4 receptor complexes are heterodimers composed of the IL-4Rα chain (p140) and the IL-2Rγ chain (γc) (1, 2). Binding of IL-4 to these receptor complexes activates the receptor-associated Janus tyrosine kinases, JAK1 and JAK3, which then phosphorylate critical tyrosine residues on the intracellular domain (ICD) of the IL-4Rα chain (3–5). These phosphorylated tyrosine residues on the ICD of IL-4Rα provide docking sites for the latent cytosolic transcription factor, STAT6 (signal transducer and activator of transcription-6) (6, 7). STAT6 binds to these docking sites via its SH2 (Src homology 2) domain and is, in turn, tyrosine-phosphorylated by the receptor-associated JAKs (8–10). Activated STAT6 then homodimerizes and translocates to the nucleus, where it binds with high affinity to STAT-binding elements (SBE) in the promoters of various IL-4/IL-13-responsive genes.
IL-4 and IL-13 induce transcription of a distinct subset of genes in human monocytes, including FcɛRIIb (CD23) (11, 12), 15-lipoxygenase (15-LO) (13, 14), IL-1 receptor antagonist (IL-1ra) (15–17), and types I and II IL-1 receptors (IL-1R) (18, 19). Type I IFNs (IFN-α and IFN-β) and type II IFN (IFN-γ) inhibit IL-4/IL-13-induced gene expression in monocytes and B cells. For example, IFN-γ suppresses IgE synthesis by IL-4-stimulated B cells (20–22). IFNs also inhibit IL-4-induced CD23 expression in both B cells (23, 24) and monocytes (25, 26). Furthermore, IFN-γ inhibits IL-4- and IL-13-induced expression of 15-LO and IL-1ra in monocytes (13, 14, 27). Moreover, we have shown recently that IFN-β and IFN-γ inhibit expression of the IL-1RI and IL-1RII genes in IL-4/IL-13-stimulated monocytes (28). The mechanism by which IFNs inhibit expression of these genes has not been defined. However, it is known that activation and nuclear translocation of STAT6 is essential for induction of gene expression by IL-4 and IL-13 (29–31).
IL-4/IL-13-response elements (i.e., STAT6-binding sites) are present in the proximal promoter regions of IL-4/IL-13-inducible genes, including Iɛ (32), IL-1ra (33), and IL-4Rα (34). The SBEs in the promoters of these genes are distinct from classical IFN-γ activation sequences (GAS elements) because they contain an intervening four-base spacer instead of a three-base spacer between the palindromic TTC/GAA nucleotides. This difference (i.e., the presence of a four-base spacer instead of a three-base spacer) makes these SBEs very selective for STAT6 (35, 36). Consequently, other STAT proteins such as STAT1α, the principal IFN-inducible STAT, do not bind to these elements and cannot compete with STAT6 for binding to these sites. Because of this, we reasoned that IFNs must inhibit IL-4-inducible gene expression through some other mechanism. In this report, we demonstrate that IFNs suppress IL-4-induced tyrosine phosphorylation and nuclear translocation of STAT6 in monocytes. This inhibitory effect could be blocked by actinomycin D and correlated temporally with expression of the JAK/STAT inhibitory gene, SOCS-1 (suppressor of cytokine signaling). Moreover, forced expression of SOCS-1 in a macrophage cell line, RAW264, markedly inhibited activation of an IL-4-inducible, STAT6-responsive reporter gene. Thus, IFNs induce synthesis of a negative regulatory factor (SOCS-1) that can suppress activation of STAT6. These findings define a mechanism by which IFNs can antagonize IL-4- and IL-13-inducible gene expression in human monocytes.
MATERIALS AND METHODS
Culture Medium and Reagents.
The complete medium used for monocyte culture consisted of RPMI 1640 medium (GIBCO) supplemented with 10% FCS (HyClone)/2 mM l-glutamine/50 μg/ml gentamycin. Recombinant human IL-4 (rhIL-4) and murine IL-4 (rmIL-4) were provided by Schering-Plough. Recombinant human IL-13 was obtained from BioSource International (Camarillo, CA). Recombinant human IFN-γ was provided by Genentech. Recombinant human IFN-β was provided by Chiron. Rabbit anti-STAT6 antibody was purchased from Santa Cruz Biotechnology. Rabbit anti-phospho(Tyr641)-STAT6 Ab was a kind gift from New England Biolabs.
Cells.
Normal human peripheral blood monocytes were isolated by elutriation in a Beckman JE-6B centrifugal elutriator as described previously (37). The elutriated monocyte fraction consisted of >95% monocytes as determined by histologic staining and fluorescence-activated cell sorter (FACS) analysis with the anti-CD14 mAb Leu M3 (Becton Dickinson). Monocytes were cultured routinely at 4 × 106 cells per ml in complete medium in round-bottomed, polypropylene tubes. The macrophage cell line, RAW264, was obtained from American Type Culture Collection (Manassas, VA) and cultured in complete medium.
Electrophoretic Mobility-Shift Assay (EMSA).
