Abstract
The metazoan nuclear envelope (NE) breaks down and re-forms during each cell cycle. Nuclear pore complexes (NPCs), which allow nucleocytoplasmic transport during interphase, assemble into the re-forming NE at the end of mitosis. Using in vitro NE assembly, we show that the vertebrate homologue of MEL-28 (maternal effect lethal), a recently discovered NE component in Caenorhabditis elegans, functions in postmitotic NPC assembly. MEL-28 interacts with the Nup107–160 complex (Nup for nucleoporin), an important building block of the NPC, and is essential for the recruitment of the Nup107–160 complex to chromatin. We suggest that MEL-28 acts as a seeding point for NPC assembly.
Keywords: mitosis, kinetochore, nuclear envelope assembly, nuclear pore complex, Nup107–160 complex, MEL-28
Introduction
The nuclear envelope (NE) of higher eukaryotes breaks down and re-forms during each cell cycle. Nuclear pore complexes (NPCs), which mediate nucleocytoplasmic transport, assemble both into the re-forming NE at the end of mitosis and into the interphase NE. Postmitotic NPC assembly occurs in an orderly, only partly understood manner and involves the recruitment of soluble and membrane-associated NPC components and also membranes to the decondensing chromatin in telophase (Hetzer et al, 2005; Prunuske & Ullman, 2006).
A screen in Caenorhabditis elegans for factors essential for normal nuclear morphology led to the identification of MEL-28 (maternal effect lethal), a protein localized to the NE in interphase and kinetochores during mitosis (Galy et al, 2006). A detailed study of its function in worms showed that mel-28 mutation or depletion of MEL-28 by RNA interference (RNAi) impairs nuclear integrity and leads to an abnormal distribution of both NE and NPC proteins (Fernandez & Piano, 2006; Galy et al, 2006). Sequence analysis showed that C. elegans MEL-28 has orthologues in most multicellular eukaryotes, suggesting that its role in nuclear organization might be evolutionarily conserved. This prompted us to examine human and Xenopus laevis MEL-28 in more detail. The Xenopus egg extract system allows nuclear assembly in vitro (Lohka & Masui, 1983; for review see Gant & Wilson, 1997) and thus biochemical dissection of nuclear dynamics. Several steps in nuclear assembly have been identified. First, sperm DNA is decondensed by nucleoplasmin (Philpott et al, 1991). Next, membranes and a subcomplex of the NPC, the Nup107–160 complex (Nup for nucleoporin), bind to chromatin (Walther et al, 2003a). These processes can be separated experimentally, although they are likely to be coordinated during NE assembly in vivo. Subsequent events include fusion of membranes to form a closed NE, addition of further NPC components, import of lamins and nuclear growth accompanied by further chromatin decondensation.
Here, we show that vertebrate MEL-28 is an NE protein in interphase and redistributes to kinetochores in mitosis. It shares this behaviour with certain nucleoporins (proteins that constitute the NPC). Interestingly, immunoprecipitation experiments showed that MEL-28 interacts with the Nup107–160 complex, which is an important building block of the vertebrate nuclear pore. During nuclear assembly in vitro, MEL-28 is recruited to chromatin at an early time point. Immunodepletion of MEL-28 from extracts interferes with the formation of a functional nucleus at an early stage, leading to nuclei with a closed NE but without NPCs.
Results And Discussion
Potential homologues of C. elegans MEL-28 were identified in mammals and X. laevis, although their overall sequence conservation was low (Galy et al, 2006). The potential human MEL-28 homologue was previously identified as ELYS (embryonic large molecule derived from yolk sac) and was suggested to function as a transcription factor (Kimura et al, 2002). ELYS-deficient mice die at early stages of development (Okita et al, 2004), but the cause of death remains unclear. This prompted us to test whether the function of the protein is conserved between worms and vertebrates.
