Abstract
The eukaryotic GINS complex has an essential role in the initiation and elongation phases of genome duplication. It is composed of four paralogous subunits—Sld5, Psf1, Psf2 and Psf3—which are ubiquitous and evolutionarily conserved in eukaryotic organisms. Here, we report the biochemical characterization of the human GINS complex (hGINS). The four hGINS subunits were coexpressed in Escherichia coli in a highly soluble form and purified as a complex. hGINS was shown to interact directly with the heterodimeric human DNA primase, by using either surface plasmon resonance measurements or by immunoprecipitation experiments carried out with anti-hGINS antibodies. The DNA polymerase α-primase synthetic activity was specifically stimulated by hGINS on various primed DNA templates. The significance of these findings is discussed in view of the molecular dynamics at the human replication fork.
Keywords: DNA polymerase, DNA primase, DNA replication, genome dynamics, GINS
Introduction
The GINS complex consists of four paralogous protein subunits—Sld5 (synthetic lethality with Dpb11), Psf1 (partner of Sld5 1), Psf2 and Psf3—and the genes encoding these subunits have been identified in all sequenced eukaryotic genomes. They were originally identified by genetic analyses in Saccharomyces cerevisiae aimed at discovering interaction partners of Dpb11 (Takayama et al, 2003). Dpb11 (DNA polymerase B possible subunit 11) is an essential initiation factor responsible, together with Cdc45 (cell division control 45), for recruiting DNA polymerase (pol) α and ɛ to the replisome (Masumoto et al, 2000). The GINS complex was shown to be essential for the initiation and elongation phases of DNA replication in S. cerevisiae (Kanemaki et al, 2003; Takayama et al, 2003). Furthermore, biochemical studies on Xenopus laevis egg extracts suggested that the GINS complex has an essential role in genome duplication also in vertebrates (Kubota et al, 2003). Chromatin immunoprecipitation experiments carried out in S. cerevisiae showed that GINS, pols α and ɛ, minichromosome maintenance protein (MCM)2–7, Cdc45, and the checkpoint factors Tof1–Csm3 and Mrc1 are components of the replisome at paused DNA replication forks (Calzada et al, 2005). A similar approach was used to dissect the molecular anatomy of the replisome in X. laevis egg extracts, in which sequence-specific replication fork pausing was induced with biotin–streptavidin-modified plasmids and identified the replicative pols α, δ and ɛ, GINS, MCM2–7, Cdc45 and MCM10 as components of the vertebrate replisome (Pacek et al, 2006). A very recent proteomic study in S. cerevisiae indicated that GINS is stably associated with the progressing replication fork together with the MCM2–7 complex, Cdc45 and a specific set of other various regulatory replication factors (Gambus et al, 2006). Furthermore, in Schizosaccharomyces pombe and HeLa cells, the Psf2 subunit was found to affect chromosome segregation, perhaps through replication of the centromeres, which is critical for correct centromeric chromatin and kinetochore assembly (Huang et al, 2005). However, despite these genetic and biochemical studies, the exact molecular function of the GINS complex has not yet been explained. It has been postulated that it might coordinate the action of the multiple eukaryotic replicative polymerases at the replication fork during the elongation phase (Kubota et al, 2003; Takayama et al, 2003). In addition, the GINS complex has been proposed to act as an activator of the MCM2–7 helicase function in association with Cdc45 (Kubota et al, 2003; Takahashi et al, 2005; Pacek et al, 2006). Conversely, in mammalian cells, the biochemical features and physiological role of the GINS homologues have been poorly investigated so far. It has been reported that the gene coding for human Psf2 is upregulated in liver cancer cells (Obama et al, 2005), and the mouse Psf1 protein has been shown to be essential for cell proliferation at an early stage of embryonic development (Ueno et al, 2005). Here, we report that the recombinant human GINS complex (hGINS) can physically interact with the primase subunits of pol α-primase and can specifically stimulate its polymerase activity.