Nuclear protein extracts were prepared from cytokine-treated cells by using a modification (38) of the original method described by Dignam et al. (39). A double-stranded oligonucleotide (IL-1ra SBE1) based on a DNA sequence present in the promoter of the human IL-1ra gene was used as a probe in the gel-shift assays (33). This oligonucleotide contains an N4-type GAS element that can bind STAT6 but not STAT1α (33). Binding reactions were performed as described previously (38). A portion of each binding reaction mixture (8 μl per sample) was electrophoresed on nondenaturing, 6% polyacrylamide gels (Novex) by using 0.25× TBE buffer (22 mM Tris⋅HCl, pH 8.0/22 mM borate/0.5 mM EDTA). The gels then were dried and visualized by autoradiography.
Western Blots.
After cytokine treatment, the cells were washed three times with Dulbecco’s PBS, and cytosolic and nuclear protein extracts were prepared as described previously (38). Equal amounts (75 μg per sample) of these extracts were resolved by electrophoresis on 8% polyacrylamide Tris-glycine gels and then transferred onto polyvinylidene difluoride membranes (Novex). Immunoblot analysis was performed by using rabbit anti-phospho-STAT6 antibody or anti-STAT6 antibody at a final dilution of 1:1,000 in TTBS containing 3% nonfat dry milk (TTBs = 20 mM Tris, pH 7.6/137 mM NaCl/0.1% Tween-20). The membranes were then incubated with goat anti-rabbit IgG–horseradish peroxidase (HRP) (Amersham) diluted 1:5,000. After washing, the membranes were incubated with the HRP substrate, SuperSignal (Pierce). STAT6 was detected by enhanced chemiluminescence (ECL) according to the manufacturer’s protocol (Pierce).
Northern Blots.
Total RNA was isolated from cultured monocytes by the acid/guanidinium thiocyanate/phenol/chloroform extraction method as described previously (40). Equivalent amounts of RNA (10 μg per lane) were size-fractionated by electrophoresis in 1% agarose gels containing 0.66 M formaldehyde. The RNA then was transferred onto Nytran membranes and cross-linked by exposure to UV light. The membranes then were hybridized and washed according to standard procedures. The SOCS-1 probe was a 724-bp XbaI fragment of a full-length murine SOCS-1 cDNA (41). Gel-purified insert DNA was radiolabeled by the random-primer method of Feinberg and Vogelstein (42).
Construction of the SOCS-1 Expression Plasmids.
The SOCS-1 expression plasmid was kindly provided by Hiroyuki Mano (43). The Flag-tagged version of SOCS-1 was prepared by PCR. The full-length cDNA was amplified by using the following primers: 5′-CGC GGG GGG ATC CTT GTA GCA CAC AAC CAG GTG GCA GCC-3′ (sense) and 5′-GCG CCC GGA ATT CGG TCA AAT CTG GAA GGG GAA GGA GCT CAG-3′ (antisense). The SOCS-1 mutant, bearing a single amino acid change in the SH2 domain (R105A), was generated by using two-step PCR cloning and the following primers: 5′-GGC ACC TTC CTG GTG GCC GAC AGC CGC CAG CGG-3′ (sense) and 5′-CCG CTG GCG GCT GTC GGC CAC CAG GAA GGT GCC-3′ (antisense). The PCR products were cloned into the BamHI and EcoRI sites of a pcDNA3 derivative carrying a Flag epitope tag. The integrity of the DNA fragments was confirmed by DNA sequence analysis.
Transfections.
The IL-4-responsive reporter construct, C/EBP-N4, has been described previously (44). The IL-4 response element in this construct is derived from a sequence in the promoter of the human Ig heavy chain germ-line ɛ gene that contains adjacent binding sites for C/EBP and STAT6 (44). Transient transfections were performed by using the DEAE-Dextran method as described previously (44). Briefly, 1 × 106 RAW264 cells were incubated in serum-free DMEM containing 100 μg/ml DEAE-dextran solution, 10 μg of the IL-4 luciferase reporter, and 10 μg of the SOCS-1 expression plasmids (wild type or mutant) or vector control for 1 hr at 37°C. DMSO was added at a final concentration of 10% for 1 min. This medium then was washed off and replaced with fresh medium. Each transfection was carried out in triplicate. Eighteen hours after transfection, the cells were stimulated with 10 ng/ml recombinant murine IL-4 for 6 hr. Luciferase activity was measured by using luciferase assay kits obtained from Promega. Luciferase units were normalized to the amount of total protein.
RESULTS
IL-4 Activates STAT6 in Human Monocytes.
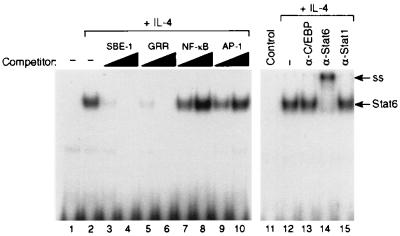
Expression of IL-4-inducible genes is regulated, in part, via activation and nuclear translocation of STAT6 (29–31). To examine the effects of IFNs on STAT6 activity, we used an oligonucleotide probe derived from a DNA sequence in the promoter of the human IL-1 receptor antagonist gene (33). This probe, denoted SBE1, contains a GAS-like sequence that binds STAT6 with high affinity, but does not bind other STAT proteins such as STAT1α or STAT3 (33). As shown in Fig. 1, nuclear extracts from IL-4-stimulated monocytes generated a single DNA-binding complex when coincubated with radiolabeled SBE1 probe (Fig. 1, lane 2). This IL-4-inducible DNA-binding activity was inhibited competitively by cold SBE1 or GRR oligonucleotides, but not by equivalent amounts of NF-κB or AP-1 oligonucleotides. The IL-4-inducible DNA-binding activity also could be supershifted by an anti-STAT6 antibody (Fig. 1, lane 14) but not by an anti-STAT1α antibody (Fig. 1, lane 15) or an antibody directed against an unrelated transcription factor, C/EBPβ (Fig. 1, lane 13).
