Abstract
Immunoglobulin gene rearrangement in avian B cell precursors generates surface Ig receptors of limited diversity. It has been proposed that specificities encoded by these receptors play a critical role in B lineage development by recognizing endogenous ligands within the bursa of Fabricius. To address this issue directly we have introduced a truncated surface IgM, lacking variable region domains, into developing B precursors by retroviral gene transfer in vivo. Cells expressing this truncated receptor lack endogenous surface IgM, and the low level of endogenous Ig rearrangements that have occurred within this population of cells has not been selected for having a productive reading frame. Such cells proliferate rapidly within bursal epithelial buds of normal morphology. In addition, despite reduced levels of endogenous light chain rearrangement, those light chain rearrangements that have occurred have undergone variable region diversification by gene conversion. Therefore, although surface expression of an Ig receptor is required for bursal colonization and the induction of gene conversion, the specificity encoded by the prediversified receptor is irrelevant and, consequently, there is no obligate ligand for V(D)J-encoded determinants of prediversified avian cell surface IgM receptor.
The rearrangement of chicken Ig genes generates minimal antibody diversity. At the light chain (L) locus, the unique, functional VL1 gene rearranges to the unique JL segment in all B cells (1). Similarly, at the heavy chain (H) locus, unique VH1 and JH genes undergo rearrangement with a family of highly conserved DH elements to form a VDJH complex of limited diversity (2, 3). Junctional diversity is limited further because of the lack of N nucleotide additions in chicken V(D)J junctions (4). A diverse repertoire of B cell specificities is generated by somatic gene-conversion events in which VDH and VL sequences in functionally rearranged VDJH and VJL genes are replaced by homologous sequences from upstream VL and VH pseudogenes (ΨVL and ΨVH) (2, 5–7).
The bursa of Fabricius is the primary site of B cell lymphopoiesis in avian species, and surgical or chemical ablation of the chicken bursa profoundly disrupts the normal progression of B cell development and the generation of diversity by gene conversion (reviewed in refs. 7–10). The bursa is colonized by a single wave of B cell precursors during embryogenesis starting at about embryonic day 8 (E8) and lasting about a week (11). Within the bursa, B cell precursors first are observed in the mesenchymal tissue, and 20,000–40,000 precursors subsequently migrate across the bursal epithelial basement membrane and begin to proliferate in oligoclonal clusters or epithelial buds from which the discrete follicles of the mature bursa are derived (12, 13).
Surface Ig (sIg)+ B cell precursors first are observed by about E9–E10, and the frequency of such cells increases rapidly during the embryonic period (14). Although Ig gene rearrangement is not intrinsically biased toward productive rearrangement, only those B cells that have undergone productive rearrangement are selected within the bursa (15). Because there is no detectable clonal expansion of B cell precursors in bursal follicles before the expression of sIg (13), transit across the basement membrane and subsequent precursor expansion in epithelial buds require sIg expression and represent a critical checkpoint in B lymphopoiesis.
The colonization of epithelial buds by B cell precursors occurs at a time when those precursors exhibit minimal diversity, i.e., before the onset of gene conversion. This has led to considerable speculation that the specificity encoded by the prediversified sIg receptor plays a critical role in B cell development (2, 3, 15–19).
In this study we have used retroviral gene transfer in vivo to address directly whether an interaction between V(D)J-encoded determinants of the prediversified sIg receptor and ligand is required for the normal progression of chicken B cell development. We show that a truncated μ receptor (Tμ), lacking all V(D)J-encoded determinants, is expressed on developing bursal cells after retroviral gene transfer. Bursal cells expressing this receptor productively colonize bursal follicles despite lacking endogenous sIg receptors, demonstrating that recognition of an endogenous ligand by the prediversified sIg receptor is not required for the progression of B cell development in the avian embryo.
MATERIALS AND METHODS
Oligonucleotides.
The following oligonucleotides were synthesized by Sheldon Biolabs (McGill University, Montreal): VHL, 5′-ATGAGCCCACTCGTCTCCTCCCTCCTGCTCCTGGCCGCCCTGCCAGGGCTGATGGCGGCCGTG-3′; Cμ2, 5′-CTTGATCGGCCGTGACGACCCCTAATGGCATCCCCCTTTTCGTCACC-3′; mCμ3′, 5′-CCTTGATTTCGAAGTGGAGAAGACGTCGGGAGGTGGAGA-3′; VL5′, 5′-ACGCGTCAGGTACTCGTTGCGCCTGGTC-3′; VL3′, 5′-ACCATCAGCTGCTCCTTGCACTGGCAGG-3′; JL3′, 5′-ATCGATTCACCTAGGACGGTCAGGGTTG-3′; JCI, 5′-CTCGGG CAC ATT TTC TGG TCAA-3′; VJI, 5′-GTATGGTCTGAATTCTCTGTGCTGTT-3′; VH5′, 5′-GCTCCGTCAGCGCTCTCTGTCCTTC-3′; JH3′, 5′-GGGTTGAAGACTGTCCGGAGGAGACG-3′; VH3′, 5′-TTCAGCGCCTTGGGTTGCAACGGTG-3′; DH5′, 5′-ACGAATT-CGTGGCCGCTCAAGCAGCAACC-3′; RAG5′, 5′-TGTCACTGCAGATGGTATCAGC-3′; RAG3′, 5′-CGTGGGACAGGAATCTCTTCGGC-3′; JL, 5′-GGGGCCGGGACAACCCTGACCG-3′; RAGI, 5′-TCCCTGCTGAATTTTGATGGGCAC-3′; VLfr3, 5′-CACATTAACCATCACTGG GGT CC-3′.
