Abstract
Hepatitis B viruses specifically target the liver, where they efficiently infect quiescent hepatocytes. Here we show that human and avian hepatitis B viruses can be converted into vectors for liver-directed gene transfer. These vectors allow hepatocyte-specific expression of a green fluorescent protein in vitro and in vivo. Moreover, when used to transduce a type I interferon gene, expression of interferon efficiently suppresses wild-type virus replication in the duck model of hepatitis B virus infection. These data suggest local cytokine production after hepatitis-B-virus-mediated gene transfer as a promising concept for the treatment of acquired liver diseases, including chronic hepatitis B.
Persistent viral infections can be viewed as acquired genetic diseases and therefore as a major challenge for the application of gene therapy. Generally, treatment will require transient rather than permanent effector gene expression, such as stimulation of the immune system against the infectious agent (1). Appropriate gene-delivery systems should specifically target the infected tissue or cell type, should allow for control of duration and strength of effector gene expression, and, ideally, should be administered by injection into the bloodstream (1, 2). To treat infectious diseases of the liver, none of the currently available vector systems meets all of these criteria (3).
Hepatitis B viruses, or hepadnaviruses, are small enveloped DNA viruses with distinct features that make them attractive candidates as vectors for gene therapy of acquired liver diseases. They selectively target the liver after inoculation into the bloodstream, and they efficiently infect quiescent hepatocytes. Viral gene expression is directed by hepatocyte-specific promoter-enhancer elements (4, 5), and, in contrast to retroviruses, genome replication via reverse transcription does not require integration of the hepadnaviral DNA into the host genome (6); rather it establishes a stable episomal transcription template. The feasibility, however, of a hepadnaviral vector system has not yet been demonstrated.
Chronic viral hepatitis affects approximately 800 million people and is the principal cause in the world of chronic liver disease, liver cirrhosis, and hepatocellular carcinoma (7). Currently, the only therapy for chronic hepatitis that has a lasting beneficial effect is systemic treatment with interferon (IFN)-α; a sustained response is achieved in only one-third of patients with chronic hepatitis B and in only one-fifth of patients with chronic hepatitis C (7). Nucleoside analogues provide a therapeutic alternative leading to a rapid decrease in serum HBV DNA levels and to histologic improvement of liver disease (8). However, short-term treatment leads to a rapid relapse of disease and long-term treatment often results in the selection of resistant viral variants (7); these outcomes emphasize the need for novel therapeutic approaches (7, 9). New concepts include gene therapy and the use of defective or attenuated viruses to block wild-type viral infection (3). Immunomodulatory cytokines such as IL-12, IFN-γ or tumor necrosis factor-α potently suppress hepatitis B virus (HBV) replication in an HBV transgenic mouse model (10, 11), whereas IL-12 and the Th1 cytokines IFN-γ and IL-2 seem to play an important role for viral clearance in chronically infected patients (12). However, systemic application of cytokines is limited by severe side effects (7, 9). Local production of cytokines after liver-directed gene transfer should provide a more efficient and better-tolerated alternative, and hepatitis B virus-based vectors might be particularly suitable for this approach.
Here we show that in vitro infection of primary human hepatocytes with recombinant HBV carrying a gene coding for a green fluorescent protein (GFP) (13) leads to clearly detectable GFP expression. Because the availability and infectibility of these cells is limited (14, 15), and no feasible in vivo infection system exists for the human virus, we took advantage of the duck hepatitis B virus (DHBV) model, which readily allows for infection studies with primary hepatocytes and whole animals (16). We demonstrate in vivo infection of hepatocytes after injection of recombinant DHBV-GFP into a peripheral vein, and we prove that cells preinfected with the wild-type virus can be superinfected with the recombinant virus. As a first step toward therapeutically useful hepadnavirus vectors, we constructed a recombinant DHBV carrying the duck homologue of IFN-α, which efficiently suppressed wild-type virus replication.
MATERIALS AND METHODS
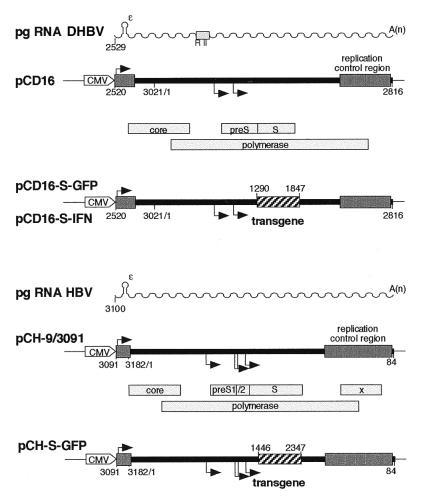
Plasmid Constructs. HBV/DHBV constructs contain, under control of the cytomegalovirus immediate early promoter-enhancer, a terminally redundant genome of HBV, subtype ayw 1 (pCH-9/3091, HBV nucleotides 3091 to 3182-1 to 3182/1 to 84, numbering from the core initiation codon) (17), or a terminally redundant genome of DHBV, subtype 16 (18) (pCD16, DHBV nucleotides 2520 to 3021/1 to 2816) (19, 20), respectively (Fig. 1). The respective helper constructs pCH3142 for HBV (21) and pCD4 for DHBV (20) are nearly identical except that they lack part of the 5′-proximal RNA packaging signal ɛ, which makes them encapsidation-deficient (Fig. 1).