Figure 1.
IL-4 induces STAT6 activity in human monocytes. Monocytes (4 × 106 cells per ml) were incubated at 37°C in the presence (lane 2) or absence (lane 1) of recombinant human IL-4 (rhIL-4; 10 ng/ml) for 30 min. The cells then were harvested, and nuclear protein extracts were prepared. The nuclear extracts then were assayed by EMSA using radiolabeled IL-1ra SBE1 probe. In lanes 3–10, 5 μg of the nuclear extract from IL-4-stimulated monocytes (lane 2) was preincubated with a 10- or 50-fold molar excess of unlabeled SBE1, GRR, NF-κB, or AP-1 oligonucleotide. In lanes 11–15, 5 μg of the nuclear extract from IL-4-stimulated monocytes (lane 2) was preincubated with 1 μl of rabbit antisera specific for C/EBPβ, STAT6, or STAT1α as indicated. ss, super shift.
IFN-β and IFN-γ Inhibit Activation of STAT6 by IL-4.
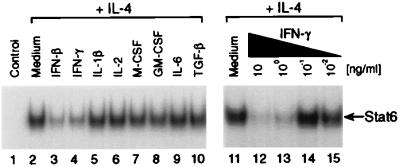
IFN-β and IFN-γ inhibit induction of several genes by IL-4 in human monocytes (13, 14, 25–28). To determine whether IFNs and/or other cytokines can inhibit activation of STAT6 by IL-4, we pretreated monocytes for 1 hr at 37°C with a panel of cytokines that included IFN-β, IFN-γ, IL-1β, IL-2, macrophage colony-stimulating factor (M-CSF), granulocyte/macrophage colony-stimulating factor (GM-CSF), IL-6, and transforming growth factor β (TGF-β). IL-4 then was added, and the cultures were incubated for an additional 30 min. At the end of the incubation period, the cells were harvested and nuclear protein extracts were prepared. The extracts then were assayed for STAT6 activity by EMSA by using radiolabeled IL-1Ra SBE1 probe. As shown in Fig. 2, pretreatment with IFN-β (Fig. 2, lane 3) or IFN-γ (Fig. 2, lane 4), but none of the other cytokines tested, inhibited induction of STAT6 activity by IL-4. Furthermore, the ability of IFN-γ to inhibit IL-4-induced STAT6 activity was dose-dependent and demonstrable when IFN-γ was used at a final concentration of 10 or 1 ng/ml (Fig. 2, lanes 11–15).
Figure 2.
Effect of pretreatment with selected cytokines on induction of STAT6 activity by IL-4 in monocytes. Monocytes (4 × 106 cells per ml) were preincubated for 1 hr at 37°C with medium alone, IFN-β, IFN-γ, IL-1β, IL-2, M-CSF, GM-CSF, IL-6, or TGF-β (10 ng/ml each). At the end of this primary incubation period, IL-4 (1 ng/ml) was added (lanes 2–10), and the cultures were incubated for an additional 30 min. Nuclear protein extracts were prepared and assayed by EMSA for STAT6 activity by using radiolabeled IL-1ra SBE1 probe. In lanes 11–15, monocytes were preincubated with medium alone (lane 11) or IFN-γ at the indicated concentrations for 1 hr. At the end of this initial incubation period, IL-4 (1 ng/ml) was added, and the cultures were incubated for an additional 30 min. Nuclear extracts then were prepared and assayed for STAT6 activity.
Inhibition of STAT6 Activation by IFNs Is Transcription-Dependent.
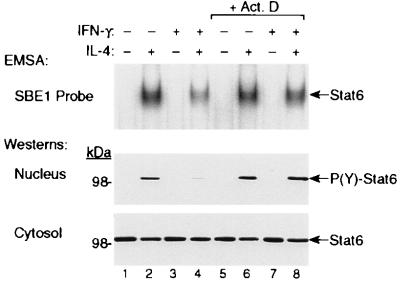
To determine whether the inhibition by IFN is mediated by a gene that is newly transcribed, we tested whether actinomycin D could block inhibition of STAT6 activity by IFN-γ. As shown in Fig. 3 Top, pretreatment with IFN-γ suppressed induction of STAT6 activity by IL-4 in control cells (Fig. 3, lane 4), but did not inhibit STAT6 activity in cells that were cotreated with actinomycin D (Fig. 3, lane 8). These differences were even more apparent when the nuclear and cytosolic protein extracts were analyzed by Western blotting with anti-phospho-STAT6 (Fig. 3, Middle) and anti-STAT6 Abs (Fig. 3, Bottom), respectively. Reduced STAT6 DNA-binding activity was associated with decreased tyrosine phosphorylation and nuclear translocation of STAT6 in cells that were pretreated with IFN-γ (Fig. 3, lane 4), and this reduction did not occur when cells were cotreated with actinomycin D (Fig. 3, lane 8).