First, we generated antibodies against human and Xenopus MEL-28. The human antibody recognized a single major band of the expected size of 250 kDa in HeLa nuclear extracts and a minor, probably crossreacting band (Fig 1A), whereas the frog antibody recognized a doublet in Xenopus extracts. Both in HeLa (Fig 1B) and Xenopus XL177 cells (supplementary Fig 1 online), the protein localized mainly to the NE in interphase, as indicated by co-staining with monoclonal antibody (mAb)414, an antibody that recognizes a subset of nucleoporins. This colocalization of MEL-28 and nucleoporins that can be stained with mAb414 could also be seen at higher resolution on the surface of the NE of human U2OS cells (Fig 1C), indicating that MEL-28 localizes to NPCs. In addition to the NE staining, the protein showed nucleoplasmic staining (Fig 1B). During mitosis, vertebrate MEL-28 was detected on the chromatin surface and at kinetochores. Thus, the localization of the vertebrate protein was similar to that of C. elegans MEL-28.
Figure 1.
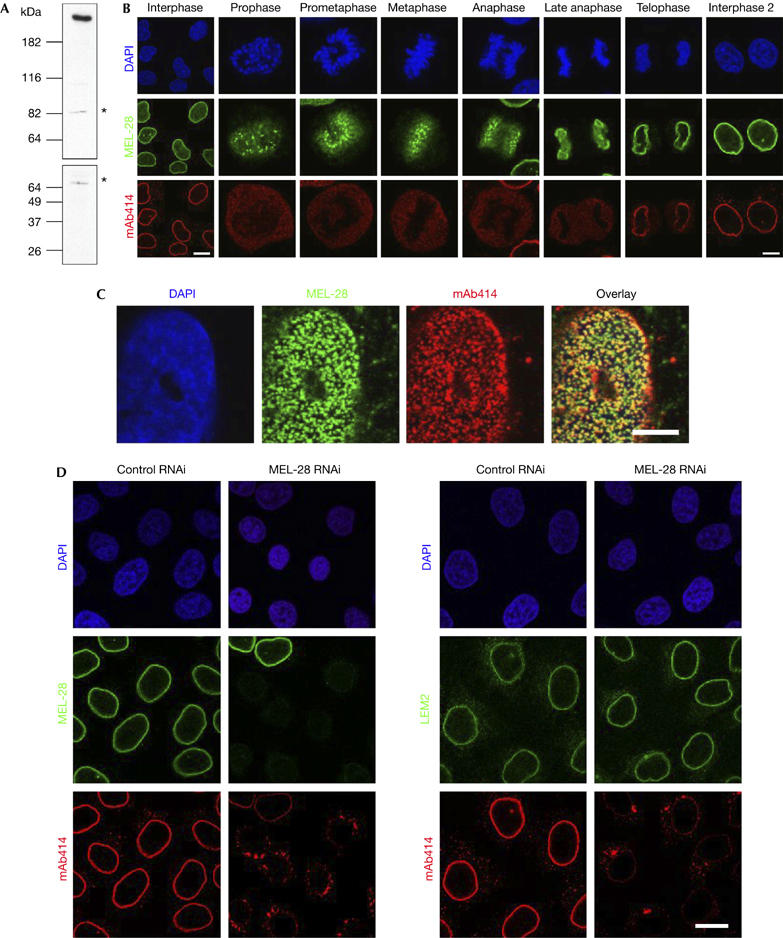
Characterization of MEL-28 in human cells. (A) Antibodies against human MEL-28 recognize a high-molecular-weight protein in HeLa nuclear extracts. Asterisks indicate a crossreacting band. (B) Immunolocalization of MEL-28 throughout the cell cycle in HeLa cells: chromatin stained with DAPI is shown in the upper row, MEL-28 (green) in the middle row and NPCs visualized with mAb414 (red) in the lower row. Scale bars, 15 μm (left column) and 5 μm (all others). (C) MEL-28 colocalizes with NPCs: a surface image of the NE from a U2OS nucleus is shown using the same colour coding as in (B). Scale bar, 8 μm. (D) RNA interference depletion of MEL-28: HeLa cells, transfected with either control or MEL-28 siRNA duplexes, were fixed after 72 h and immunostained with MEL-28 antibodies (green; left columns), LEM2 antibodies (green; right columns) or mAb414. DAPI staining is shown in blue. Scale bar, 15 μm. DAPI, 4,6-diamidino-2-phenylindole; LEM, lamina-associated polypeptide-emerin-MAN1; MEL, maternal effect lethal; NE, nuclear envelope; NPC, nuclear pore complexes; siRNA, short interfering RNA.