Generation of RCAS-Tμ.
The IgVH leader sequence (VHL) containing a naturally occurring EagI site at the 3′ end was synthesized and cloned into the SmaI site of the CLA-12 adapter plasmid (20). Cμ sequences from the Cμ1/2 exon junction to 66 nt of 3′ untranslated sequence were PCR-amplified from a λZap II phage containing a full-length membrane μ insert by using the oligonucleotide combination Cμ2/mCμ3′ and cloned into pCR II (Invitrogen) to flank the membrane Cμ sequences (mCμ) by EagI sites derived from the Cμ2 primer at the 5′ end and the pCR II vector at the 3′ end. The mCμ insert was excised with EagI and cloned into the CLA-12:VHL plasmid at the EagI site. The resulting VHL-mCμ sequence encoding Tμ was excised with ClaI and cloned into the ClaI site of RCAS(BP)B (20) to yield the RCAS-Tμ plasmid.
RCAS-Tμ Virus Production and in Vivo Gene Transfer.
Chicken embryo fibroblasts (CEFs) were prepared from line 0 embryos (Regional Poultry Research Laboratories, East Lansing, MI), which contain no endogenous retroviral loci (21), and cultured in Iscove’s modified Dulbecco’s medium (GIBCO) supplemented as described elsewhere (22). RCAS(BP)B or RCAS-Tμ plasmids were transfected into CEFs by calcium phosphate precipitation. Because transfected CEFs produce productive, infectious virus, within 5 days essentially all fibroblasts expressed cytoplasmic viral proteins and Tμ in the case of the RCAS-Tμ transfectants as determined by flow cytometry and Western blotting. Day 3 SC line embryos (Hy-Line International, Dallas Center, IA) were inoculated with 1 × 106 RCAS- or RCAS-Tμ-transfected fibroblasts by injection with a 1-inch needle through the top of the egg. At the time of inoculation into day 3 embryos, greater than 98% of RCAS-Tμ-transfected fibroblasts expressed the Tμ protein.
Antibodies and Flow Cytometry.
11C6 (anti-chicken IgL), LT14, 21–1A4 (anti-ChB6.1), and Fu5.11G2 (anti-ChB6.2) were affinity-purified and used for flow cytometry as described previously (22–24). Hy18 (anti-chicken μ; obtained from E. Humphries) was used as tissue culture supernatant. Binding of primary antibodies was detected by using FITC, phycoerythrin, or biotin-conjugated goat anti-mouse Ig isotype-specific secondary reagents, the latter being detected with streptavidin conjugated to spectral red (Southern Biotechnology Associates). Viable cells were analyzed on either a FACScan or a FACS Vantage (Becton Dickinson) by gating on forward and side scatter. For analysis of cellular DNA content, stained viable cells were sorted by using a FACS Vantage, pelleted, and resuspended in Vindelov’s solution (25). DNA content of the resulting nuclei was analyzed on a FACScan.
PCR Amplification and Cloning of V Gene Sequences. Aliquots of 1,500 cells stained as described above were sorted by using a FACS Vantage directly into PCR tubes containing 14.5 μl of deionized H2O and 0.5 μl of 10 mg/ml proteinase K. Sorted cells were incubated at 50°C for 1 hr and then 85°C for 20 min and stored at −30°C until use. PCR primer combinations used to amplify Ig sequences were as follows: VJL, VL5′/JCI; unrearranged VL locus, VJI/JCI; VDJH, VH5′/JH3′; DJH, DH5′/JH3′; germ-line VH1, VH5′/VH3′. All amplification reactions also contained the RAG5′/RAG3′ combination of primers to amplify RAG2 sequences from genomic DNA as an internal standard. PCRs were cycled 30 times in a Hypercell Biological thermal cycler (MJ Research, Cambridge, MA) as described elsewhere (26). VJL sequences were cloned into pCR II (Invitrogen) and sequenced by the Sheldon Biolabs (McGill University).
PCR products were electrophoresed, blotted, and probed as described elsewhere (27). The following γ-32P end-labeled oligonucleotides were used: VJL and the unrearranged VL1 locus, JL; VDJH and DJH, JH3′; and VH1, VH3′. Radioactivity was determined by phosphoimagery (Molecular Dynamics) that was analyzed with imagequant software. Membranes subsequently were stripped and hybridized with the end-labeled RAGI oligonucleotide to quantitate RAG2 intensity.
CDR3 Length Determination.
One microliter of diluted VJL PCR product (1:60 for products from μ+L+ cells or 1:3 for products from μ+L− cells) was used as template for second-round PCRs by using the VL5′/JL3′ oligonucleotide combination for 24 cycles as described above. One microliter of the product was added to 5 pmol [γ-32P] end-labeled VLfr3 oligonucleotide/0.2 mM dNTPs/10 mM Tris⋅HCl, pH 9.0/5 mM KCl/0.1% Triton X-100/1.5 mM MgCl2/0.4 unit Taq DNA polymerase in 10 μl. The reaction was cycled 10 times through denaturation (94°C for 30 sec), annealing (60°C for 30 sec), and elongation (72°C for 30 sec) steps before a final elongation at 72°C for 5 min. Five-microliter aliquots of the reaction were run on sequencing gels as described elsewhere (28, 29).
Immunohistochemistry.