Figure 3.
Inhibition of IL-4-induced STAT6 activity by IFN-γ is abrogated by the transcription inhibitor, actinomycin D. Monocytes (4 × 106 cells per ml) were preincubated with medium alone or IFN-γ (10 ng/ml) in the presence or absence of actinomycin D (Act. D) for 90 min at 37°C. The cells then were treated with or without IL-4 (1 ng/ml) for an additional 30 min at 37°C. At the end of the second incubation period, nuclear extracts were prepared and assayed for STAT6 activity by EMSA using the IL-1ra SBE1 probe (Top). The nuclear and cytosolic protein extracts also were analyzed by immunoblotting with rabbit anti-P(Y)-STAT6 (Middle) and rabbit anti-STAT6 (Bottom) antibodies, respectively.
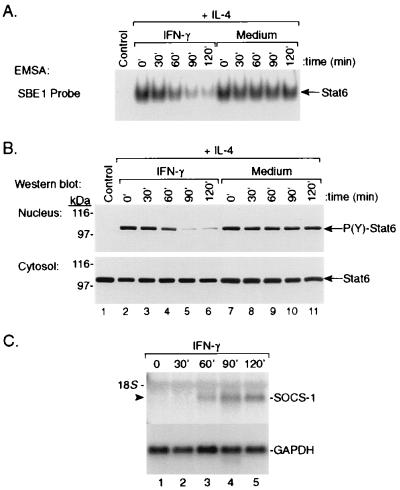
Inhibition of STAT6 Activity by IFN Is Temporally Associated with Expression of the SOCS-1 Gene. Temporal analysis of the effect of IFN-γ on IL-4-induced STAT6 activity showed that the cells need to be preincubated with IFN at 37°C for at least 60 min before IL-4 stimulation (Fig. 4A). More complete inhibition was attained when cells were preincubated with IFN for 90–120 min. As shown in Fig. 4B, reduced STAT6 DNA-binding activity was associated with decreased tyrosine phosphorylation and nuclear translocation of STAT6 as determined by immunoblot analysis of the nuclear and cytosolic protein extracts using rabbit anti-P(Y)-STAT6 and anti-STAT6 Abs, respectively.
Figure 4.
(A) Kinetic analysis of the induction of inhibition by IFN-γ in monocytes. Monocytes (4 × 106 cells per ml) were preincubated with IFN-γ (10 ng/ml) (lanes 2–6) or medium (lanes 7–11) for 0, 30, 60, 90, or 120 min at 37°C. IL-4 (1 ng/ml) then was added, and the cultures were incubated for an additional 30 min. Nuclear extracts then were prepared and assayed by EMSA for STAT6 activity. (B) The nuclear and cytosolic protein fractions also were analyzed by Western blotting with anti-P(Y)-STAT6 and anti-STAT6 antibodies, respectively. (C) Monocytes were incubated with rhIFN-γ for 0–2 hr at 37°C. At the indicated time points, the cells were harvested and total RNA was isolated. The levels of SOCS-1 and glyceraldehyde-3-phosphate dehydrogenase mRNA then were examined by Northern blotting.
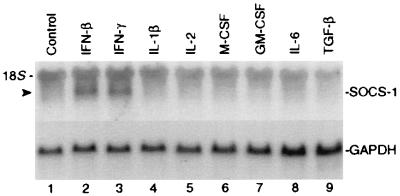
The SOCS genes constitute a family of cytokine-inducible, SH2-containing proteins that inhibit JAK/STAT-dependent gene expression (41, 45, 46). One of these genes, SOCS-1 (also known as JAB or SSI-1), is rapidly inducible by multiple cytokines, including type I and II IFNs, in the leukemic murine myeloid cell line M1 (41, 47). To determine whether this gene is IFN-inducible in human monocytes, we treated monocytes with IFN-γ (10 ng/ml) for 0–2 hr and then measured SOCS-1 mRNA levels by Northern blotting. As shown in Fig. 4C, IFN-γ induced SOCS-1 expression in these cells. Peak levels of SOCS-1 mRNA occurred approximately 90 min posttreatment. The appearance of SOCS-1 mRNA in monocytes correlated temporally with the suppression of IL-4-induced STAT6 activity (Fig. 4 A and B). Furthermore, as shown in Fig. 5, analysis of the ability of multiple cytokines to induce SOCS-1 gene expression in monocytes demonstrated that only those cytokines that induced SOCS-1 expression (i.e., IFN-β and IFN-γ) inhibited IL-4-induced activation of STAT6 (Fig. 2).
Figure 5.