To study the function of human MEL-28, we first carried out RNAi experiments in HeLa cells using a short interfering RNA (siRNA) oligonucleotide specific for human MEL-28 or control oligonucleotides. The expression of MEL-28 was progressively reduced at 48 h (not shown) and 72 h after transfection, as measured by staining the nuclear rim using a MEL-28-specific antibody (Fig 1D). By contrast, LEM2 (LEM for lamina-associated polypeptide-emerin-MAN1), an inner NE protein (Brachner et al, 2005), showed normal localization at the rim and no obvious reduction in antibody staining. mAb414 staining at the nuclear rim was reduced and cytoplasmic aggregates appeared (Fig 1D), which corresponded to annulate lamellae, when analysed by transmission electron microscopy (TEM; supplementary Fig 2A online). This effect was even more striking in a second human cell line, U2OS (supplementary Fig 2B online), and similar results were obtained in both cell lines with a second anti-MEL-28 siRNA (data not shown). This phenotype is reminiscent of that reported after depletion of nucleoporins of the Nup107–160 complex (Walther et al, 2003a) and suggests that depletion of MEL-28 affects NPC assembly.
To obtain a more detailed picture of MEL-28 function, we characterized the role of the protein in NE and NPC assembly in vitro in X. laevis egg extracts. Here, nuclear assembly is initiated by the addition of cytosol and membranes to sperm heads, which are free of endogenous MEL-28 (supplementary Figs 1B, 4E online). First, we investigated at what point during NE formation MEL-28 is recruited to chromatin (Fig 2A). Sperm head chromatin was decondensed with membrane-free cytosol for 5 min followed by the addition of membranes. The reaction was stopped by fixation at the indicated time points and samples were processed for immunofluorescence. MEL-28 was recruited to the chromatin early in nuclear assembly, diffusely staining the chromatin surface 5 min after the addition of cytosol. The binding of MEL-28 preceded detectable recruitment of the Nup107–160 complex, which is known to bind to chromatin early during assembly (Harel et al, 2003; Walther et al, 2003a). The Nup107–160 complex signal on chromatin increased progressively, while mAb414 antigens became detectable after 20 min (Fig 2A). At this time point, focal MEL-28 and Nup160 staining at the chromatin surface colocalized with mAb414. During the course of nuclear assembly, both signals increased and remained colocalized with mAb414.
Figure 2.
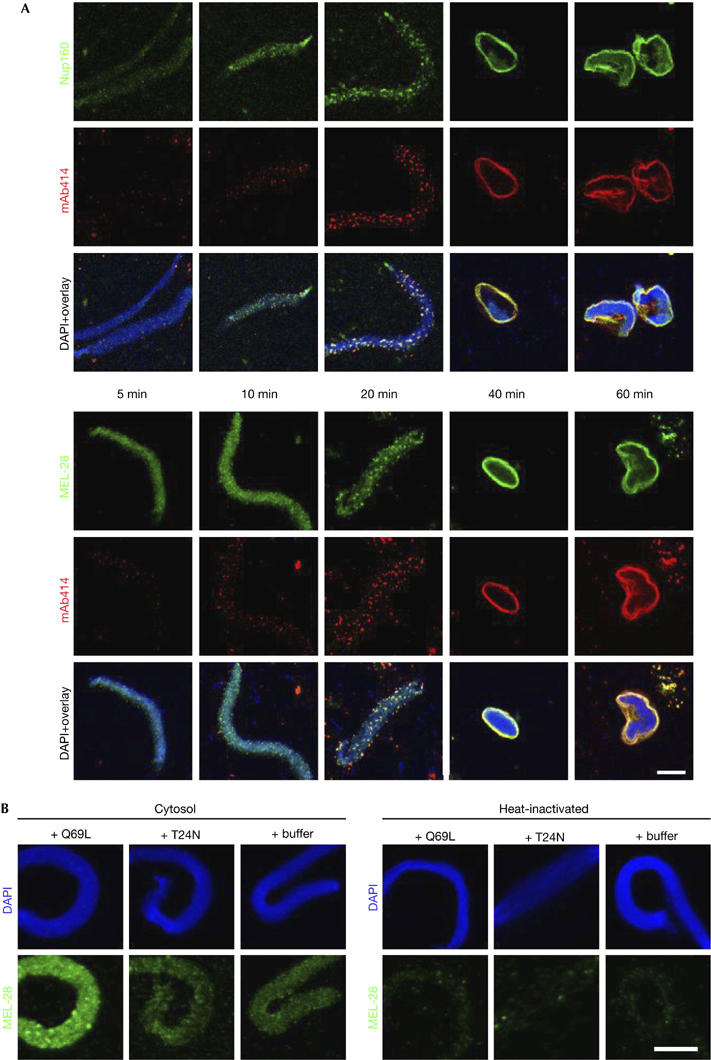