Neonatal bursae were frozen on dry ice, sectioned, and stained essentially as described elsewhere (24) with mouse mAbs to ChB6 (23) followed by biotinylated goat anti-mouse IgG1 (Southern Biotechnology Associates). Biotinylated antibodies were detected by using the ABC kit (Vector Laboratories), revealed with diaminobenzidine substrate to which NiCl2 was added according to the manufacturer’s instructions.
RESULTS
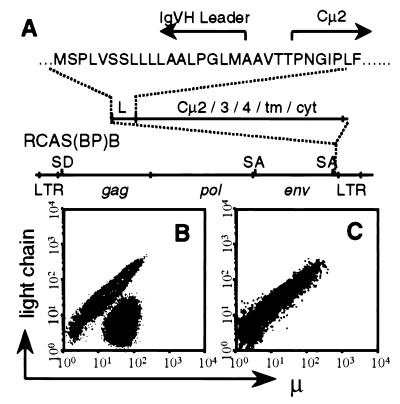
A truncated form of the chicken sIgμ (Tμ) comprising the chicken IgH leader sequence fused to the Cμ2 domain was constructed and cloned into RCAS(BP)B, a productive chicken retroviral vector that allows efficient gene transfer both in vitro and in vivo (30, 31) (Fig. 1A). Tμ-encoding transcripts initiate in the 5′ long terminal repeat (LTR) and terminate in the 3′ LTR of RCAS(BP)B with Tμ mRNA resulting from RNA splicing between the splice donor in the 5′ end of the gag gene and the splice acceptor 5′ to the env gene. The Tμ protein was stably expressed in RCAS-Tμ transfected fibroblasts in the absence of Ig light chains consistent with the elimination of the VDJH and Cμ1 domains required for light chain association (data not shown).
Figure 1.
μ+L− cells in the bursa of RCAS-Tμ-infected neonatal chicks. (A) Schematic representation of the RCAS-Tμ construct. SD and SA indicate splice donor and splice acceptor sites, respectively. (B and C) Contour plots of 10,000 bursal cells from neonatal chicks infected as day 3 embryos with RCAS-Tμ (B) or RCAS(BP)B (C) stained for μ and L expression.
Chicks expressing Tμ were generated by inoculation of SC line E3 embryos with RCAS-Tμ transfected fibroblasts. Bursal cells from neonatal normal chicks or chicks infected with an empty RCAS(BP)B virus (lacking a transgene) contain equivalent numbers of μ+ and L+ cells (Fig. 1C), reflecting the expression of endogenously encoded sIgM on bursal B cells. In contrast, age-matched RCAS-Tμ-infected chickens contained a substantial proportion of μ+L− cells (Fig. 1B). The proportion of μ+L− cells varied from 15 to 70% in 26 chicks assayed. Bursal lymphocyte cellularity, as judged by gross bursal weight and recovered lymphocyte numbers, was normal in RCAS-Tμ-infected chicks. Therefore, Tμ expression allows the maintenance of a population of cells within the neonatal bursa that fail to express normal levels of IgL.
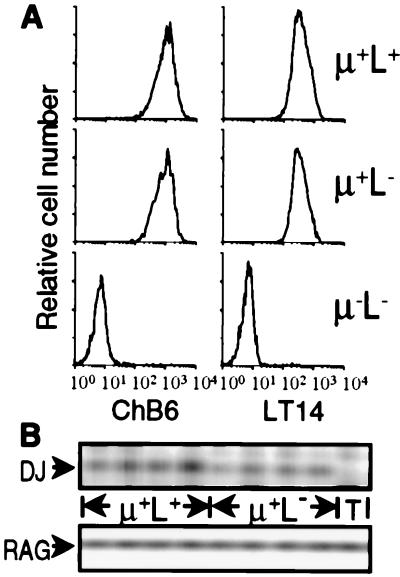
The phenotype of μ+L− bursal cells from RCAS-Tμ-infected chicks was indistinguishable from that of μ+L+ cells (expressing endogenous sIgM receptors) from the same chicks. Identical staining profiles were obtained when comparing μ+L+ and μ+L− bursal cells for the expression of LT14, a marker restricted to bursal B lineage cells (22), and the ChB6 (Bu-1) B cell surface antigen (23) (Fig. 2A). Additionally, by using a further panel of 15 mAbs specific for a variety of antigens expressed on B lineage cells, no differences were observed between the μ+L+ and μ+L− populations (data not shown).
Figure 2.
μ+L− cells in the bursa of RCAS-Tμ-infected neonatal chicks are B lineage cells. (A) Bursal cells from neonatal chicks infected as day 3 embryos with RCAS-Tμ were stained for the expression of μ, IgL, and ChB6 or LT14. Histograms of 10,000 cells gated on μ and L expression and stained for ChB6 or LT14 are shown. (B) Bursal cells from neonatal chicks infected as day 3 embryos with RCAS-Tμ were stained and FACS-sorted based on the expression of μ and L. Typical purity of the sorted populations was >98%. Genomic DNA was PCR-amplified from 1,500 sorted μ+L+ and μ+L− bursal cells from four individual RCAS-Tμ-infected chicks and from unstained thymocytes (T) for the presence of DJH rearrangements and RAG2.
In chickens, DJH rearrangements are restricted to B lineage cells (32). DJH rearrangement was detected by PCR in μ+L− bursal cells from RCAS-Tμ-infected chicks at levels comparable to those observed in the μ+L+ populations from the same chicks (Fig. 2B). Thus, the μ+L− bursal cells from RCAS-Tμ-infected chicks are committed B lineage cells.