Figure 1.
Plasmid constructs used for the production of recombinant hepadnaviruses. The parental plasmids pCH-9/3091 (HBV) and pCD16 (DHBV) are based on terminally redundant hepadnavirus genomes (thick black lines) functionally mimicking the circular DNA genomes formed by reverse transcription of the RNA pregenomes [sinuous lines with A(n) representing the poly(A) tails]. Numbers refer to nucleotide positions. The replication control regions (heavy black lines), encompassing HBV nucleotides 2360 to 40 and DHBV nucleotides 2100 to 2800, include cis signals for pregenomic RNA synthesis and maturation, and for RNA encapsidation and reverse transcription. These are continuous on the authentic circular viral genomes and partially duplicated here to create the terminal elements required for replication of the linearized genomes. Transcription start sites are indicated by the attached arrows, authentic viral genes by the open bars with the gene designations inside. The positions of the transgenes in the recombinant plasmids are shown by the hatched boxes. Synthesis of the pregenomic RNAs is driven by a cytomegalovirus-IE enhancer/promoter element (marked CMV), whereas subgenomic RNAs, which encode the preS/S and the S envelope proteins and, for HBV, the X protein, are produced from internal promoters. In the RNA pregenomes, ɛ denotes a 5′-proximal stem-loop that, in the case of DHBV, acts together with a second region (box marked R II) as an encapsidation signal. The 5′-terminal part of ɛ (HBV up to nucleotide 3142, DHBV up to nucleotide 2579) is deleted in the helper constructs used to provide missing gene products in trans. In pCH-S-GFP, a fragment encompassing the S gene was replaced by a DNA fragment encoding GFP fused to the first three amino acids of S. In pCD16-S-GFP and pCD16-S-IFN, DNA fragments encoding GFP and duck IFN, respectively, replace the KpnI to BstEII fragment encompassing the DHBV S gene.
Marker constructs pCH-S-GFP and pCD-S-GFP were obtained by replacing DNA fragments containing the small envelope (S) gene (in pCH-S-GFP from XhoI, position 1409, to NsiI, position 2347; in pCD-S-GFP from KpnI, position 1290, to BstEII, position 1847) with a PCR fragment (733 nt) encoding a fluorescence-enhanced, red-shifted GFP prepared from plasmid pTR-UF5 (13) (kindly provided by N. Muzyczka) with expression expected to be driven by the S promoter (see Fig. 1). pCD-S-IFN was obtained analogously by inserting a PCR-derived fragment (591 nt) encoding the complete duck type I IFN gene (22) (kindly provided by P. Staeheli and U. Schultz). For production of recombinant duck IFN protein, the IFN gene was cloned into a pUC-based cytomegalovirus-IE promoter-controlled expression vector (pCDuIFN).
Production of Recombinant (r-) Virus Stocks.
For the production of rHBV and rDHBV, human hepatoma HuH7 cells (23) and chicken hepatoma LMH cells (24), respectively, were cotransfected at 30–40% confluence by using the calcium-phosphate method with 50 μg of the respective expression construct and 25 μg of the helper construct per each 15-cm dish. Cell culture medium containing recombinant virions was collected from days 3 to 6 posttransfection; it was then concentrated 10- to 50-fold by precipitation with 6.5% polyethylene glycol 20,000/0.9% NaCl at 0°C and stored in PBS/10% glycerol at −20°C until further use. Wild-type HBV was produced by transfecting 25 μg of plasmid pCH-9/3091 accordingly. Virus titers, measured as DNA-containing enveloped viral particles (vp), were determined by density-gradient centrifugation and dot-blot analysis relative to an HBV- or DHBV-DNA standard (19).
Isolation of Primary Hepatocytes.
Primary hepatocytes were isolated by a standard two-step collagenase perfusion and subsequent differential centrifugation (50 × g). Surgical human liver biopsies were obtained (after informed consent of the donor) and, after sealing of smaller vessels, perfused via a large branch of the portal vein. Two- to three-week-old Peking ducks and 16- to 20-week-old CH57BL/6 mice were perfused via the portal vein. Primary human and mouse hepatocytes (2.5 × 105 cells per cm2 and 4 × 105 cells per cm2, respectively) were seeded onto collagen type I (Sigma Aldrich) coated tissue culture plates in a maintenance medium (25) with 10% FCS and maintained with 5% FCS. Primary duck hepatocytes (PDHs) (2.5 × 105 cells per cm2) were seeded and maintained without FCS on untreated cell culture dishes (25). DHBV-infected duck hepatocytes were obtained from ducks experimentally infected the first day after hatching with 100 μl of duck serum containing 109 DHBV16 virions, resulting in infection of virtually all hepatocytes (26). Sinusoidal endothelial and Kupffer cells in the hepatocyte cultures were identified by their receptor-mediated uptake of fluorescence-labeled acetylated low-density lipoprotein [Di-I-AcLDL, Paesel and Lorei, Duisburg, Germany (27)] and by phagocytosis (28) of Texas red-labeled Saccharomyces cerevisiae (Molecular Probes), respectively.
