Abstract
Chromosomes in eukaryotes are linear, whereas those of most, but not all, prokaryotes are circular. To explore the effects of possessing a linear genome on prokaryotic cells, we linearized the Escherichia coli genome using the lysogenic λ-like phage N15. Linear genome E. coli were viable and their genome structure was stable. There were no appreciable differences between cells with linear or circular genomes in growth rates, cell and nucleoid morphologies, genome-wide gene expression (with a few exceptions), and DNA gyrase- and topoisomerase IV-dependent growth. However, under dif-defective conditions, only cells with a circular genome developed an abnormal phenotype. Microscopy indicated that the ends of the linear genome, but not the circular genome, were separated and located at each end of a new-born cell. When tos—the cis-element required for linearization—was inserted into different chromosomal sites, those strains with the genome termini that were more remote from dif showed greater growth deficiencies.
Keywords: Escherichia coli , linear genome, N15 phage, TelN protein, dif
Introduction
There are two types of linear bacterial genome (Volff & Altenbuchner, 2000). One type, represented by the filamentous soil bacteria Streptomyces species, has a protein-designated terminal protein that is covalently joined to the 5′ ends of both termini of the genome, which is approximately 8-Mb. The terminal regions of the Streptomyces linear genome are unstable and frequently give rise to amplifications, large deletions and even circular chromosomes that are very unstable (Volff & Altenbuchner, 2000). The second type of linear bacterial genome, exemplified by the spirochete Borrelia burgdorferi (Ferdows & Barbour, 1989), has telomeres with covalently closed hairpin structures at their termini. The ends of linear Borrelia chromosomes are similar to those of the linear Borrelia plasmid, the Escherichia coli phage N15 (Rybchin & Svarchevsky, 1999) and certain animal viruses such as the poxvirus (Stuart et al, 1992).
The linearization mechanism of the Borrelia type genome has been established by analysing the linearization process of the N15 phage genome (Ravin, 2003). N15 is similar to the λ phage in many aspects, such as genome size and in having cohesive ends (Rybchin & Svarchevsky, 1999). However, unlike λ, N15 is lysogenized as a linear plasmid with hairpin termini. On infection, the injected linear genome circularizes through cohesive ends, as with the λ phage. For the linearization to lysogenize, two components—the tos site and protelomerase (TelN) protein—are required. As shown in Fig 1C–F, an almost perfect palindromic sequence, termed telLR (56 bp) at the centre of the tos site (312 bp) on the circular DNA (Fig 1C), is cut at staggered positions by the TelN protein (Fig 1D). Next, the two exposed single-stranded DNA regions are self-annealed (Fig 1E), and finally the remaining nicks are sealed by the TelN protein, producing two termini with hairpin structures (Fig 1F; Deneke et al, 2000). By using this N15 lysogenic system, we linearized the E. coli genome, and have compared the two strains with linear and circular genomes.
Figure 1.

Linearization process of a circular Escherichia coli chromosome mediated by the TelN protein. (A) The circle indicates the E. coli circular genome, in which oriC (replication origin) and the dif site are shown. The two arrows indicate bidirectional replication. (B) Expansion of the dif site from (A). The positions of the dif, tos and kan genes are shown as closed circles, grey and white boxes, respectively. The locations of the two probes used in hybridization experiments and the pair of primers used for the genome linearity PCR assay are shown beneath. The tos site (392 bp) shown in (B) is expanded in (C), and a part of tos (telRL: 56 bp) is expanded in (D). The almost perfect palindromic sequences are shown as a pair of dotted arrows in (D). TelN protein binds to the telRL site and cuts at the staggered positions indicated by vertical arrows in (D), producing two separate ends with long single-stranded regions, each of which can be self-annealed (E), and TelN seals the nicks, producing two ends with hairpin structure (F).
Results And Discussion
The tos sequence and a kan gene, which was used as a selection marker, were inserted into a site (−3 kb from the dif site) in the replication termination region of the E. coli chromosome by using the linear transformation method (see Methods; Fig 1A,B). Next, the tos–kan site was transduced into two wild-type strains—MG1655 and W3110—using the P1vir phage to establish two tos–kan strains. The telN gene was then separately inserted in-frame downstream of the pBAD24 arabinose promoter (see Methods) and the resulting plasmid (pBAD-telN) was transduced into two wild-type (tos) strains, generating MG(tos)(pBAD-telN) and W(tos)(pBAD-telN).
After confirming the presence of TelN activity in extracts of the MG(tos)(pBAD-telN) strain grown in the presence of arabinose, as expected (Deneke et al, 2000), genomic DNA was extracted from these cells and the above-described derivatives were grown under different conditions. On digestion with Streptopmyces phaeochromogenes (SphI) and after agarose gel electrophoresis, Southern hybridization experiments were carried out using a mixture of the two DNA fragments flanking the tos site as probes (Fig 1B). The circular genome digested with SphI yielded a single 6.71 kb DNA fragment (Fig 1B), whereas the linear genome was expected to produce two discrete smaller fragments of 5.25 and 1.46 kb. As shown in Fig 2A, lanes a and b, only a single band of DNA approximately 6.7 kb appears in MG1655 (tos)(+ or − pBAD24) grown in the presence of arabinose. By contrast, MG1655 (tos)(pBAD24-telN) (Fig 2A, lane c) grown in the presence of arabinose did not yield the 6.7 kb DNA band but instead two new bands of about 5.3 and 1.5 kb. However, the same cells grown in medium containing glucose (Fig 2A, lane d) yielded not only the original 6.7 kb fragment but also the two 5.3 and 1.5 kb bands derived from the linear genome, suggesting leaky expression of telN.