Retroviral gene transfer of the Tμ protein was mediated by inoculating chicken embryos with viable RCAS-Tμ-infected fibroblasts. We have formally excluded the possibility that the μ+L− bursal cells detected in such chicks are derived from the transferred fibroblasts. The ChB6 (Bu-1) antigen is polymorphic (23), and the ChB6.1 and ChB6.2 alleles segregate within the SC line embryos that were inoculated with the RCAS-Tμ fibroblasts. In ChB6.1 homozygous SC line chicks, both μ+L+ and μ+L− populations expressed exclusively the ChB6.1 allele. Conversely, in ChB6.2 homozygous hatchmates infected with the same stock of RCAS-Tμ-infected fibroblasts, both μ+L+ and μ+L− populations expressed exclusively the ChB6.2 allele (data not shown).
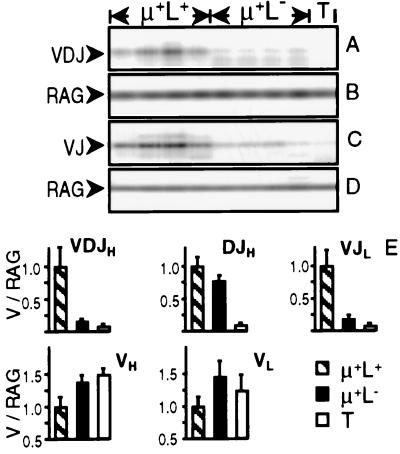
Expression of endogenous sIgM therefore was not required for the presence of B lineage bursal cells at hatch. Nonetheless, it remained possible that μ+L− cells observed neonatally were selected initially into the bursal microenvironment on the basis of productive Ig rearrangement, leading to the expression of endogenous sIgM receptors that subsequently were lost by, for example, nonproductive gene-conversion events. We, therefore, determined by PCR whether endogenous Ig gene rearrangement in μ+L− cells had progressed beyond the DJH rearrangements observed above. Very low levels of VDJH rearrangement were detected in sorted μ+L− cells from RCAS-Tμ-infected chicks as compared with μ+L+ cells expressing endogenous receptors from the same chicks (Fig. 3A). Similarly, at the light chain locus in these chicks, very low levels of VJL rearrangement were detected in sorted μ+L− cells as compared with the μ+L+ cells (Fig. 3C).
Figure 3.
V gene rearrangements in μ+L− cells from the bursa of neonatal RCAS-Tμ-infected chicks. (A–D) Genomic DNA was PCR-amplified from 1,500 FACS-sorted μ+L+ and μ+L− bursal cells from four individual RCAS-Tμ-infected neonatal chicks and from thymocytes (T) for the presence of VDJH (A) or VJL (C) rearrangements. The same samples were coamplified with RAG2 primers B and D, respectively. (E) Genomic DNA was PCR-amplified from sorted μ+L+ and μ+L− bursal cells and thymocytes from four individual RCAS-Tμ-infected chicks for VDJH, DJH, and VJL rearrangements and for germ-line VH1 and VL1 loci. RAG2 sequences were coamplified with all samples. Ig signal intensity was normalized to RAG2 signal intensity for each sample. The normalized Ig signal intensity was set arbitrarily to 1.0 for the μ+L+ bursal cell population, and the μ+L− and thymocyte populations were calculated relatively, with each bar showing the mean ± SD of four individual samples.
To estimate the proportion of μ+L− cells containing V(D)J rearrangements we performed semiquantitative PCR on genomic DNA from FACS-sorted cell populations. After PCR amplification, samples were assayed by gel filtration, Southern blotting, and hybridization. Ig-specific signal intensity was normalized to a single-copy gene (RAG2) coamplified with an appropriate and noninterfering primer combination. Relative levels of VDJH, DJH, and VJL rearrangement in μ+L+ cells expressing endogenous sIgM from RCAS-Tμ-infected chicks were indistinguishable from those of normal bursal cells. Levels of DJH rearrangement in μ+L− cells approached 75% of levels seen in normal B cells. In contrast, levels of V gene rearrangement were reduced substantially in μ+L− cells to approximately 15% at the heavy chain locus and 20% at the light chain locus (Fig. 3E).
Increased levels of the unrearranged VH1 gene were detected among μ+L− cells, as compared with the μ+L+ population, with levels close to those observed in sorted T cells (which contain both VH1 alleles in germ-line configuration). Similarly, at the light chain locus, increased levels of unrearranged VL1 genes were detected in the μ+L− populations as compared with μ+L+ cells (Fig. 3E). Thus, the low level of V gene rearrangement observed among μ+L− cells was not a consequence of the loss of rearranged Ig V genes from the genome of these cells.
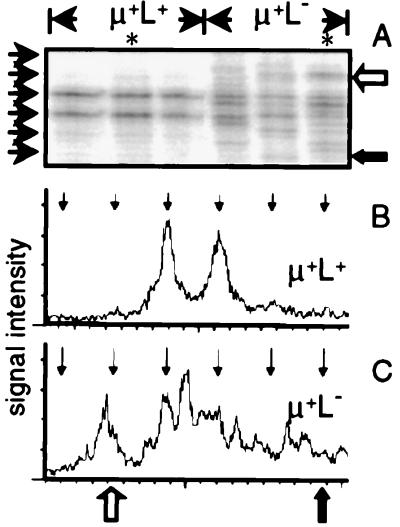
To obviate the possibility that the low level of V(D)J rearrangement seen in μ+L− cells could be implicated in the selection of these cells within the bursa, we determined whether VJL rearrangements from these cells had been selected for productivity. In-frame rearrangements can be distinguished from nonproductive, out-of-frame rearrangements by using PCR run-off assays as described previously (28, 29) and by using an oligonucleotide designed to assay all VJL CDR3 sequences independent of whether they had undergone sequence divergence by gene conversion. Selection for in-frame VJL junctions was observed clearly in the μ+L+ populations from RCAS-Tμ-infected chicks with two bands, separated in size by 3 nt, dominating the VJL junctions (Fig. 4), consistent with the rapid elimination of out-of-frame rearrangements in the chicken embryo bursa (15). In contrast, such restriction of CDR3 length was not observed in μ+L− cells from the same chicks, which showed the heterogeneity of VJL junction length expected from B cell populations before selection for endogenous sIg expression (Fig. 4).