Xenopus MEL-28 is recruited early to chromatin in a Ran-dependent manner. (A) Demembranated sperm chromatin was preincubated with Xenopus egg extract for 5 min and membranes were added to the reaction. Assembly reactions were stopped by fixation at the indicated time points after the addition of cytosol and analysed by confocal microscopy after immunofluorescence using Nup160 antibodies (green, upper panel), MEL-28 antibodies (green, lower panel) or mAb414 (red). Chromatin was stained with DAPI (blue in the overlays). Scale bar, 10 μm. (B) Membrane-free cytosol was preincubated on ice for 15 min with 5 μM RanQ69L, RanT24N or buffer and then added to sperm head chromatin. The reactions were stopped by fixation after 10 min and samples were processed for immunofluorescence with MEL-28 antibodies. Non-specific binding of the antibodies to chromatin is shown after incubation with heat-inactivated extracts, which were free of the respective antigens, that is, conditions that allow only decondensation of sperm DNA. Scale bar, 10 μm. DAPI, 4,6-diamidino-2-phenylindole; mAb; monoclonal antibody; MEL, maternal effect lethal; Nup, nucleoporin.
Several aspects of nuclear assembly are controlled by the small GTPase Ran (Walther et al, 2003b). Previous observations in C. elegans suggested that MEL-28 localization is affected by components of the Ran cycle (Fernandez & Piano, 2006). To test whether the recruitment of MEL-28 to chromatin is triggered by RanGTP, we incubated sperm heads for 10 min with membrane-free cytosol in the presence of 5 μM Ran mutants or buffer (Fig 2B). The addition of RanQ69L, a constitutively active form of Ran, caused a strong recruitment of MEL-28 to chromatin as expected if RanGTP regulates this interaction. Adding RanT24N, which blocks the exchange of GTP onto endogenous RanGDP, did not alter MEL-28 levels on chromatin, suggesting that some MEL-28 can bind independently of Ran.
Next, we depleted MEL-28 from the extracts. As shown in Fig 3A, the Xenopus protein runs as a doublet and can be efficiently removed by passing the cytosol twice over an antibody resin without co-depleting nucleoporins Nup160, Nup107 or Nup93. When these depleted extracts were used for in vitro assembly, nuclei formed, as indicated by membrane staining (Fig 3B), but they were smaller than control nuclei and mAb414 staining was absent (Fig 3B). Two different nucleoporin depletion phenotypes that lead to loss of mAb414 staining have been described. In the absence of Nup155, pom121 (pore membrane protein) or NDC1 (nuclear division cycle protein), mAb414 staining is reduced or absent. Membranes dock to chromatin, but do not fuse to form a closed NE (Antonin et al, 2005; Franz et al, 2005; Mansfeld et al, 2006). After depletion of the Nup107–160 complex, mAb414 staining is also absent, but the membranes form a closed NE lacking NPCs (Harel et al, 2003; Walther et al, 2003a). Analysis by TEM of the nuclei assembled in the MEL-28-depleted extract showed that the chromatin remained highly condensed, whereas a closed NE that lacked NPCs formed on the chromatin surface (Fig 3C).
Figure 3.