Analysis of the ability of multiple cytokines to induce SOCS-1 gene expression in monocytes. Monocytes (4 × 106 cells per ml) were cultured at 37°C for 2 hr in the presence of medium alone, IFN-β, IFN-γ, IL-1β, IL-2, M-CSF, GM-CSF, IL-6, or TGF-β (10 ng/ml each). At the end of the incubation period, the cells were harvested, RNA was extracted, and SOCS-1 mRNA levels were measured by Northern blotting.
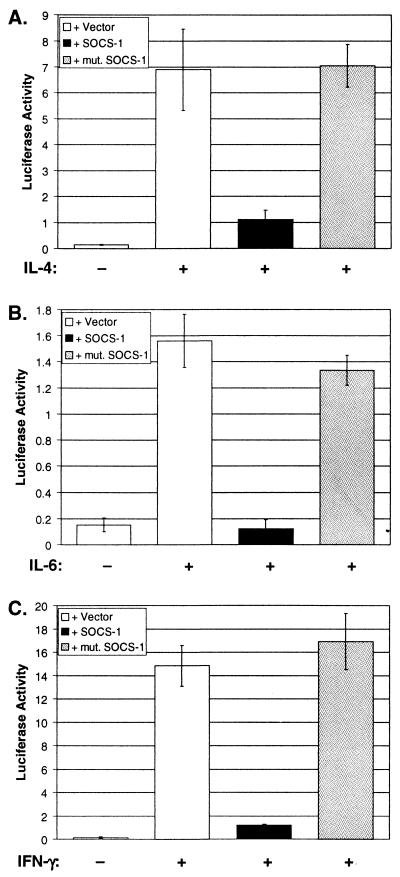
To determine whether forced expression of the SOCS-1 gene in a myeloid cell line can suppress IL-4-inducible promoter activity, we transfected the macrophage cell line, RAW264, with an IL-4-inducible reporter gene, C/EBP-N4, in the presence or absence of a wild-type or mutant SOCS-1 expression plasmid. We then measured reporter gene activity 6 hr after stimulating these cells with IL-4. The reporter gene used to assess IL-4 responsiveness contains a prototypical N4 STAT6-binding site (44). As shown in Fig. 6A, IL-4 induction of control RAW264 cells transfected with the reporter gene alone resulted in a 42-fold increase in basal luciferase activity. In contrast, these levels were diminished markedly (≈84% suppression) in cells that were cotransfected with the reporter gene plus the wild-type SOCS-1 expression plasmid. The SOCS proteins contain central SH2 domains that enable them to bind to phosphorylated tyrosine residues on the intracellular domains of cytokine receptors or the receptor-associated JAKs (41, 45, 46). These SH2 domains appear to be essential for the biological activity of the SOCS proteins. To examine the role of the SOCS-1 SH2 domain in mediating inhibition of IL-4-induced reporter gene activity, we introduced an Arg → Ala mutation in the phosphotyrosine-binding pocket of the SOCS-1 SH2 domain. This mutation abrogated the ability of SOCS-1 to inhibit IL-4-induced reporter gene activity in RAW264 cells.
Figure 6.
(A) Forced expression of SOCS-1 in the macrophage cell line, RAW264, inhibits IL-4-induced trans-activation of a STAT6-responsive reporter gene. RAW264 cells were transfected with an IL-4-inducible reporter, C/EBP-N4-Luc, in the presence or absence of a wild-type SOCS-1 expression plasmid or a SOCS-1 mutant (mut. SOCS-1) containing a single amino acid change (Arg105 → Ala) in the SH2 domain. Control cells were transfected with the reporter gene plus the plasmid vector alone. Eighteen hours after transfection, the cells were stimulated with recombinant murine IL-4 (rmIL-4; 10 ng/ml) for 6 hr. Luciferase activity then was measured and normalized to total protein. Values represent the mean ± SD of three independent experiments. (B) Forced expression of SOCS-1 in RAW264 cells inhibits IL-6-induced trans-activation of a STAT3-responsive reporter, IRF-1-Luc. (C) Forced expression of SOCS-1 in RAW264 cells inhibits IFN-γ-induced trans-activation of a STAT1-responsive reporter (IRF-1-Luc).
To determine whether forced expression of SOCS-1 in RAW264 cells also inhibits activation by other cytokines that utilize the JAK/STAT signaling pathway, we cotransfected the SOCS-1 expression construct together with a reporter gene (IRF-1-Luc) that can be activated by either IL-6 or IFN-γ. As shown in Fig. 6 B and C, SOCS-1 markedly inhibited trans-activation of this reporter gene by both IL-6 and IFN-γ. Therefore, once expressed, SOCS-1 can inhibit signal transduction mediated via multiple cytokine receptors.
DISCUSSION
Type I and type II IFNs inhibit expression of several IL-4/IL-13-inducible genes in monocytes, including CD23 (25, 26), 15-lipoxygenase (13, 14), IL-1ra (27), IL-1RI, and IL-1RII (28). The promoter regions of these genes contain functional STAT6-binding sites through which IL-4 and IL-13 induce (or amplify) their expression. The present studies were conducted to determine the mechanism(s) by which IFNs suppress IL-4- and IL-13-inducible gene expression in primary human monocytes. We found that pretreatment of monocytes with type I or II IFNs inhibits IL-4-induced activation and nuclear translocation of STAT6. Inhibition of STAT6 activation required preincubation with IFN for at least 1 hr and could be blocked by actinomycin D. Furthermore, the onset of inhibition correlated temporally with expression of the JAK/STAT inhibitory gene, SOCS-1. Moreover, forced expression of SOCS-1 in the macrophage cell line, RAW264, markedly suppressed IL-4-induced trans-activation of a STAT6-dependent reporter gene. These findings define a mechanism by which IFNs may suppress expression of multiple IL-4/IL-13-inducible genes.