Infection of Primary Hepatocytes and Gene Transfer by Recombinant Viruses.
Primary human hepatocytes were incubated with either rHBV-S-GFP or wild-type HBV, diluted in maintenance medium at a multiplicity of infection (moi) of 10 to 500 vp per cell for 24 hr on day 1 after plating. PDHs were incubated accordingly with either rDHBV or wild-type DHBV from a DHBV16-positive duck serum at the desired moi on day 2 after plating. GFP expression was monitored by fluorescence microscopy with a standard FITC filter set with excitation by blue light (488 nm). For in vivo infections, 1-day-old ducklings were inoculated via a foot vein with 109 rDHBV-GFP virions. On day 7 postinfection (p.i.), animals were anesthetized and perfused via the portal vein with cold 4% paraformaledhyde/0.25% glutaraldehyde. Livers were removed, postfixed for 24 hr in perfusion buffer, saturated with 30% sucrose, and sectioned serially (10–15 μm) on a freezing microtome. Primary hepatocytes were also isolated and analyzed as described above.
Coinfection of DHBV-Positive PDHs with Recombinant DHBV-IFN.
DHBV-negative PDHs were simultaneously infected with serum-derived DHBV (moi of 25) and either rDHBV-IFN (moi of 50) or rDHBV-GFP (moi of 50). DHBV-positive PDHs were infected according to the same procedure. Cell lysates were analyzed for intracellular DHBV proteins by Western blot analysis (see below), and release of progeny DHBV virus into the cell culture medium was quantitatively determined by DHBV-DNA dot-blot analysis. As a positive control, DHBV-infected PDHs were incubated with a diluted preparation of recombinant duck IFN-α protein at a dose sufficient (as proven in previous experiments) to maximally inhibit DHBV replication. Recombinant duck IFN was obtained as cell culture supernatant of LMH cells transfected with plasmid pCDuIFN. IFN protein was added on day 3 p.i., at which time transgene expression from rDHBV-GFP was first detectable.
Immunofluorescence Staining and Western Blot Analysis.
For immunodetection with an appropriate fluorescence-labeled secondary antibody of intracellular viral antigens, we used (i) polyclonal rabbit antisera against the HBV (29) or (ii) polyclonal rabbit antisera against the DHBV (30) core protein, or (iii) monoclonal antibody 7C.12 (31) recognizing the DHBV S protein (kindly provided by J. C. Pugh). For direct detection of intracellular viral proteins, 106 PDHs were lysed by the addition of 250 μl of protein sample buffer (25) after removal of the cell culture medium. In addition, cytoplasmic lysates from 107 cells were incubated with rabbit antiserum against DHBV preS protein (32) or they were incubated with rabbit antiserum against GFP (CLONTECH); immunoprecipitated proteins were released by boiling the beads in 50 μl of sample buffer (32). From each solution, 25 μl was separated by 10% SDS-PAGE, blotted to a positively charged nylon membrane, immunostained with polyclonal antiserum against DHBV core-S or preS protein (30, 32), or against GFP, and visualized with the ECL system (Amersham), essentially as described (25).
RESULTS
Production of Recombinant Hepadnaviruses. As a basis for constructing recombinant hepatitis B virus genomes carrying the GFP or the IFN gene, we used plasmids pCH-9/3091 (HBV) and pCD16 (DHBV), which, on transfection, give rise to the production of infectious HBV or DHBV particles (19, 23) (Fig. 1). Because of experience with initial studies of recombinant HBV (33), care was taken not to exceed the authentic genome size and not to affect cis-acting control elements, such as the replication control region that directs synthesis, packaging, and reverse transcription of the RNA pregenome (4, 6, 34), the internal promoters or enhancers, and the several less well-defined control elements primarily involved in RNA maturation (19, 35–37). These control elements comprise approximately one-third of the viral genome (Fig. 1). Despite these precautions, among the several constructs in which different genome segments were replaced, only substitution of the small envelope (S) gene by foreign sequences turned out to be successful (U.P., M.N., and H.S., unpublished work).