Results And Discussion
Production of hGINS in a soluble recombinant form
The aim of this study was to analyse the biochemical properties of the hGINS subunits' homologues. We initially attempted to express the corresponding genes in E. coli individually and found that the recombinant proteins were poorly soluble irrespective of the growth conditions and the vector systems used. We therefore reasoned that coexpression of the four genes might improve protein solubility, resulting in the association of the GINS subunits to form a soluble complex in the E. coli recombinant cells. We constructed an expression vector that contained the four open reading frames in tandem as a polycistronic unit, each preceded by a ribosome-binding site, in the order Psf3–Psf1–Sld5–Psf2 (see supplementary information online; Fig 1A). The Psf3 open reading frame was fused to an amino-terminal hexahistidine tag to facilitate the purification procedure. Using this system, we were able to obtain the four hGINS subunits in highly soluble form. The four polypeptides co-purified by three subsequent chromatographic steps, suggesting that they truly associate to form a stable complex (see Fig 1B). The identity of the polypeptides in the purified sample was confirmed by N-terminal sequencing of each of the four protein bands. This analysis allowed us to determine that the four bands were present in the complex in a stoichiometric ratio of 1:1:1:1, as also tested by densitometric analysis (data not shown). This result is in agreement with the subunit composition reported for the S. cerevisiae and X. laevis GINS complex (Kubota et al, 2003; Takayama et al, 2003). In addition, the four hGINS subunits showed a mobility on SDS–polyacrylamide gels that was lower than expected on the basis of their predicted molecular size. The X. laevis GINS homologues showed a similar anomalous electrophoretic behaviour (Kubota et al, 2003).
Figure 1.
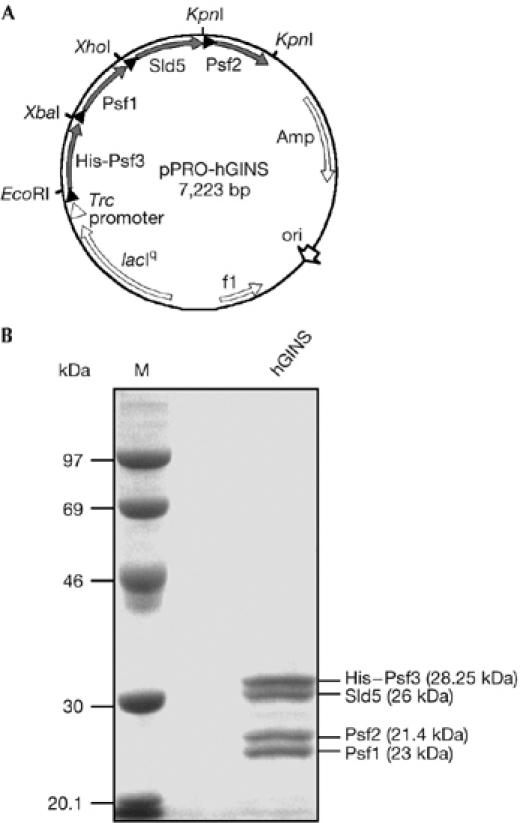
Production of human GINS in Escherichia coli. (A) Schematic representation of the pPRO-EX HTa vector derivative (pPRO-hGINS) used to express hGINS in E. coli. The construction of this plasmid is described in detail in the supplementary information online. Closed triangles represent ribosome binding sites of E. coli. (B) Electrophoretic analysis of purified recombinant hGINS (5 μg) on a 10% SDS–polyacrylamide gel with Coomassie blue staining. The identity of the four protein bands as GINS subunits was determined by amino-terminal sequencing after electrophoretic transfer onto a polyvinylidene difluoride membrane. Their predicted molecular weight is reported in parentheses. Amp, ampicillin; hGINS, human GINS; M, molecular weight marker; ori, origin of replication.
hGINS physically interacts with pol α-primase
Direct physical interaction was identified between the hGINS complex and pol α-primase by using the surface plasmon resonance technique. For these analyses, the recombinant human heterodimeric DNA primase (composed of the p48 and p58 subunits) was used. Fig 2A shows typical overlaid sensorgrams obtained by testing four different concentrations of human DNA primase (25–200 nM), which were passed over an hGINS-immobilized sensor chip. These results indicated that hGINS physically interacts with the DNA primase. Quantitative analysis of this interaction showed a KD of 3.1 × 10−6 M. The physical association was then confirmed by immunoprecipitation experiments carried out using protein A Sepharose beads conjugated with antibodies raised against the purified recombinant hGINS, as shown in Fig 2B. In summary, these data gave evidence that hGINS could bind to the DNA primase subunits of the heterotetrameric pol α-primase complex.
Figure 2.