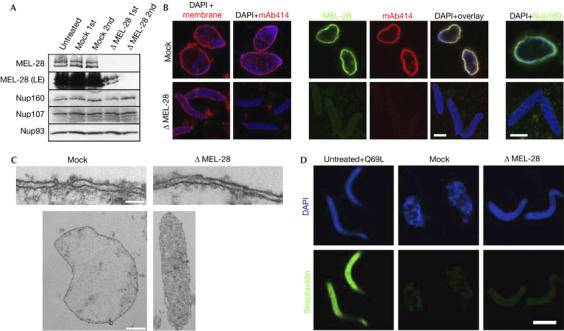
Removal of MEL-28 inhibits nuclear pore complex but not nuclear envelope formation. (A) MEL-28 was depleted from the Xenopus nuclear reconstitution system by two rounds of incubation with immobilized MEL-28 antibodies: western blot analysis of mock-depleted and MEL-28-depleted extracts using the antibodies indicated on the left. (B) MEL-28-depleted extracts assemble nuclei without NPCs: nuclei were assembled for 90 min. Samples were fixed with 4% PFA and 0.5% glutaraldehyde and analysed for membrane staining (DiIC18, left column) or the presence of NPCs by immunofluorescence with mAb414. Samples in the other four columns were fixed with 4% PFA and analysed by immunofluorescence with the indicated antibodies. Chromatin was stained with DAPI (blue). Scale bars, 10 μm. (C) Transmission electron microscopy of nuclei assembled from mock-depleted (left side) or MEL-28-depleted (right side) extracts. Scale bar, 100 nm (upper images) and 2 μm (lower images). (D) Nuclei assembled in MEL-28-depleted extracts have a closed NE: an exclusion assay on chromatin incubated in RanQ69L-treated, mock-depleted or MEL-28-depleted extracts was carried out. Labelling of chromatin with Oregon green–streptavidin conjugate (green) indicates the absence of a closed NE. Scale bar, 10 μm. 1st, first round; 2nd, second round; DAPI, 4,6-diamidino-2-phenylindole; LE, Long exposure; mAb, monoclonal antibody; MEL, maternal effect lethal; NE, nuclear envelope; NPC, nuclear pore complexes; Nup, nucleoporin; PFA, paraformaldehyde.
To ensure that the NEs were closed in the absence of MEL-28, we used a nuclear exclusion assay (see the Methods for details and supplementary information online for comprehensive characterization). In brief, sperm chromatin was decondensed with recombinant nucleoplasmin and loaded with biotinylated histones. Nuclear assembly reactions were initiated on these chromatin templates by the addition of membranes and cytosol. At the end of the reaction, fluorescently labelled streptavidin was added, which can reach the chromatin-bound histones only if the NE is not closed. After quenching, the reactions were stopped by fixation and the nuclei were re-isolated and analysed by fluorescence microscopy. In control nuclei assembled in the mock-depleted extract, the fluorescently labelled streptavidin did not reach the chromatin, as indicated by the absence of a green signal (Fig 3D) with 99% of chromatin substrates excluding streptavidin. By contrast, when 10 μM RanQ69L was added to the reaction to prevent closed NE formation (Hetzer et al, 2000), the chromatin was stained intensely (with only 1% of chromatin templates excluding streptavidin). Nuclei assembled in MEL-28-depleted extracts showed no streptavidin labelling on the chromatin (98% of chromatin substrates excluded streptavidin), indicating that the NEs were uniformly closed (Fig 3D) and also confirming the light and electron microscopic data given above.
The depletion phenotype indicated that MEL-28 is important early in NPC assembly. To assess further the molecular mechanism by which MEL-28 acts, we set out to identify and characterize its interaction partners. For this, cytosol was passed over a MEL-28 immunoaffinity resin. After intensive washing, several proteins were eluted with MEL-28 and identified by mass spectrometry as Nup160, Nup133, Nup96, Nup107 (all members of the Nup107–160 complex) and importin-β (known to associate with this complex; Fig 4A).
Figure 4.