Plasmids pCH-S-GFP and pCD-S-GFP elicited strong GFP fluorescence 36 to 48 hr after transfection into appropriate hepatoma cells, thus demonstrating functional insertion of the foreign gene (data not shown). Because S gene replacement destroys the surface protein and polymerase ORFs, it was necessary for the generation of recombinant virus that the corresponding gene products be trans-complemented by cotransfection (24, 38) with the respective encapsidation-deficient helper construct (38, 39). The result was the production of enveloped recombinant HBV (rHBV) at titers between 108 and 109 vp/ml and recombinant DHBV (rDHBV) at titers between 3 × 107 and 2.5 × 108 vp/ml in different experiments, comparable to the production of wild-type HBV and DHBV obtained by transfection (19). Virus could be concentrated up to 50-fold without loss of infectivity by precipitation with polyethylene glycol.
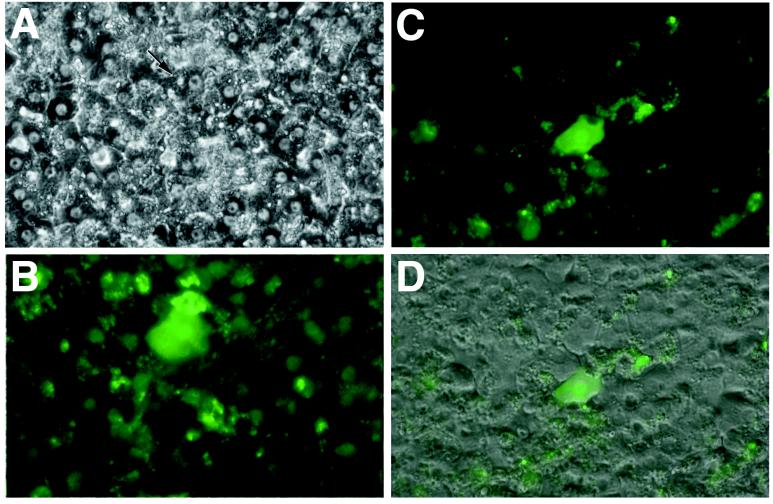
Hepatocyte Infection by Recombinant Hepadnaviruses. Infectivity of recombinant virus particles was demonstrated by incubating primary human hepatocytes with equal amounts of rHBV-GFP or of wild-type HBV. By using an moi of 100 vp per cell, 1/102 hepatocytes was found, by specific immunofluorescence staining for HBV core protein (data not shown), to be infected with virus at day 6 p.i. We therefore assume that infectivity of the recombinant virus is comparable to that of wild-type virus. However, because of the high autofluorescence background in human hepatocyte cultures, weakly green fluorescent cells could not be unequivocally identified, a problem much less significant in cultured duck hepatocytes (see below). Therefore, GFP fluorescence was clearly detectable in only 1/104 hepatocytes upon reaching its maximum at day 12 p.i. (Fig. 2). Because of the technical limitation in GFP detection, the core assay is probably a more reliable measure of infection efficiency.
Figure 2.
Transduction of primary human hepatocytes by recombinant HBV. Primary human hepatocytes were infected with rHBV-GFP, a recombinant HBV that carries a GFP gene under control of the HBV S promoter (Fig. 1). Phase contrast of hepatocyte cultures (A) and GFP expression (B) in a transduced cell (marked by the arrow) is shown at ×200 magnification. Transduction of another cell is demonstrated by an overlay of the fluorescence (C) with the phase contrast of the same field (D) (44).
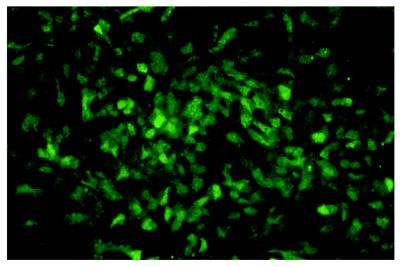
Because of the known variability in infecting isolated human hepatocytes (14, 15), we decided to use the duck model for more quantitative analyses and for in vivo experiments. To establish the dose dependence of transduction, cultured PDHs were incubated with rDHBV-GFP at moi ranging from 2 to 250 vp per cell. Three days p.i., a faint green fluorescence became detectable. This signal increased markedly until day 5, reaching a maximum at day 8 p.i. The proportion of fluorescent cells was clearly dose-dependent with an moi of 200 vp per cell resulting in ≥90% of GFP-positive cells (Fig. 3). Stable GFP expression was observed for up to 3 weeks, with some variation in the intensity of green fluorescence between hepatocytes. In addition, synthesis of DHBV core protein and GFP after infection of duck hepatocytes with rDHBV was confirmed by Western blot analysis of cellular lysates. GFP-specific antibodies revealed two closely spaced bands of approximately 30 kDa, probably representing GFP and a DHBV-S/GFP fusion. An additional RNA, initiating from the preS promoter, might serve for the expression of a preS/GFP fusion protein (Fig. 1). However, no larger products corresponding to such a fusion protein were detected (data not shown).
Figure 3.