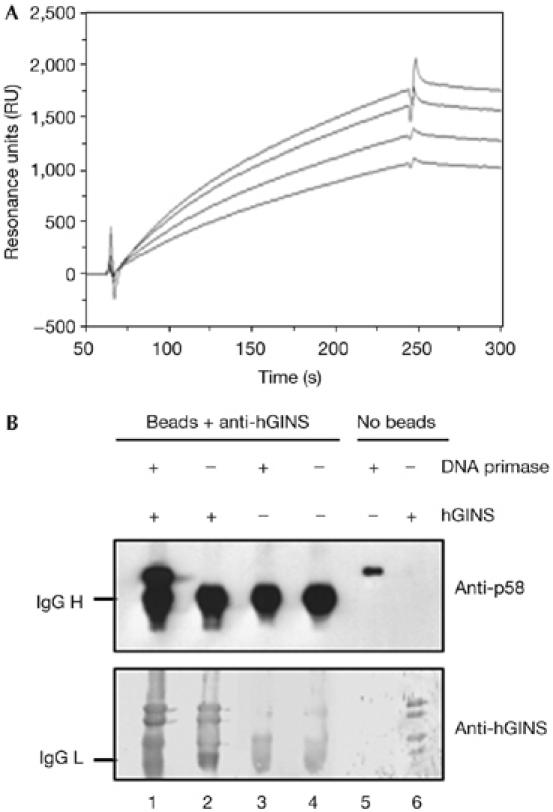
Physical interaction between human GINS and human DNA primase. (A) An overlaid plot of sensorgrams was obtained by fluxing the human heterodimeric DNA primase at various concentrations over an hGINS-immobilized sensor chip, as described in the Methods (lower to upper curve: the DNA primase was at 25, 50, 100 and 200 nM, respectively). (B) Immunoprecipitation experiments were carried out using protein A Sepharose beads conjugated with anti-hGINS antibodies and the following samples: a mixture of purified recombinant hGINS and the heterodimeric DNA primase (lane 1), hGINS alone (lane 2) and human DNA primase alone (lane 3), as described in the Methods. A sample of beads conjugated with antibodies (lane 4), human DNA primase (100 ng, lane 5) and hGINS (200 ng, lane 6) were run as control. Western blot analysis was carried out by using anti-human primase p58 and anti-hGINS antibodies, as described in the Methods. hGINS, human GINS; RU, resonance units.
hGINS stimulates DNA synthesis by pol α-primase
Next, we wanted to test whether the binding of hGINS had any effect on the catalytic activity of pol α-primase. Fig 3 shows that hGINS can dose-dependently and specifically stimulate the polymerase activity of pol α. This effect was observed on either activated calf thymus DNA (sevenfold; Fig 3A) or poly(dA)-oligo(dT) (six- to sevenfold; Fig 3B). Conversely, no stimulation of the primase activity by the GINS complex was seen when tested on single-stranded M13 DNA, in the presence of the four ribonucleoside triphosphates (data not shown). Furthermore, no effect was observed by hGINS on pol β (Fig 3C), pol δ (Fig 3D) or pol λ (Fig 3F), whereas a slight but reproducible stimulation was found on pol ɛ (1.5- to 2-fold; Fig 3E). Next, a synthesis product analysis was carried out with singly primed M13 single-stranded DNA (ssDNA). As shown in Fig 4A, in the presence of hGINS, total DNA synthesis by pol α-primase was markedly increased: an approximate tenfold increase in stimulation was measured when synthesis products longer than 125 nucleotides were quantified (Fig 4B). Together, these data suggested that the stimulation by hGINS on pol α-primase was specific.
Figure 3.