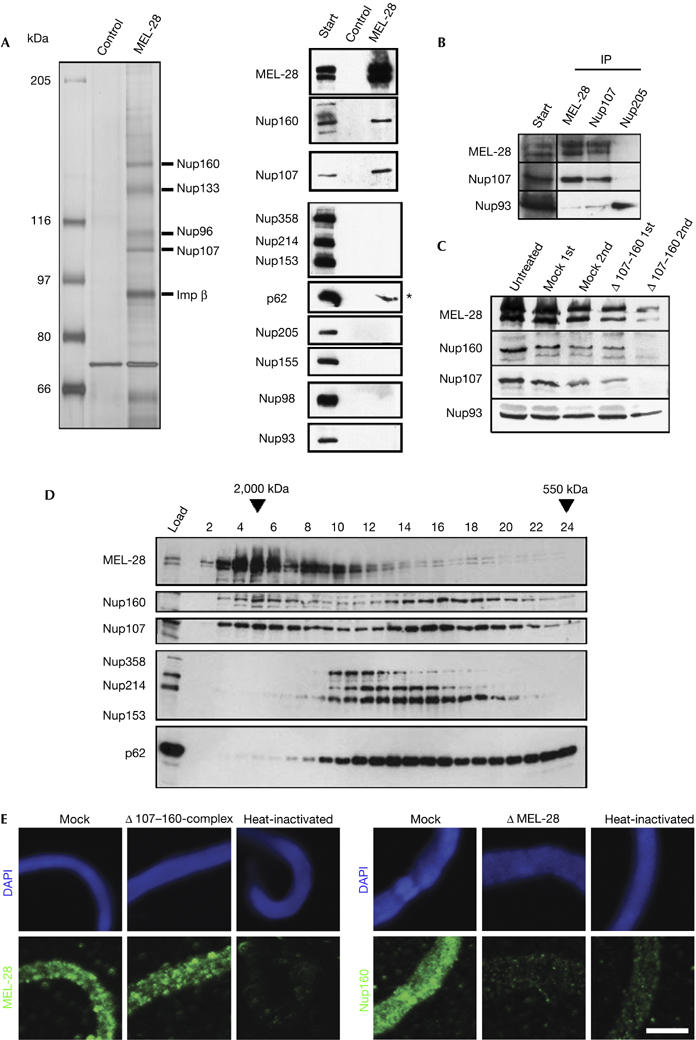
MEL-28 interacts with the Nup107–160 complex and is required for Nup107–160 recruitment to chromatin. (A) MEL-28 antibodies co-immunoprecipitate the Nup107–160 complex. The indicated proteins were identified by mass spectrometry after silver staining (left) or by western blotting with respective antisera (right) after immunoprecipitation from cytosol. Two high-molecular-weight bands that stain weakly on the silver gel represent MEL-28 (not shown). The asterisk indicates a background signal for p62. (B) Co-immunopreciptation of Nup107 and MEL-28: immunoprecipitations were performed from cytosol with antibodies against MEL-28, Nup107 or Nup205 as a control. The indicated proteins were identified by western blotting with respective antisera. (C) Depletion of the Nup107–160 complex reduces but does not fully deplete MEL-28: extracts were depleted of the Nup107–160 complex by two passages over an affinity resin. Untreated starting material, mock-depleted extracts and Nup107–160-depleted extracts after each round of depletion were analysed by western blotting with the indicated antisera. (D) Both MEL-28 and the Nup107–160 complex exist as free pools: extracts were separated on a Superose 6 gel filtration column and analysed by western blotting for the indicated proteins. Fraction 1 corresponds to the void; elution of dextran 2000 and ferritin markers for 2,000 and 550 kDa, respectively, are indicated. (E) MEL-28 is recruited to chromatin in the absence of the Nup107–160 complex, but not vice versa: sperm heads were incubated for 20 min in mock-depleted, Nup107–160-depleted, MEL-28-depleted or heat-inactivated extracts, fixed and analysed by immunofluorescence. Chromatin (upper row, blue) is stained with DAPI. MEL-28 or Nup160 (green) is shown in the lower row. Scale bar, 10 μm. 1st, first round; 2nd, second round; DAPI, 4,6-diamidino-2-phenylindole; MEL, maternal effect lethal; Nup, nucleoporin.