Transduction of PDHs by recombinant DHBV. PDHs were infected with rDHBV-GFP, a recombinant DHBV in which the S gene was replaced by a GFP gene (see Fig. 1). GFP expression demonstrates transduction of 80 to 90% of the hepatocytes (resulting from infection for 24 hr at an moi of 100 vp per cell) at day 6 p.i. (×100).
To demonstrate liver-directed gene transfer by hepadnaviral vectors in vivo, we injected six ducklings intravenously with 109 rDHBV-GFP particles each. At day 3, 7, and 14 p.i., liver-tissue sections and isolated hepatocytes from these animals were analyzed by immunofluorescence microscopy. At day 7 and day 14, 20 to 25 per 106 hepatocytes were clearly GFP positive, indicating successful in vivo gene transfer. In control animals infected with wild-type DHBV, no GFP-positive cells were detected (data not shown).
Hepadnaviral Gene Transfer Is Hepatocyte-Specific.
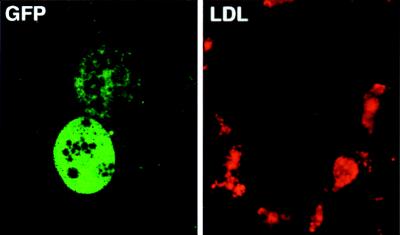
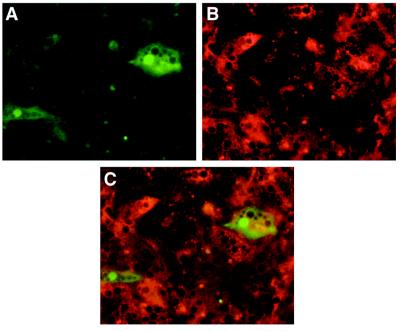
To test whether hepadnaviral vectors selectively target hepatocytes, we analyzed cell-type specificity in vitro. Primary liver cell cultures prepared by collagenase perfusion and differential sedimentation are known to contain 3–20% nonparenchymal liver cells, mainly sinusoidal endothelial cells, which are identified by receptor-mediated uptake of red-fluorescent acetylated low-density lipoprotein and Kupffer cells which, in turn, are identified by their ability to phagocytose particles >2 μm labeled with Texas-red (40). As demonstrated in Fig. 4 for endothelial cells by confocal microscopy, neither Kupffer cells nor sinusoidal endothelial cells, which accounted for approximately 10% of the total cell population in our cultures, expressed GFP. This also held true for infection with rDHBV-GFP at moi levels that resulted in GFP transduction into nearly all hepatocytes. Incubation of human hepatocytes with rDHBV-GFP, or of duck hepatocytes with rHBV-GFP, or of mouse hepatocytes with either of the recombinant hepadnaviruses, did not result in GFP expression. These data indicate that transgene expression from recombinant hepadnaviruses is both hepatocyte- and species-specific.
Figure 4.
Hepatocyte-specific transduction by recombinant hepadnaviruses. (Left) Infection of PDH cultures with rDHBV-GFP at an moi of 100 vp per cell for 24 hr. Transduced hepatocytes were detected by GFP expression at day 6 p.i. (Right) Sinusoidal endothelial cells surrounding the same hepatocytes were identified by receptor mediated uptake of red-fluorescent acetylated low density lipoprotein (LDL). Hepatocyte specificity is shown by the absence of GFP expression in the sinusoidal endothelial cells. (Confocal microscopy, ×63 lens.)
IFN Gene Transfer Blocks the Establishment of Wild-Type Virus Infection.
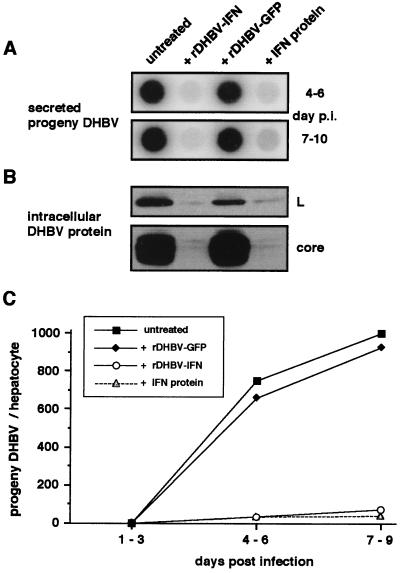
The duck homologue of IFN-α has recently been cloned, and the recombinant IFN was shown to inhibit DHBV replication in cultured duck hepatocytes (22). We therefore asked whether this secretory protein, expressed from a corresponding hepadnaviral vector, would similarly interfere with viral replication. As shown in Fig. 5, release of progeny DHBV from PDHs infected with replication-competent wild-type DHBV was reduced approximately 20-fold (14- to 24-fold in different experiments) upon co-infection with rDHBV-IFN. This inhibition was equivalent to the maximal inhibition obtained by treatment with recombinant IFN (16- to 26-fold) added at day 3 p.i., at which time expression of IFN from rDHBV-IFN was expected to start. Likewise, intracellular levels of DHBV core and L protein were strongly suppressed (Fig. 5A). In contrast, progeny virus release was not affected by co-infection with rDHBV-GFP (Fig. 5B). These data demonstrate that IFN expression mediated by hepadnaviral gene-transfer specifically interferes with the establishment of a hepadnaviral infection.