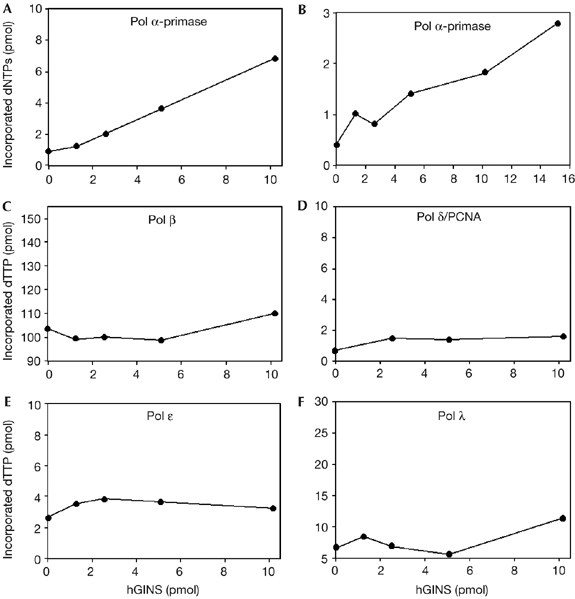
Effect of human GINS on the synthetic activity of five different human DNA polymerases. The DNA synthesis activities of various human polymerases were measured in the presence of increasing concentrations of hGINS. Polymerase (pol) α-primase was assayed on activated calf thymus DNA (144 fmol; A) and on poly(dA)-oligo(dT)(522 fmol; B). Pol β (5 fmol; C), pol δ/PCNA (152 fmol; D), pol ɛ (50 fmol; E) and pol λ (750 fmol; F) were assayed on poly(dA)-oligo(dT). Enzymatic assays were carried out at least in triplicate and each plot reports the results of a typical experiment. hGINS, human GINS; PCNA, proliferating cell nuclear antigen.
Figure 4.
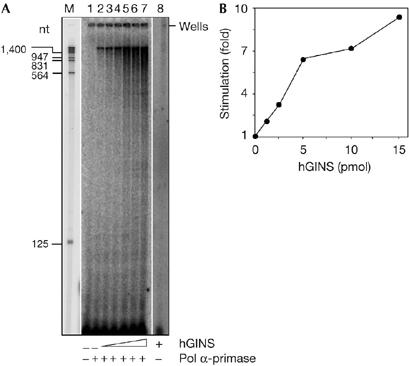
Human GINS stimulates DNA synthesis by polymerase α-primase on singly primed M13 mp18 single-stranded DNA. (A) Reaction mixtures (volume 10 μl) contained singly primed M13mp18 single-stranded DNA (25 fmol), 18 μM dATP, 18 μM dGTP, 18 μM dTTP, 6 μM dCTP and [α-32P]dCTP (1.25 μCi) and, where indicated, polymerase (pol) α-primase (70 fmol). Lane 1, no enzyme control; lane 2, pol α-primase alone; lanes 3–7, pol α-primase in the presence of 1.27, 2.55, 5.1, 10.2 and 15.3 pmol of hGINS, respectively. Samples were incubated at 37°C for 15 min. Reaction products were run on a 6% polyacrylamide sequencing gel, which was analysed using a phosphorimager apparatus. Lane M contains EcoRI–HindIII-digested λ DNA as the size marker (the exposure of this lane was shorter for identifying the position of the marker). Lane 8 contains a control reaction carried out with 1.5 μg of hGINS without pol α-primase and run on a different gel. (B) Reaction products longer than 125 nucleotides detected in lanes 2–8 of the gel shown in (A) were quantified by using a phosphorimager, as described in the Methods. The intensity of the signal in each lane was normalized to the value calculated for the reaction carried out in the absence of hGINS. hGINS, human GINS.
Previous genetic and biochemical analyses of S. cerevisiae and X. laevis replisome composition showed that the GINS complex is important in the establishment and progression of replication forks (Kanemaki et al, 2003; Kubota et al, 2003; Takayama et al, 2003). In yeast, it was found that GINS associates with chromatin at the replication origins together with Cdc45 and Dpb11, which are required for recruiting pol α-primase and pol ɛ to the replisome (Masumoto et al, 2000). Our biochemical analysis showed that, in the human system, the GINS complex directly interacts with the DNA primase subunits of the heterotetrameric pol α-primase and produces a marked stimulation of the pol α-primase DNA synthetic function, and it does not activate the priming function. Nevertheless, we failed to co-immunoprecipitate hGINS and the pol α-primase complex from cell extracts tested under different conditions. These experiments were carried out on either logarithmically growing HeLa cells or S-phase bladder tumour human cells, by using either polyclonal anti-hGINS antibodies or the monoclonal anti-human primase p58 subunit (data not shown). In agreement with our findings, DNA polymerases have not been found as components of the so-called ‘replisome progression complexes' in a proteomic analysis of the factors interacting with GINS in S. cerevisiae (Gambus et al, 2006). Furthermore, in X. laevis only GINS, Cdc45 and MCM2–7, and not any of the replicative polymerases, are found to accumulate at replication pausing sites in the presence of aphidicolin, an inhibitor of pols α, δ and ɛ that produces uncoupling of the DNA synthesis with DNA unwinding in cell-free DNA replication systems (Pacek et al, 2006).