Western blot analysis using antisera specific for several nucleoporins showed that components of other NPC subcomplexes such as Nup205, Nup155, Nup98 or Nup93 were not co-immunoprecipitated in these experiments (Fig 4A). Nup107 antibodies co-precipitated MEL-28, confirming the interaction (Fig 4B). As shown above, depletion of MEL-28 led to a similar phenotype in NE assembly as depletion of the Nup107–160 complex (Harel et al, 2003; Walther et al, 2003a). However, removal of MEL-28 did not deplete the Nup107–160 complex from the cytosol (Fig 3A). Depletion of the Nup107–160 complex removed some MEL-28 from extracts, but did not remove it completely (Fig 4C). This suggests that a fraction of MEL-28 binds to the Nup107–160 complex, but there are independent free pools of MEL-28 and of the Nup107–160 complex in the cytosol. This was confirmed by size-exclusion chromatography: extracts were separated on a Superose 6 gel filtration column and the fractions were analysed by western blotting (Fig 4D). A fraction of both MEL-28 and constituents of the Nup107–160 complex co-eluted early after the void (peak in fractions 4–6). However, both MEL-28 and the Nup107–160 complex also eluted in additional, nonidentical peaks in fractions 8–10 and 14–18, respectively (Fig 4D), indicating that free pools of MEL-28 and of the Nup107–160 complex exist.
As depletion of either MEL-28 or the Nup107–160 complex resulted in a similar NE phenotype and both bind early to chromatin, we examined whether recruitment of one would be dependent on the other. When MEL-28-depleted extracts were used in a chromatin-binding reaction using membrane-free cytosol (Walther et al, 2003a), no recruitment of Nup160 was detected after 20 min as compared with mock-depleted extracts (Fig 4E). By contrast, when the Nup107–160 complex was depleted from extracts, MEL-28 still bound to chromatin (Fig 4E). This indicates that MEL-28 is necessary for the recruitment of the Nup107–160 complex to chromatin and thus acts earlier than the Nup107–160 complex in NPC assembly.
Conclusion
In summary, we have demonstrated that vertebrate MEL-28 localizes to the nuclear rim in interphase and on kinetochores during mitosis; it is important for NPC assembly both in mammals and Xenopus. We have shown that MEL-28 interacts with the Nup107–160 complex, an important building block of NPCs. When MEL-28 was depleted from Xenopus extracts, nuclei formed with a closed NE but with highly condensed chromatin, and NPCs were absent, a phenotype similar to that reported for the depletion of the Nup107–160 complex. However, the similar phenotypes were not due to co-depletion of MEL-28 and the Nup107–160 complex. Instead, MEL-28 was shown to be required for the recruitment of the Nup107–160 complex to chromatin. We suggest that MEL-28 acts as an anchoring platform for the Nup107–160 complex on chromatin and serves as a seeding point for NPC formation on the chromatin surface. MEL-28 association with chromatin is therefore the earliest event in NPC assembly identified so far.
Methods
Molecular cloning and protein purification. A full-length complementary DNA of Xenopus MEL-28 was generated by reverse transcription–PCR using X. laevis oocyte RNA as template and primers designed according to Xenopus expressed sequence tags. Human MEL-28 was PCR amplified from HeLa cDNA with primers designed according to the published ELYS sequence (Kimura et al, 2002). Imageclones IMAGp998D1112243Q3, IMAGp998N1114482Q3 and IMAGp998L218243Q1 for Xenopus Nup93, Nup205 and Nup160 were used as templates, respectively (Imageclones, RZPD, Deutsches Ressourcenzentrum für Genomforschung, Berlin, Germany).
Fragments of human MEL-28 (aa 1,572–2,266) and Xenopus MEL-28 (aa 1,602–2,120), Nup93 (aa 1–230), Nup160 (aa 1–414) and Nup205 (aa 1–230) were cloned into pET28a and expressed as a hexahistidine fusion protein in Escherichia coli strain BL21 (DE3) Rosetta or BL21 (DE3), purified using standard protocols and used for antibody production in rabbits. Antibodies against Nup107 and human LEM2 have been described previously (Walther et al, 2003a; Ulbert et al, 2007). mAb414 was purchased from Covance Research Products (Münster, Germany).