Figure 5.
Recombinant DHBV transferring a IFN gene interferes with the establishment of DHBV infection. PDHs were infected with wild-type DHBV, or they were coinfected with DHBV-IFN or rDHBV-GFP as a negative control. Success of infection was monitored for release of progeny DHBV into cell culture medium by DNA dot-blot (A) and by detection of structural DHBV proteins in cell lysates by Western blot (B) in a representative experiment (C). Quantitative evaluation of DHBV-DNA by dot-blot analysis. Coinfection with rDHBV-IFN interfered with the establishment of a productive DHBV infection as effectively as did IFN protein added at a dose causing maximal inhibition.
Recombinant Hepadnaviruses Superinfect Wild-Type Virus-Infected Hepatocytes.
To test whether hepadnavirus vectors are able to transduce hepatocytes infected with the homologous wild-type virus, we used cultured hepatocytes that were fully DHBV-infected, as demonstrated by immunofluorescence staining for DHBV S protein (Fig. 6). Incubation with rDHBV-GFP at moi levels ranging from 25 to 100 vp per cell resulted in 1–4% GFP-positive hepatocytes. Unequivocal evidence for superinfection of hepatocytes with resident wild-type virus was obtained by the detection of GFP and S protein in the same cell, because S protein is not encoded by the recombinant rDHBV-GFP particles (Figs. 1 and 6). However, efficiency of GFP transduction was approximately 20-fold lower than that observed with uninfected hepatocyte cultures (Fig. 3), indicating interference with the homologous wild-type virus, probably because of receptor down-regulation (K. Breiner, S. Urban, H.S., unpublished work).
Figure 6.
Recombinant DHBV superinfects wild-type DHBV-infected hepatocytes. Productively DHBV-infected hepatocytes (see Materials and Methods) were incubated with rDHBV-GFP (moi of 50) overnight. After 6 days, cells were investigated for GFP fluorescence (A) and stained for DHBV S protein by using a red-fluorescent TRITC-labeled secondary antibody (B). As confirmed by the overlay (C), GFP-expressing cells also stained positive for DHBV S protein. Because the S protein is expressed only from DHBV wild-type and not from rDHBV-GFP (see Fig. 1), co-expression of GFP and S proves double infection with both viruses.
IFN Gene Transfer Suppresses an Established Hepadnaviral Infection.
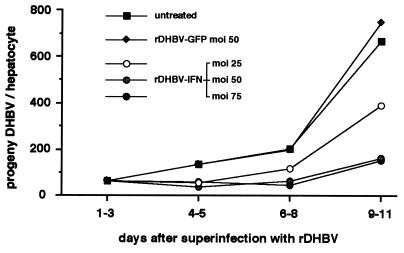
To test whether hepadnaviral cytokine gene transfer was principally suited for gene therapy of chronic hepatitis B disease, we superinfected DHBV-positive hepatocytes with rDHBV-IFN and monitored the release of progeny DHBV as described above. DHBV production was decreased (see Fig. 7) relative to untreated controls, in a dose-dependent fashion, between 1.7-fold (moi of 25 vp per cell) and 4.6-fold (moi of 75 vp per cell), comparable to the effect observed in treatment with the IFN protein at a dose showing maximal effect (4.1-fold reduction). In contrast, no change in DHBV progeny production was seen on superinfection with rDHBV-GFP, indicating that inhibition was caused by the transduced IFN gene.
Figure 7.
Therapeutic gene transfer by rDHBV. DHBV preinfected hepatocytes were superinfected at various moi levels with rDHBV-IFN or with rDHBV-GFP as a negative control. The time course of progeny DHBV release is shown (see Fig. 5).
DISCUSSION
The data presented show the principal practicability of a hepadnavirus-based vector system, and they illustrate several of its distinct advantages. We demonstrate (i) that it is possible to generate high titers of rHBV particles carrying a functional transgene of at least 800 bp; (ii) that these particles infect their target cells with the same high hepatocyte-specificity as the parental virus and lead to effective expression of the foreign gene; and (iii) that a virus-transduced IFN gene blocks establishment of hepadnavirus infection and also considerably reduces virus production from preinfected hepatocytes.
The data presented here go much beyond earlier studies reporting the generation of defective DHBV particles carrying deletions or small inserts of noncoding foreign DNA (41). In more recent attempts to produce HBV recombinants, even insertion of the very small HIV-1 tat gene (276 bp) reduced the yield of mature enveloped virus particles by >95% (42); larger inserts were not tolerated at all unless compensatory deletions were introduced (33). Importantly, none of those studies demonstrated functional expression of a foreign gene upon transduction of hepatic cells by the recombinant hepadnavirus.