Our findings are consistent with a recent report on the archaeal DNA replication machinery in which the GINS homologue has been shown to interact directly with the heterodimeric eukaryotic-like DNA primase and with the MCM-like complex (Marinsek et al, 2006). Nonetheless, we were unable to detect any direct physical association between purified recombinant hGINS and human MCM2–7 or MCM4/6/7 complexes using the surface plasmon resonance technique to detect protein–protein interaction (Y. Ishimi & F.M. Pisani, unpublished data). In fact, it is likely that the association of the eukaryotic GINS and MCM complexes requires other replication factors, such as Cdc45 and MCM10, as recently reported for the Drosophila embryo system (Moyer et al, 2006). How hGINS activates pol α-primase can only be speculated. hGINS might act at the single–double-stranded DNA junction of the replication forks promoting the synthesis of Okazaki fragments by pol α-primase on the lagging strand and by coordinating this function with DNA unwinding by replicative DNA helicase on the leading strand, as proposed for the archaeal system (Marinsek et al, 2006). Furthermore, it should be noted that association of pol α-primase with the replisome was found to be dependent on MCM10 (Ricke & Bielinsky, 2004) and Cdc45 (Mimura & Takisawa, 1998), and S. pombe MCM10 was reported to interact with the pol α-primase p180 catalytic subunit and to increase its primer utilization in vitro (Fien et al, 2004). The biochemical characterization of the human counterparts of these replication factors will help to understand the molecular dynamics at the human DNA replication forks in more detail, and to discover the differences and similarities between human and yeast systems.
Methods
Enzymes. Human pols α, δ and ɛ were purified as described by Weiser et al (1991). Recombinant pols β and λ were isolated as described by Toueille et al (2004) and by Ramadan et al (2003), respectively. The human recombinant heterodimeric DNA primase was produced in E. coli by using the construct pET-Hp48-HisHp58 as described previously (Schneider et al, 1998).
Surface plasmon resonance measurements. Real-time interactions of the GINS complex and the recombinant heterodimeric DNA primase were monitored by using the surface plasmon resonance biosensor system Biacore 2000 (Biacore, Uppsala, Sweden). hGINS (11,500 resonance units (RU)) was coupled to the surface of a CM5 sensor chip in 10 mM sodium acetate buffer (pH 3.6), according to the manufacturer's instructions. To collect sensorgrams, the indicated protein was passed over the sensor surface at a flow rate of 10 μl/min at various concentrations. Recorded sensorgrams were normalized to a baseline of 0 RU and analysed using the BIA Evaluation software (version 3.2).
Immunoprecipitation experiments. Protein A Sepharose CL-4B resin (250 μg) was resuspended in binding buffer (50 mM Tris–HCl pH 8.0, 40 mM NaCl, 10 mM MgCl2) and conjugated with anti-hGINS antibodies. Mixtures with a final volume of 20 μl contained 10 μg of hGINS and 5 μg of heterodimeric DNA primase, and either 10 μg of hGINS or 5 μg of DNA primase in binding buffer. Protein A Sepharose resin conjugated with anti-hGINS antibodies (40 μl) was added to each sample. After incubation for 1 h at 22°C with gentle shaking, the resin of each mixture was washed with 5 ml of washing buffer (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM MgCl2, 1% Triton X-100) and then resuspended in 60 μl of SDS–polyacrylamide gel electrophoresis sample buffer 1 × (50 mM Tris–HCl pH 6.8, 10% glycerol, 200 mM 2-mercaptoethanol, 0.5% SDS, 0.01% blue bromophenol). Samples were run on a 10% SDS–polyacrylamide denaturing gel and transferred to a polyvinylidene difluoride membrane. The upper part of the membrane was analysed with anti-human primase p58 rat monoclonal antibodies and anti-rat IgG antibodies conjugated with horseradish peroxidase using the ECL+ system (GE Healthcare, Uppsala, Sweden). The lower half of the membrane was analysed with anti-hGINS and anti-rabbit IgG antibodies conjugated with alkaline phosphatase using a standard colorimetric procedure.