Cell culture, RNA interference and immunofluorescence. HeLa K and U2OS cells were kind gifts from Jan Ellenberg and Ulrike Kutay, respectively. For RNAi experiments, we used the siRNAs (sense sequences) MEL-28 (AATATCTACATAATTGCTCTT) and control (AATCGAAGTATTCCGCGTACG) at 50 nM final concentration and transfected using HiPerfect (Qiagen, Hilden, Germany).
Exclusion assay. For an assembly reaction, 1,500 sperm heads were incubated with 10 μg recombinant nucleoplasmin core (Antonin et al, 2005) and 0.5 μg biotinylated core histones (prepared using core histones (Upstate, Millipore, Schwalbach, Germany) and sulpho-NHS-LC-Biotin (Pierce, Perbio Science, Bonn, Germany)) for 30 min at 20°C. Floated membranes were bound to the chromatin template for 10 min. Nuclear assembly was initiated by the addition of 15 μl cytosol, 20 mg/ml glycogen and an ATP-regenerating system. After 2 h, 0.05 μg Oregon green-labelled streptavidin was added for 10 min. The reaction was quenched by the addition of 20 μl quenching buffer (100 mM KAc, 3 mM MgAc, 5 mM EGTA, 20 mM HEPES, 150 mM sucrose, 0.2 mg/ml biotinylated insulin; Sigma-Aldrich, Munich, Germany) for 30 min on ice. Samples were fixed and processed as for immunofluorescence.
Miscellaneous. Generation of an affinity resin, preparation of sperm heads and floated membranes, nuclear assembly reactions and TEM were carried out as described (Franz et al, 2005). For immunoprecipitation, high-speed interphase extracts were diluted 1:1 with cold PBS and then spun for 20 min at 100,000 r.p.m. in a TLA 100.4 rotor. The supernatant was applied to the affinity resin, bound for 2 h, washed with 0.5 M NaCl in PBS, and eluted with 0.2 M glycine and 0.15 M NaCl (pH 2.3).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank J. Ellenberg, M. Wilm and all members of the Mattaj lab for critical discussion of the manuscript.
References
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW (2005) The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17: 83–92 [DOI] [PubMed] [Google Scholar]
- Brachner A, Reipert S, Foisner R, Gotzmann J (2005) LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J Cell Sci 118: 5797–5810 [DOI] [PubMed] [Google Scholar]
- Fernandez AG, Piano F (2006) Network analysis of C. elegans early embryogenesis identifies MEL-28 as a central coordinator of chromatin maintenance and nuclear envelope function. Curr Biol 16: 1757–1763 [DOI] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW (2005) Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 24: 3519–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW (2006) MEL-28, a novel nuclear envelope and kinetochore protein essential for zygotic nuclear envelope assembly in C. elegans. Curr Biol 16: 1748–1756 [DOI] [PubMed] [Google Scholar]
- Gant TM, Wilson KL (1997) Nuclear assembly. Annu Rev Cell Dev Biol 13: 669–695 [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Bilbao-Cortés D, Walther TC, Gruss OJ, Mattaj IW (2000) GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell 5: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Walther TC, Mattaj IW (2005) Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 21: 347–380 [DOI] [PubMed] [Google Scholar]
- Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, Nakashima K, Nobuhisa I, Taga T (2002) Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells 7: 435–446 [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y (1983) Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science 220: 719–721 [DOI] [PubMed] [Google Scholar]
- Mansfeld J et al. (2006) The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell 22: 93–103 [DOI] [PubMed] [Google Scholar]
- Okita K, Kiyonari H, Nobuhisa I, Kimura N, Aizawa S, Taga T (2004) Targeted disruption of the mouse ELYS gene results in embryonic death at peri-implantation development. Genes Cells 9: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH, Laskey RA (1991) Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell 65: 569–578 [DOI] [PubMed] [Google Scholar]
- Prunuske AJ, Ullman KS (2006) The nuclear envelope: form and reformation. Curr Opin Cell Biol 18: 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S, Antonin W, Platani M, Mattaj IW (2007) The inner nuclear membrane protein Lem2 is critical for normal nuclear envelope morphology. FEBS Lett 580: 6435–6441 [DOI] [PubMed] [Google Scholar]
- Walther TC et al. (2003a) The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M (2003b) RanGTP mediates nuclear pore complex assembly. Nature 424: 689–694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