These previous data, as well as our own failure to generate the corresponding core gene recombinants, indicate that replacement of the small envelope gene is particularly suited to allow for the production of high-titer hepadnavirus vectors carrying functional transgenes. With titers of at least 108 enveloped virus particles per ml of cell culture supernatant and the possibility of further concentrating virus stocks without loss of infectivity, recombinant hepadnaviruses compare favorably with other vector systems such as retro- or parvoviruses (1, 2). An obvious disadvantage is the size constraint of HBV-based vectors. The deletion of additional coding sequences should allow for inserts larger than 1 kb. But even without such further improvements, many potentially useful effector genes coding for specific antisense-RNAs, for most immunomodulatory cytokines, or for dominant-negative protein variants such as variant viral capsid proteins (43), fit into hepadnaviral vectors.
The data we obtained with cultured hepatocytes suggest that the infectivity of recombinant virus particles is similar to that of the respective wild-type viruses. The low absolute number of transduced primary human hepatocytes observed was mainly because of the notoriously low permissivity for HBV of these cells in culture (14, 15). Notably, our data provide direct evidence that gene transfer into cultured hepatocytes by an HBV vector is possible. Modified cell culture conditions improving infection with wild-type HBV will probably also improve the transduction rates attainable with recombinant HBV.
For gene therapy of infectious liver diseases, HBV-based vectors must be able to superinfect a liver with an established infection. Interference would particularly be expected if the vector and the resident pathogen was an HBV. It is therefore important that a significant fraction of wild-type DHBV-infected hepatocytes were superinfected with the recombinant virus, although the infection rate was about 20-fold reduced in comparison with that of uninfected cells. Successful superinfection was corroborated by the suppression of wild-type virus replication by a hepadnaviral IFN vector in preinfected cells.
In vivo, recombinant virus particles successfully transduced hepatocytes after injection into a peripheral vein. Although the apparently low transduction rate might limit the applicability of hepadnaviral vectors for somatic gene therapy, there are several arguments for a cytokine gene transfer into the liver for the treatment of infectious liver diseases in general and chronic HBV infection in particular. First, optimized conditions for production of recombinant hepadnavirus should allow inoculation with a dose well above that presently used—approximately one virus particle per hepatocyte—and varying the route of application might further enhance transduction efficiency of hepatocytes. Second, the secretory nature of IFN and other cytokines obviates the need to transduce every infected cell. Third, coinfection of recombinant and wild-type virus in HBV-infected individuals might result in a limited replication of the recombinant virus with the wild-type acting as a helper, providing the lacking gene products in trans. Thus we are confident that local cytokine production in the liver provides a feasible concept for the treatment of infectious diseases of the liver, including chronic hepatitis B.
Acknowledgments
The authors thank Andrea Frank for expert technical assistance, Ernst Klar and Walter Hofmann for their co-operation in obtaining human liver tissue, Bärbel Glass and Heiko Vogel for preparation of primary hepatocytes, and Karin Coutinho for editorial assistance. This work was supported by the Bundesministerium für Bildung, Wissenschaft, Forschung, und Technologie (01KV9516/3) and by the Fonds der Chemischen Industrie.
ABBREVIATIONS
- DHBV
duck hepatitis B virus
- GFP
green fluorescent protein
- HBV
hepatitis B virus
- IFN
interferon
- moi
multiplicity of infection
- PDH
primary duck hepatocytes
- p.i.