Figure 4.
VJL rearrangements from neonatal μ+L− bursal cells have not been selected for productive reading frame. (A) VJL rearrangements in neonatal μ+L+ and μ+L− bursal cells from RCAS-Tμ-infected chicks. Short arrows along the left correspond to in-frame rearrangements (confirmed by sequencing). Arrows to the right of A correspond to the indicated positions on the scanned profiles shown in B and C. (B and C) Radioactivity from lanes marked with a ∗ in A from μ+L+ (B) and μ+L− (C) cells was scanned by phosphoimagery. Vertical arrows at the top of B and C correspond to the positions of in-frame rearrangements.
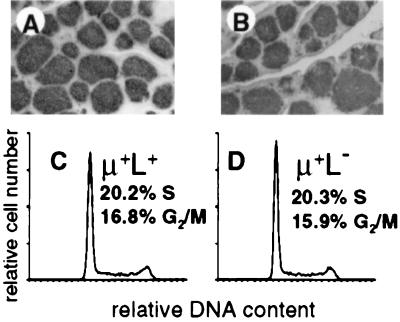
Bursal B cell development in the normal chicken embryo occurs in the microenvironment of epithelial buds. Colonization of epithelial buds in RCAS-Tμ-infected chicks was normal as detected by immunohistochemistry using anti-ChB6 (pan-B cell) antibodies (Fig. 5). In addition, both μ+L+ and μ+L− populations of bursal cells from neonatal RCAS-Tμ-infected chicks showed indistinguishable proportions of cells in S, G2, and M phases of the cell cycle, both populations showing the high rates of proliferation characteristic of neonatal bursal cells (Fig. 5).
Figure 5.
μ+L− bursal cells in the neonatal bursa of RCAS-Tμ-infected chicks are proliferating in epithelial buds. (A and B) Frozen sections of bursal tissue stained with anti-ChB6 from neonatal chicks infected as day 3 embryos with RCAS-Tμ (A) or RCAS (B). The RCAS-Tμ bursa shown contained 18.5% μ+L− cells. (C and D) Histograms from 10,000 μ+L+ (C) or μ+L− (D) cells purified by FACS cell sorting and analyzed for cellular DNA content by propidium iodide staining.
Eighty rearranged VJL sequences were cloned from neonatal μ+L− bursal cells from RCAS-Tμ-infected chicks. Among 38 sequences that had not undergone gene conversion at the VJL junction, 60% contained out-of-frame rearrangements, confirming that these rearrangements and the cells containing them had not been selected on the basis of endogenous light chain gene expression. All 80 sequences contained gene-conversion events as exemplified in Fig. 6. Clone 6.131 contains a single gene-conversion event, spanning codons 22–53, using ΨVL12 as donor sequence (5). Clone 7.26 contains three gene-conversion events; codons 21–25 derive from ΨVL4, codons 27–35 derive from ΨVL18, and codons 74–78 have several potential donors (ΨVL 2, 5, 7, 10, or 12). Both clones contain out-of-frame VJL junctions and P nucleotide additions.
Figure 6.
VJL sequences in μ+L− bursal cells from neonatal RCAS-Tμ-infected chicks have undergone gene conversion. Nucleotide changes from the germ-line VL1 sequence are indicated.
DISCUSSION
The bursa of Fabricius is critical to normal avian B lineage development. Under normal circumstances, productive colonization of the bursa by B cell precursors requires those cells to express sIgM, resulting in oligoclonal expansion within epithelial buds and the generation of repertoire diversity by gene conversion. In the bursa of RCAS-Tμ-infected chicks all B lineage cells express either endogenous sIgM (μ+L+) or the Tμ receptor (μ+L−). This is consistent with the rapid elimination of out-of-frame V(D)J junctions that occurs selectively in the embryo bursa (15) and precedes the onset of repertoire generation by gene conversion (3). Bursal cells from RCAS-Tμ-infected chicks that lack μ chains are not B lineage cells because they lack characteristic B cell markers (Fig. 2) and include stromal and/or structural cells of the bursa. This provides further strong evidence that expression of a sIg complex is required for the initiation of B cell development in the bursa at a time when the bursal sIg receptor has not diversified by gene conversion.
RCAS-based viruses infect a wide range of target cells (31). Consequently, RCAS-Tμ transfected fibroblasts express the Tμ protein and neonatal RCAS-Tμ-infected chicks contain non-B lineage cells that express Tμ. Typically, 0.5–3% of CD4/CD8+ thymocytes coexpress Tμ (data not shown). Because all Tμ+ bursal cells are B lineage (Fig. 2), expression of a sIg complex therefore is necessary but not sufficient for productive bursal colonization; cells must also be B lineage-restricted.
Selection within the bursa for cells expressing a sIg complex is not limited to initial colonization of the bursa. In the juvenile bursa, ≈95% of developing bursal cells die in situ and cells that lose the expression of sIg die by apoptosis (24). In normal chickens at this time the sIg receptor is undergoing substantial diversification by gene conversion. Therefore, the ability of the bursa to select for B lineage cells that express a sIg receptor complex is maintained independent of the specificity of the sIg receptor.