A family of genes known as the SOCS or SSI (STAT-induced STAT inhibitor) genes recently has been identified (41, 45, 46). The proteins encoded by these genes are relatively small molecules (20–40 kDa) that can interact physically via their central SH2 domains with either phosphotyrosine residues in the JAK kinases and/or the intracellular domains of certain cytokine receptors and, thereby, inhibit cytokine-induced STAT activation and STAT-dependent gene expression. We considered whether inhibition of IL-4-induced STAT6 activity by IFNs might be mediated via the induction of one or more of these genes. Recent studies have shown that SOCS-1 expression in the leukemic murine myeloid cell line M1 can be markedly up-regulated by treatment with various cytokines, including IFNs (47). In addition, both type I and type II IFNs have been shown to induce SOCS-1 expression in murine bone marrow stem cells (41). However, to date, no published studies have examined whether IFNs can induce SOCS gene expression in primary human cells. We found that IFN treatment induces high levels of SOCS-1 expression in monocytes and that expression of this gene coincides with decreased STAT6 activity.
Inhibition of STAT6 activity by IFNs required that the cells be preincubated with IFN-β or IFN-γ for at least 60 min before IL-4 stimulation. More complete suppression was attained when monocytes were preincubated with IFN for 90–120 min. This lag period appears to be necessary for de novo expression of SOCS-1. Inhibition could be blocked by actinomycin D, demonstrating that it is newly transcribed. In normal resting cells, the SOCS proteins do not exist as preformed latent molecules in the cytosol. However, their expression can be induced rapidly by multiple cytokines, including type I and type II IFNs (41, 47). We found that IFN-β and IFN-γ induce high levels of SOCS-1 expression in monocytes and that induction of SOCS-1 mRNA coincides with the suppression of IL-4-induced STAT6 activity. Other cytokines such as IL-1, IL-2, M-CSF, GM-CSF, IL-6, and TGF-β did not induce SOCS-1 mRNA and did not inhibit IL-4-induced activation of STAT6. Forced expression of the SOCS-1 gene in the macrophage cell line, RAW264, markedly inhibited IL-4-induced trans-activation of a STAT6-responsive reporter gene. Thus, IFNs inhibit IL-4-induced STAT6 activity and STAT6-dependent gene expression, at least in part, by inducing SOCS-1 gene expression.
Although preincubation of monocytes with IFNs suppressed IL-4-induced tyrosine phosphorylation of STAT6, decreased STAT6 phosphorylation actually may be a secondary consequence of decreased JAK1 activity. JAK1 is constitutively associated with the intracellular domain of the IL-4Rα chain in both type I and type II IL-4R complexes, and both IL-4 and IL-13 induce tyrosine phosphorylation of JAK1 (3–5). Furthermore, studies have shown that JAK1-deficient cell lines are unable to transduce IL-4-mediated signals (5, 48). Although the precise mechanism by which SOCS proteins inhibit cytokine-induced signaling has not been fully defined, studies have shown that SOCS-1 can bind via its SH2 domain to the tyrosine kinase domain (JH1 domain) in all four JAKs and, thereby, inhibit their activity (45–47). It is likely, therefore, that IFN-induced SOCS-1 targets primarily JAK1 and that phosphorylation of STAT6 is decreased only as a secondary consequence of decreased activation of JAK1. Unfortunately, IFN-β and IFN-γ themselves induce tyrosine phosphorylation of JAK1, so it is difficult to determine directly whether IL-4-induced JAK1 activity is decreased in IFN-treated monocytes because the basal levels of tyrosine-phosphorylated JAK1 are elevated significantly by IFN pretreatment. However, a recent report showed that stable expression of SOCS-1 in the murine B cell line, M12, markedly inhibits IL-4-induced activation of JAK1 (49).
The ability of IFNs to antagonize IL-4- or IL-13-induced STAT6 activity is not limited to monocytes because we have found that IFN-β and IFN-γ also inhibit activation of STAT6 in fibroblasts. These findings demonstrate that this inhibitory pathway is inducible in both hematopoietic and nonhematopoietic cells and may be a common mechanism by which IFNs can antagonize IL-4- and IL-13-induced signaling in multiple cell types. Studies have shown that IFN-γ can inhibit IL-4-induced proliferation by Th2-type T cell clones (50). In addition, a number of earlier reports showed that pretreatment of primary B cells or B cell lines with type I or II IFNs inhibits IL-4-induced Ig isotype switching to IgE (20–22). A functional STAT6-binding site is present in the proximal promoter region of the Iɛ gene, and binding of activated STAT6 to this site is essential for IL-4-induced gene expression (44). Furthermore, recent studies show that IFN-γ also inhibits activation of STAT6 in certain B cell lines (49, 51). It is possible, therefore, that this is also a mechanism by which IFNs inhibit IgE synthesis in B cells.