postinfection
- r
recombinant
- vp
DNA-containing enveloped viral particles
References
- 1.Anderson W F. Nature (London) 25–30. 1998. [DOI] [PubMed] [Google Scholar]
- 2.Kay M A, Liu D, Hoogerbrugge P M. Proc Natl Acad Sci USA. 1997;94:12744–12746. doi: 10.1073/pnas.94.24.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Weizsäcker F, Wieland S, Köck J, Offensperger W B, Offensperger S, Moradpour D, Blum H E. Hepatology. 1997;26:251–255. doi: 10.1002/hep.510260237. [DOI] [PubMed] [Google Scholar]
- 4.Ganem D. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott-Raven; 1996. pp. 2703–2737. [Google Scholar]
- 5.Nassal M, Schaller H. Trends Microbiol. 1993;1:221–228. doi: 10.1016/0966-842x(93)90136-f. [DOI] [PubMed] [Google Scholar]
- 6.Nassal M, Schaller H. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 7.Hoofnagle J H, di Bisceglie A. N Engl J Med. 1997;336:347–536. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 8.Lai C L, Chien R N, Leung N W, Chang T T, Guan R, Tai D I, Ng K Y, Wu P C, Dent J C, Barber J, et al. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 9.Main J, McCarron B, Thomas H C. Antiviral Chem Chemother. 1998;9:449–460. doi: 10.1177/095632029800900601. [DOI] [PubMed] [Google Scholar]
- 10.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 11.Cavanaugh V J, Guidotti L G, Chisari F V. J Virol. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossol S, Marinos G, Carucci P, Singer M V, Williams R, Naoumov N V. J Clin Invest. 1997;99:3025–3033. doi: 10.1172/JCI119498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galle P R, Hagelstein J, Kommerell B, Volkmann M, Schranz P, Zentgraf H. Gastroenterology. 1994;106:664–673. doi: 10.1016/0016-5085(94)90700-5. [DOI] [PubMed] [Google Scholar]
- 15.Gripon P, Diot C, Theze N, Fourel I, Loreal O, Brechot C, Guguen-Guillouzo C. J Virol. 1988;62:4136–4143. doi: 10.1128/jvi.62.11.4136-4143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schödel F, Weimer T, Fernholz D, Schneider R, Sprengel R, Wildner G, Will H. In: Molecular Biology of the Hepatitis B Virus. McLachlan A, editor. Boca Raton, FL: CRC; 1991. pp. 53–80. [Google Scholar]
- 17.Nassal M, Junker-Niepmann M, Schaller H. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- 18.Mandart E, Kay A, Galibert F. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obert S, Zachmann-Brandt B, Deindl E, Tucker W, Bartenschlager R, Schaller H. EMBO J. 1996;15:2565–2574. [PMC free article] [PubMed] [Google Scholar]
- 20.Bartenschlager R. Ph.D. thesis. Heidelberg, Germany: University of Heidelberg; 1990. [Google Scholar]
- 21.Junker-Niepmann M, Bartenschlager R, Schaller H. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz U, Kock J, Schlicht H J, Staeheli P. Virology. 1995;212:641–649. doi: 10.1006/viro.1995.1522. [DOI] [PubMed] [Google Scholar]
- 23.Chang C M, Jeng K S, Hu C P, Lo S J, Su T S, Ting L P, Pfaff E, Salfeld J, Schaller H. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condreay L D, Aldrich C E, Coates L, Mason W S, Wu T T. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hild M, Weber O, Schaller H. J Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jilbert A R, Miller D S, Scougall C A, Turnbull H, Burrell C J. Virology. 1996;226:338–345. doi: 10.1006/viro.1996.0661. [DOI] [PubMed] [Google Scholar]
- 27.Irving M G, Roll F J, Huang S, Bissell D M. Gastroenterology. 1984;87:1233–1247. [PubMed] [Google Scholar]
- 28.McCuskey R S, McCuskey P A, Urbaschek R, Urbaschek B. Infect Immun. 1984;45:278–280. doi: 10.1128/iai.45.1.278-280.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birnbaum F, Nassal M. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlicht H J, Salfeld J, Schaller H. J Virol. 1987;61:3701–3709. doi: 10.1128/jvi.61.12.3701-3709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugh J C, Di Q, Mason W S, Simmons H. J Virol. 1995;69:4814–4822. doi: 10.1128/jvi.69.8.4814-4822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlicht H J, Kuhn C, Guhr B, Mattaliano R J, Schaller H. J Virol. 1987;61:2280–2285. doi: 10.1128/jvi.61.7.2280-2285.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang P W. Ph.D. thesis. Heidelberg, Germany: Univ. of Heidelberg; 1992. .P. W. [Google Scholar]
- 34.Seeger C, Hu J. Trends Microbiol. 1997;5:447–450. doi: 10.1016/s0966-842x(97)01141-4. [DOI] [PubMed] [Google Scholar]
- 35.Huang J, Liang T J. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang M, Summers J. J Virol. 1994;68:1564–1572. doi: 10.1128/jvi.68.3.1564-1572.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvert J, Summers J. J Virol. 1994;68:2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlicht H J, Radziwill G, Schaller H. Cell. 1989;56:85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- 39.Bartenschlager R, Junker-Niepmann M, Schaller H. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston D E, Jasuja R. Hepatology. 1994;20:436–444. [PubMed] [Google Scholar]
- 41.Horwich A L, Furtak K, Pugh J, Summers J. J Virol. 1990;64:642–650. doi: 10.1128/jvi.64.2.642-650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaisomchit S, Tyrrell D L J, Chang L-J. Gene Ther. 1997;4:1330–1340. doi: 10.1038/sj.gt.3300544. [DOI] [PubMed] [Google Scholar]
- 43.Nassal M. Arch Virol. 1997;142:611–628. doi: 10.1007/s007050050107. [DOI] [PubMed] [Google Scholar]
- 44.Niederau C, Heintges T, Lange S, Goldmann G, Niederau C M, Mohr L, Haussinger D. N Engl J Med. 1996;334:1422–1427. doi: 10.1056/NEJM199605303342202. [DOI] [PubMed] [Google Scholar]