Chicken Ig gene rearrangement leads to sIg expression but results in a very limited diversity of combining sites on bursal cells before the onset of gene conversion. This has led to considerable speculation that productive bursal colonization requires an interaction between the prediversified sIg receptor and a ligand within the bursa. However, we show here that B cell precursors expressing a truncated surface μ receptor colonize the bursa despite the lack of endogenous sIg receptors. Thus, bursal colonization and the induction of gene conversion among developing B cells does not require an interaction between V(D)J-encoded determinants of sIgM and a bursal ligand.
The chicken embryo develops free of environmental antigens, and gene conversion has diversified the sIg receptor by the time of hatch. Therefore, there can be no requirement for recognition of foreign antigen by the prediversified receptor complex. Because we show here that there is no requirement for recognition of a self-antigen by the prediversified sIg receptor, the structure of the prediversified receptor cannot be subject to constraints imposed by antigen-binding requirements. Consistent with this, the unique functional chicken Ig VH1 and VL1 gene segments are highly polymorphic with 5 VH1 and 9 VL1 alleles identified among 15 partially inbred chicken flocks (27). Polymorphisms resulting in multiple amino acid changes were observed in the complementarity determining regions of both VH1 and VL1 alleles. Consequently, we propose that the only evolutionary constraints imposed on the sequence of the VH1 and VL1 genes are those of having the potential for formation of a functional, prediversified sIg receptor whose specificity is irrelevant but nonetheless can provide an efficient substrate for the introduction of gene-conversion events.
The results presented here demonstrate that a specific interaction between the prediversified receptor and endogenous ligand(s) present in the developing embryo is not required for normal bursal B cell development. Although an analysis of VJL junctions in the Muscovy duck has been interpreted in terms of such a requirement (19), the restriction in VJL junctions observed may reflect selection for IgL that can efficiently form a functional complex with IgH, thereby allowing sIg expression. This would be analogous to the VH usage in murine B cell development that can be restricted as a consequence of a required association of VDJHμ with the VpreB/λ5 components of the mammalian pre-B cell receptor (33–36).
It is important to appreciate that we have demonstrated large numbers of μ+L− cells in the bursa of chicks infected with RCAS-Tμ under circumstances in which we have not otherwise blocked the development of B cells expressing endogenously encoded sIg. Thus, bursal cells expressing the truncated μ receptor appear to be under no disadvantage with respect to bursal colonization and the induction of gene conversion when competing with B cells expressing endogenous sIgM. This is in clear contrast to murine B cells rendered anergic by exposure to soluble antigen. Under these circumstances, anergic B cells persist unless normal B cells are present as competition for limited physiological space (37). In this regard, B lineage progression has been observed in mice transgenic for truncated Ig μ receptors that lack either VDJH (38) or VDJH, Cμ1, and half of Cμ2 (39) and are unable to associate with the VpreB/λ5 components of the pre-B cell receptor. Although these results demonstrate that murine B cell development can progress in the absence of surface pre-B cell receptor expression, it remains unclear as to how efficiently this progression occurs relative to that of B cell precursors that can express a normal pre-B receptor complex.
Although expression of V(D)J-encoded determinants of the sIg receptor is not required for bursal B cell development, expression of a sIg complex is essential. The transmembrane and cytoplasmic domains of chicken sIgM show extensive homology to their mammalian counterparts, and evolutionary conservation of signal transduction pathways through the sIgM complex (40–42) predict the existence of a chicken homologue to the mammalian Igα/β heterodimer (43–45). If there is an extracellular ligand for the sIgM complex on bursal B cells, this ligand must recognize either the Tμ chain (Cμ2-Cμ4 domains) or the extracellular domains of the predicted Igα/β heterodimer. Conversely, there may be no required extracellular ligand for sIgM in the developing bursa. Under these circumstances, productive rearrangement leading to sIgM expression would bring IgM as well as the Igα/β heterodimer to the plasma membrane surface into proximity with components of the downstream-signaling cascade, inducing B cell progression. Such a model has been proposed recently to explain the required expression of the pre-T cell receptor in the developing murine thymus, because expression of a truncated pre-Tα transgene is sufficient to allow thymocytes to progress to the next stage of development (46).
Tμ expression inhibits V gene rearrangement to a greater extent than DJH rearrangement (Fig. 3). This was confirmed at the single-cell level where 9/16 v-rel-transformed clones of μ+L− cells contained a DJH rearrangement, whereas only 1/16 clones contained a VDJH rearrangement (data not shown). If Tμ expression did not inhibit DJH rearrangement, we would expect DJH levels in μ+L− cells to exceed those seen in μ+L+ cells by at least a factor of 2: VDJH rearrangement occurring in loci that already have rearranged DJH. DJH rearrangement is initiated in chicken embryos about 2 days earlier than VDJH rearrangement (32), and B cell precursors entering the bursal mesenchyme will have accumulated more DJH rearrangements than VDJH rearrangements. Consequently, our results support the conclusion that Tμ expression permits follicular entry of B cell precursors and inhibition of further endogenous gene rearrangement independent of the status of the endogenous Ig genes. These results indicate further that B cell precursor migration to the bursal mesenchyme does not require DJH rearrangement.