Further characterization of the molecular basis by which IFNs inhibit STAT6 activity may aid in the future development of specific inhibitors of STAT6. Such compounds might be very useful as therapeutic agents for treating diseases characterized by elevated IL-4 and IL-13 levels, particularly atopic disorders. This hypothesis is supported by recent reports showing that STAT6 knock-out mice are highly resistant to bronchoalveolar challenge with allergens that are known to induce IL-4 and IL-13 activity (52, 53). It is noteworthy that a recent study showed that thymocytes from SOCS-1 knock-out mice exhibit exaggerated IL-4-induced STAT6 activity (54). These findings as well as those described in this report suggest a critical role for SOCS-1 in counterregulating STAT6 activity. The ability of type I and type II IFNs to induce SOCS-1 gene expression provides a mechanism to explain how IFNs can suppress activation of IL-4- and IL-13-inducible genes.
Acknowledgments
We thank Valerie Calvert for preparing elutriated monocytes and Dr. Douglas Hilton (The Walter and Eliza Hall Institute, Parkville, Australia) and Dr. Hiroyuki Mano (Jichi Medical School, Tochigi, Japan) for providing the SOCS cDNAs. H.L.D. was the recipient of a Postgraduate Research Fellowship Award from the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the Food and Drug Administration.
ABBREVIATIONS
- EMSA
electrophoretic mobility-shift assay
- GAS
γ-interferon activation sequence
- IL-1ra
IL-1 receptor antagonist
- JAK
Janus kinase
- SBE
STAT-binding element
- SH2 domain
Src homology-2 domain
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- M-CSF
macrophage colony-stimulating factor
- GM-CSF
granulocyte/macrophage colony-stimulating factor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 2.Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R K, Paul W E, et al. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 3.Witthuhn B A, Silvennoinen O, Miura O, Lai K S, Cwik C, Liu E T, Ihle J N. Nature (London) 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 4.Keegan A D, Johnston J A, Tortolani P J, McReynolds L J, Kinzer C, O’Shea J J, Paul W E. Proc Natl Acad Sci USA. 1995;92:7681–7685. doi: 10.1073/pnas.92.17.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X H, Patel B K R, Wang L-M, Frankel M, Ellmore N, Flavell R A, LaRochelle W J, Pierce J H. J Biol Chem. 1997;272:6556–6560. doi: 10.1074/jbc.272.10.6556. [DOI] [PubMed] [Google Scholar]
- 6.Smerz-Bertling C, Duschl A. J Biol Chem. 1995;270:966–970. doi: 10.1074/jbc.270.2.966. [DOI] [PubMed] [Google Scholar]
- 7.Ryan J J, McReynolds L J, Keegan A, Wang L, Garfein E, Rothman P, Nelms K, Paul W E. Immunity. 1996;4:123–132. doi: 10.1016/s1074-7613(00)80677-9. [DOI] [PubMed] [Google Scholar]
- 8.Kotanides H, Reich N C. Science. 1993;262:1265–1267. doi: 10.1126/science.7694370. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, Schindler U, Henzel W J, Ho T C, Brasseur M, McKnight S L. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 10.Quelle F W, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben S M, Cleveland J L, Pierce J H, Keegan A D, Nelms K, et al. Mol Cell Biol. 1995;15:3336–3343. doi: 10.1128/mcb.15.6.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vercelli D, Jabara H H, Lee B W, Woodland N, Geha R S, Leung D Y M. J Exp Med. 1988;167:1406–1416. doi: 10.1084/jem.167.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie A N J, Culpepper J A, de Waal Malefyt R, Brière F, Punnonen J, Aversa G, Sato A, Dang W, Cocks B G, Menon S, et al. Proc Natl Acad Sci USA. 1993;90:3735–3739. doi: 10.1073/pnas.90.8.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad D J, Kuhn H, Mulkins M, Highland E, Sigal E. Proc Natl Acad Sci USA. 1992;89:217–221. doi: 10.1073/pnas.89.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nassar G M, Morrow J D, Roberts L J, II, Lakkis F G, Badr K F. J Biol Chem. 1994;269:27631–27634. [PubMed] [Google Scholar]
- 15.Fenton M J, Buras J A, Donnelly R P. J Immunol. 1992;149:1283–1288. [PubMed] [Google Scholar]
- 16.Vannier E, Miller L C, Dinarello C A. Proc Natl Acad Sci USA. 1992;89:4076–4080. doi: 10.1073/pnas.89.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muzio M, Re F, Sironi M, Polentarutti N, Minty A, Caput D, Ferrara P, Mantovani A, Colotta F. Blood. 1994;83:1738–1743. [PubMed] [Google Scholar]
- 18.Colotta F, Re F, Muzio M, Bertini R, Polentarutti N, Sironi M, Giri J G, Dower S K, Sims J E, Mantovani A. Science. 1993;261:472–475. doi: 10.1126/science.8332913. [DOI] [PubMed] [Google Scholar]