DNA polymerase assays. DNA polymerase assays on either activated calf thymus DNA (pol α) or poly(dA)-oligo(dT) in the presence of 120 ng proliferating cell nuclear antigen (pol δ) were carried out as described by Weiser et al (1991), on poly(dA)-oligo(dT) for pol β as described by Toueille et al (2004) and on poly(dA)-oligo(dT) in the presence of 1 mM MnCl2 for pol λ as described by Ramadan et al (2003). The assays for the five different DNA polymerases were carried out under optimal conditions for each enzyme. Each enzyme was titrated and about 10–15% of the activity was chosen for testing the effect of hGINS. For DNA synthesis, the following amounts of the five DNA polymerases were used: pol α on activated DNA (144 fmol), pol α on poly(dA)-oligo(dT) (522 fmol), pol β (5 fmol), pol δ (152 fmol), pol ɛ (50 fmol) and pol λ (750 fmol).
DNA synthesis product analysis. Singly primed ssDNA was prepared by annealing a 17-mer oligonucleotide (universal sequencing primer) to M13mp18 ssDNA in the polylinker region. Elongation assays (reaction volume 10 μl) were carried out in 20 mM potassium phosphate pH 7.5, 10 mM MgCl2, 0.1 mM EDTA, 4 mM dithiothreitol, 0.25 mg/ml BSA, 2.5 nM singly primed M13mp18 ssDNA, 18 μM dATP, 18 μM dGTP, 18 μM dTTP, 6 μM dCTP and [α-32P]dCTP (1.25 μCi) and the indicated amounts of pol α-primase and/or hGINS. Samples were incubated at 37°C for the indicated time. Reaction products were separated through a 6% polyacrylamide/8 M urea sequencing gel, which was analysed by a Molecular Dynamics Storm PhosphorImager using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA, USA).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
We are grateful to Dr H.-P. Nasheuer (Galway, Ireland) for the plasmid pET-Hp48-HisHp58 and anti-human primase p58 monoclonal antibodies, and to Dr S. Onesti (London, UK) for the hGINS subunit complementary DNA clones. E.F. and U.H. are supported by the University of Zürich and M.D.F. by a grant in aid from the UBS (im Auftrag eines Kunden). F.M.P. and M.R. are supported by a grant from ATIBB–BioTekNet (Centro Regionale di Competenza in Biotecnologie Industriali).
References
- Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K (2005) Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19: 1905–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fien K, Cho Y-S, Lee J-K, Raychaudhuri S, Tappin I, Hurwitz J (2004) Primer utilization by DNA polymerase α-primase is influenced by its interaction with Mcm10p. J Biol Chem 279: 16144–16153 [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Huang H-K, Bailis JM, Leverson JD, Gòmez EB, Fosburg SL, Hunter T (2005) Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol Cell Biol 25: 9000–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K (2003) Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–724 [DOI] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H (2003) A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD (2006) GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep 7: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H (2000) Dpb11 controls the association between DNA polymerases αand ɛ and the autonomously replicating sequence region of budding yeast. Mol Cell Biol 20: 2809–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Takisawa H (1998) Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase Cdk. EMBO J 17: 5699–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/MCM 2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication for helicase. Proc Natl Acad Sci USA 103: 10236–11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obama K, Ura K, Satoh S, Nakamura Y, Furukawa Y (2005) Upregulation of Psf2, a member of the GINS multiprotein complex, in intrahepatic cholangiocarcinoma. Oncol Rep 14: 701–706 [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hübscher U (2003) Human DNA polymerase λ possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J Mol Biol 328: 63–72 [DOI] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky A-K (2004) Mcm10 regulates the stability and chromatin association of DNA polymerase α. Mol Cell 16: 173–185 [DOI] [PubMed] [Google Scholar]
- Schneider A, Smith RWP, Kautz AR, Weissart KH, Grosse F, Nasheuer H-P (1998) Primase activity of human DNA polymerase α-primase. J Biol Chem 273: 21608–21615 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Wigley DB, Walter JC (2005) Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem Sci 30: 437–444 [DOI] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hübscher U (2004) The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase β and increases its substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res 32: 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Itoh M, Kong L, Sugihara K, Asano M, Takakura N (2005) Psf1 is essential for early embryogenesis in mice. Mol Cell Biol 25: 10528–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hübscher U (1991) Biochemical and functional comparison of DNA polymerases α, δ, and ɛ from calf thymus. J Biol Chem 266: 10420–10428 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information