Rearranged VJL genes in μ+L− neonatal bursal cells from chicks infected with RCAS-Tμ receptor have undergone gene conversion (Fig. 6). Thus, gene conversion is not initiated by an interaction between an endogenous bursal ligand and the prediversified sIg receptor. Nonproductive gene-conversion events leading to nonfunctional VJL gene sequences have been observed in the chicken B lymphoma DT40 (47, 48). However, the efficiency with which gene-conversion events maintain a functional protein product at either the H or L locus in normal bursal cells in vivo is unknown. Any gene-conversion events leading to a nonfunctional V gene product would lead to the loss of bursal cell sIg expression, and such B cells and the V gene sequences within them would be lost rapidly by apoptosis (24). In contrast, the μ+L− bursal cells from RCAS-Tμ-infected chicks reported here will provide a unique opportunity to examine the scope and efficiency of gene conversion under circumstances in which the products of those gene-conversion events do not form the basis for B cell selection in the bursa.
Productive colonization of embryonic bursal follicles leading to the onset of gene conversion represents an essential checkpoint in avian B lymphopoiesis. The maturation of cells within the bursa and their emigration to the periphery represent subsequent checkpoints that, at present, are poorly understood. The bursa has evolved as an organ in which exogenous antigens transported from the gut lumen contact lymphocytes developing in the lymphoid follicles of the bursa, a process initiated after hatch (49). Exogenous antigen therefore may modify the maturation or emigration of bursal cells after hatch, as has been suggested elsewhere (50). In this regard, the Tμ receptor does not fully support the emigration of bursal cells to the periphery after hatch; rather, Tμ+ bursal cells undergo apoptotic death in situ (C.E.S. and M.J.H.R., unpublished results). This suggests that the emigration of bursal cells expressing a diversified repertoire of B cell specificities is regulated by mechanism(s) distinct from those regulating the productive colonization of the bursa by cells expressing the prediversified sIg receptor.
In conclusion, we have demonstrated productive bursal colonization leading to the induction of V gene diversity by B cells precursors expressing sIg molecules that lack V gene-encoded determinants. Therefore, neither B cell precursor development nor the induction of repertoire diversification by gene conversion in the avian embryo bursa requires an interaction between the prediversified sIgM and ligands expressed within the bursa.
Acknowledgments
We thank S. H. Hughes for the RCAS vectors, Emilia Antecka for assistance with immunohistochemistry, Ken McDonald for flow cytometric cell sorting, and Trevor Owens for critical review of the manuscript. This work was supported by the Medical Research Council of Canada (MT10040). C.E.S. was supported by a Medical Research Council of Canada Studentship. S.L.D. was supported by a studentship from Fonds pour la Formation de Chercheurs et l’Aide à la Recherche.
ABBREVIATIONS
- sIg
surface Ig
- Tμ
truncated μ
- L
light chain
- H
heavy chain
Footnotes
References
- 1.Reynaud C A, Anquez V, Dahan A, Weill J C. Cell. 1985;40:283–291. doi: 10.1016/0092-8674(85)90142-4. [DOI] [PubMed] [Google Scholar]
- 2.Reynaud C A, Dahan A, Anquez V, Weill J C. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 3.Reynaud C A, Anquez V, Weill J C. Eur J Immunol. 1991;21:2661–2670. doi: 10.1002/eji.1830211104. [DOI] [PubMed] [Google Scholar]
- 4.McCormack W T, Tjoelker L W, Carlson L M, Petryniak B, Barth C F, Humphries E H, Thompson C B. Cell. 1989;56:785–791. doi: 10.1016/0092-8674(89)90683-1. [DOI] [PubMed] [Google Scholar]
- 5.Reynaud C A, Anquez V, Grimal H, Weill J C. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 6.Thompson C B, Neiman P E. Cell. 1987;48:369–378. doi: 10.1016/0092-8674(87)90188-7. [DOI] [PubMed] [Google Scholar]
- 7.McCormack W T, Tjoelker L W, Thompson C B. Annu Rev Immunol. 1991;9:219–241. doi: 10.1146/annurev.iy.09.040191.001251. [DOI] [PubMed] [Google Scholar]
- 8.Weill J C, Reynaud C A. Science. 1987;238:1094–1098. doi: 10.1126/science.3317827. [DOI] [PubMed] [Google Scholar]
- 9.Ratcliffe M J H. CRC Crit Rev Poult Biol. 1989;2:207–234. [Google Scholar]
- 10.Ratcliffe M J H, Paramithiotis E. Semin Immunol. 1990;2:217–226. [PubMed] [Google Scholar]
- 11.Houssaint E, Belo M, le Douarin N M. Dev Biol. 1976;53:250–264. doi: 10.1016/0012-1606(76)90227-x. [DOI] [PubMed] [Google Scholar]
- 12.Pink J R L, Vainio O, Rjinbeek A M. Eur J Immunol. 1985;15:83–87. doi: 10.1002/eji.1830150116. [DOI] [PubMed] [Google Scholar]
- 13.Ratcliffe M J H, Lassila O, Pink J R L, Vainio O. Eur J Immunol. 1986;16:129–133. doi: 10.1002/eji.1830160204. [DOI] [PubMed] [Google Scholar]