- 19.Colotta F, Re F, Muzio M, Polentarutti N, Minty A, Caput D, Ferrara P, Mantovani A. J Biol Chem. 1994;269:12403–12406. [PubMed] [Google Scholar]
- 20.Snapper C M, Paul W E. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 21.Pene J, Rousset F, Briere F, Chretien I, Bonnefoy J-Y, Spits H, Yokota T, Arai N, Arai K, Banchereau J, et al. Proc Natl Acad Sci USA. 1988;85:6880–6884. doi: 10.1073/pnas.85.18.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thyphronitis G, Tsokos G C, June C H, Levine A D, Finkelman F D. Proc Natl Acad Sci USA. 1989;86:5580–5584. doi: 10.1073/pnas.86.14.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galizzi J P, Cabrillat H, Rousset F, Menetrier C, de Vries J E, Banchereau J. J Immunol. 1988;141:1982–1988. [PubMed] [Google Scholar]
- 24.Denoroy M, Yodoi J, Banchereau J. Mol Immunol. 1990;27:129–134. doi: 10.1016/0161-5890(90)90107-b. [DOI] [PubMed] [Google Scholar]
- 25.te Velde A A, Rousset F, Peronne C, de Vries J E, Figdor C G. J Immunol. 1990;144:3052–3059. [PubMed] [Google Scholar]
- 26.Alderson M R, Armitage R J, Tough T W, Ziegler S F. Cytokine. 1994;6:407–413. doi: 10.1016/1043-4666(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 27.Sone S, Orino E, Mitzuno K, Yano S, Nishioka Y, Haku T, Nii A, Ogura T. Eur Respir J. 1994;7:657–663. doi: 10.1183/09031936.94.07040657. [DOI] [PubMed] [Google Scholar]
- 28.Dickensheets H L, Donnelly R P. J Immunol. 1997;159:6226–6233. [PubMed] [Google Scholar]
- 29.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Nature (London) 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 30.Shimoda K, van Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp R A, Chu C, Quelle F W, Nosaka T, Vignali D A, et al. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 32.Köhler I, Rieber E P. Eur J Immunol. 1993;23:3066–3071. doi: 10.1002/eji.1830231204. [DOI] [PubMed] [Google Scholar]
- 33.Ohmori Y, Smith M F, Hamilton T A. J Immunol. 1996;157:2058–2065. [PubMed] [Google Scholar]
- 34.Kotanides H, Reich N C. J Biol Chem. 1996;271:25555–25561. doi: 10.1074/jbc.271.41.25555. [DOI] [PubMed] [Google Scholar]
- 35.Seidel H M, Milocco L H, Lamb P, Darnell J E, Stein R B, Rosen J. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindler U, Wu P, Rothe M, Brasseur M, McKnight S L. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 37.Wahl L M, Katona I M, Wilder R L, Winter C C, Haroui B, Scher I, Wahl S M. Cell Immunol. 1984;85:373–383. doi: 10.1016/0008-8749(84)90251-x. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber E, Matthias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donnelly R P, Fenton M J, Kaufman J D, Gerrard T L. J Immunol. 1991;146:3431–3436. [PubMed] [Google Scholar]
- 41.Starr R, Willson T A, Viney E M, Murray L J L, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, et al. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 42.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 43.Ohya K, Kajigaya S, Yamashita Y, Miyazato A, Hatake K, Miura Y, Ikeda U, Shimada K, Ozawa K, Mano H. J Biol Chem. 1997;272:27178–27182. doi: 10.1074/jbc.272.43.27178. [DOI] [PubMed] [Google Scholar]
- 44.Mikita T, Campbell D, Wu P, Williamson K, Schindler U. Mol Cell Biol. 1996;16:5811–5820. doi: 10.1128/mcb.16.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 46.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto H, Yasukawa H, Masuhara M, Tanimura S, Sasaki A, Yuge K, Ohtsubo M, Ohtsuka A, Fujita T, Ohta T, et al. Blood. 1998;92:1668–1676. [PubMed] [Google Scholar]
- 48.Wang H Y, Zamorano J, Yoerkie J L, Paul W E, Keegan A D. J Immunol. 1997;158:1037–1040. [PubMed] [Google Scholar]
- 49.Losman J A, Chen X P, Hilton D, Rothman P. J Immunol. 1999;162:3770–3774. [PMC free article] [PubMed] [Google Scholar]
- 50.Gajewski T F, Fitch F W. J Immunol. 1988;140:4245–4252. [PubMed] [Google Scholar]
- 51.Venkataraman C, Leung S, Salvekar A, Mano H, Schindler U. J Immunol. 1999;162:4053–4061. [PubMed] [Google Scholar]
- 52.Kuperman D, Schofield B, Wills-Karp M, Grusby M J. J Exp Med. 1998;187:939–948. doi: 10.1084/jem.187.6.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akimoto T, Numata F, Tamura M, Takata Y, Higashida N, Takashi T, Takeda K, Akira S. J Exp Med. 1998;187:1537–1542. doi: 10.1084/jem.187.9.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naka T, Matsumoto T, Narazaki M, Fujimoto M, Morita Y, Ohsawa Y, Saito H, Nagasawa T, Uchiyama Y, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:15577–15582. doi: 10.1073/pnas.95.26.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]