- 14.Lydyard P M, Grossi C E, Cooper M D. J Exp Med. 1976;144:79–97. doi: 10.1084/jem.144.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack W T, Tjoelker L W, Barth C F, Carlson L W, Petryniak B, Humphries E H, Thompson C B. Genes Dev. 1989;3:838–847. doi: 10.1101/gad.3.6.838. [DOI] [PubMed] [Google Scholar]
- 16.Mansikka A, Sandberg M, Lassila O, Toivanen P. Proc Natl Acad Sci USA. 1990;87:9416–9420. doi: 10.1073/pnas.87.23.9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmonsen J, Dunon D, Skjødt K, Thorpe D, Vainio O, Kaufman J. Proc Natl Acad Sci USA. 1991;88:1359–1363. doi: 10.1073/pnas.88.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langman R E, Cohn M. Res Immunol. 1993;144:422–446. doi: 10.1016/0923-2494(93)80126-j. [DOI] [PubMed] [Google Scholar]
- 19.Pandey A, Tjoelker L W, Thompson C B. J Exp Med. 1993;177:329–337. doi: 10.1084/jem.177.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Astrin S M, Buss E G, Haywards W S. Nature (London) 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 22.Benatar T, Iacampo S, Tkalec L, Ratcliffe M J H. Eur J Immunol. 1991;21:2529–2536. doi: 10.1002/eji.1830211033. [DOI] [PubMed] [Google Scholar]
- 23.Veromaa T, Vainio O, Eerola E, Toivanen P. Hybridoma. 1988;7:41–48. doi: 10.1089/hyb.1988.7.41. [DOI] [PubMed] [Google Scholar]
- 24.Paramithiotis E, Jacobsen K A, Ratcliffe M J H. J Exp Med. 1995;181:105–113. doi: 10.1084/jem.181.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vindelov L L. Virchows Arch B Cell Pathol. 1977;24:227–242. [PubMed] [Google Scholar]
- 26.Benatar T, Tkalec L, Ratcliffe M J H. Proc Natl Acad Sci USA. 1992;89:7615–7619. doi: 10.1073/pnas.89.16.7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benatar T, Ratcliffe M J H. Eur J Immunol. 1993;23:2448–2453. doi: 10.1002/eji.1830231011. [DOI] [PubMed] [Google Scholar]
- 28.Pannetier C, Cochet M, Darche S, Casrouge A, Zöller M, Kourilsky P. Proc Natl Acad Sci USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desravines S, Hsu E. J Immunol Methods. 1994;162:210–225. doi: 10.1016/0022-1759(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 30.Petropoulos C J, Hughes S H. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petropoulos C J, Payne W, Salter D W, Hughes S H. J Virol. 1992;66:3391–3397. doi: 10.1128/jvi.66.6.3391-3397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynaud C A, Imhof B A, Anquex V, Weill J C. EMBO J. 1992;11:4349–4358. doi: 10.1002/j.1460-2075.1992.tb05534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karasuyama H, Kudo A, Melchers F. J Exp Med. 1990;172:969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsubata T, Reth M. J Exp Med. 1990;172:973–976. doi: 10.1084/jem.172.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimoto N, Kubagawa H, Ohno T, Gartland G L, Stankovic A K, Cooper M D. Proc Natl Acad Sci USA. 1991;88:6284–6288. doi: 10.1073/pnas.88.14.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ten Boekel E, Melchers F, Rolink A G. Immunity. 1998;8:199–207. doi: 10.1016/s1074-7613(00)80472-0. [DOI] [PubMed] [Google Scholar]
- 37.Cyster J G, Hartley S B, Goodnow C C. Nature (London) 1994;371:389–395. doi: 10.1038/371389a0. [DOI] [PubMed] [Google Scholar]
- 38.Corcos D, Dunda O, Butor C, Cesbron J Y, Lorès P, Bucchini D, Jami J. Curr Biol. 1995;5:1140–1148. doi: 10.1016/s0960-9822(95)00230-2. [DOI] [PubMed] [Google Scholar]
- 39.Shaffer A L, Schlissel M S. J Immunol. 1997;159:1265–1275. [PubMed] [Google Scholar]
- 40.Ratcliffe M J H, Tkalec L. Eur J Immunol. 1990;20:1073–1078. doi: 10.1002/eji.1830200519. [DOI] [PubMed] [Google Scholar]
- 41.Jiang A, Craxton A, Kurosaki T, Clark E A. J Exp Med. 1998;188:1297–1306. doi: 10.1084/jem.188.7.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto A, Okada H, Jiang A, Kurosaki M, Greenberg S, Clark E A, Kurosaki T. J Exp Med. 1998;188:1287–1295. doi: 10.1084/jem.188.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams G T, Peaker C J, Patel K J, Neuberger M S. Proc Natl Acad Sci USA. 1994;91:474–478. doi: 10.1073/pnas.91.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grupp S A, Mitchell R N, Schreiber K L, McKean D J, Abbas A K. J Exp Med. 1995;181:161–168. doi: 10.1084/jem.181.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pleiman C M, Chien N C, Cambier J C. J Immunol. 1994;152:2837–2844. [PubMed] [Google Scholar]
- 46.Irving B A, Alt F W, Killeen N. Science. 1998;280:905–908. doi: 10.1126/science.280.5365.905. [DOI] [PubMed] [Google Scholar]
- 47.Buerstedde J M, Reynaud C A, Humphries E H, Olson W, Ewert D L, Weill J C. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Humphries E H, Tjoelker L, Carlson L, Thompson C B. Mol Cell Biol. 1990;10:3224–3231. doi: 10.1128/mcb.10.6.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorvari T, Sorvari R, Ruotsalainen P, Toivanen A, Toivanen P. Nature (London) 1975;253:217–219. doi: 10.1038/253217a0. [DOI] [PubMed] [Google Scholar]
- 50.Paramithiotis E, Ratcliffe M J H. Eur J Immunol. 1993;23:96–102. doi: 10.1002/eji.1830230116. [DOI] [PubMed] [Google Scholar]