Figure 2.
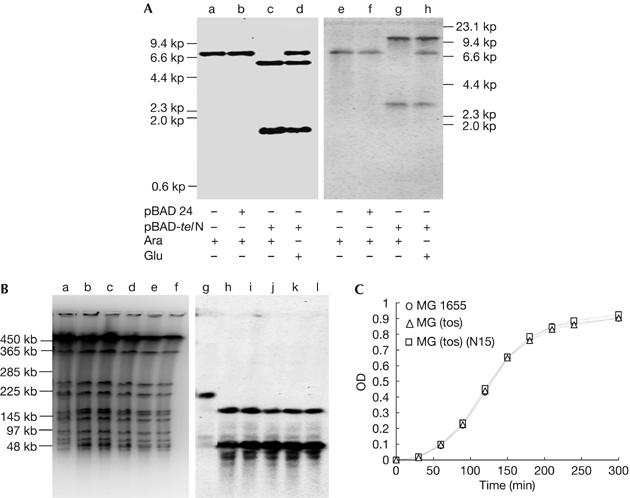
Linearization of the Escherichia coli genome, stability of the linearized genome and growth of the linear genome strain. Linearization assay using agarose gel electrophoresis under neutral and alkaline conditions. (A) Linearization assay using Southern hybridization of chromosomal DNA of MG1655(tos) and its derivatives extracted from overnight cultures, digested with SphI and separated by agarose gel electrophoresis under neutral (a–d) and alkaline (e–h) conditions. Probes were a mixture of probe 1 and 2 shown in Fig 1B. (a,e) MG(tos) in arabinose, (b,f) MG(tos)(pBAD24) in arabinose, (c,g) MG(tos)(pBAD24-telN) in arabinose and (d,h) MG(tos)(pBAD24-telN) in glucose. The positions of HindIII-digested λ phage markers are shown to the right and left of the gels. (B) Stability of the linear genome in the MG1655(tos)(N15) strain. Southern hybridizations were carried out after pulse-field gel electrophoresis of agarose-embedded cells grown from a single colony using a mixture of probes 1 and 2 shown in Fig 1B. (a–f) Ethidium bromide staining and (g–l) Southern hybridization patterns. (a,g) A parental MG1655 clone (30 generations), (b,h) an MG (tos)(N15) clone (30 generations), (c,d,i,j) two independent MG (tos)(N15) clones (84 generations) and (e,f,k,l) two independent clones (165 generations). (C) Linear genome strain growth curves. Overnight cultures (0.1 ml) of MG1655, MG1655 (tos) and MG1655(tos)(N15) were inoculated into fresh LB broth (10 ml) and shaken at 37°C; their optical densities (ODs) were measured by spectrophotometer at 770 nm at different time intervals.
Next, we examined whether the cleaved tos site had a hairpin structure. The same samples used in Fig 2A, lanes a–d, were analysed in the same way but under alkaline conditions (Fig 2A, lanes e and f; Deneke et al, 2000). Comparison of lanes d and h shows that the 6.7 kb fragment migrated in a similar manner under both conditions, whereas the 5.3 and 1.5 kb fragments migrated more slowly under alkaline conditions because they are twice as large—10.5 and 2.9 kb—as the bands seen under neutral conditions. This indicates that the terminal tos sites have been covalently closed. The difficulty in detecting the 6.7 kb DNA band in MG1655 (tos)(pBAD-telN) grown in the presence of arabinose suggests that almost all the genomes in the culture are present in a linear form. The same results were obtained using the W3110 strain (data not shown); therefore, we conclude that E. coli cells with linear genomes are viable.
Utilization of pBAD-telN as a TelN protein supplier, however, has two disadvantages: (i) controlling the arabinose concentration in the cell, and (ii) the presence of a mixture of replicons, both linear (chromosome) and circular (plasmid), in a single cell. The latter is a serious problem when investigating the effect of topoisomerase IV (topoIV) deficiency on cells with linear genomes. To avoid these problems, we used the N15 prophage as a TelN supplier. The N15 lysogen of MG1655 (tos) was isolated and the chromosomal DNA was extracted and analysed. The Southern hybridization pattern was similar to that in Fig 2A (data not shown), indicating that the MG1655 (tos) N15 lysogen genome is linear in both log and stationary phases. Furthermore, the pulse-field gel electrophoresis patterns in Fig 2B, lanes a and g, show that Anabaena variabilis (AvrII) digestion of MG1655 (N15) produced a single DNA band (199 kb), whereas the same digestion of MG1655 (tos) (N15) separated the original DNA fragment into two smaller bands (144 and 55 kb; Fig 2B, lanes b,h). The gel patterns of Fig 2B, lanes b–f,g–l (EtBr staining and DNA hybridization), remained unchanged for at least 170 generations, suggesting that not only linearity but also the whole genomic structure is stable. Therefore, in the following experiments, we used N15 prophage as the TelN supplier.
To measure the ratio of circular to linear genome in the sample used in Fig 2B, lane b, we carried out PCR across the tos site. This was unsuccessful with the linear strain, but successful with the circular genome strain (supplementary Fig 1 online); therefore, we could estimate that the amount of circular genomic DNA was at least 2 × 10−8-fold less than the linear form.
We next examined the growth rates of circular and linear genome strains under different culture conditions. Both strains showed similar growth rates in L broth (Fig 2C) and some other media (minimal (ME) glucose- and ME-glycerol medium) at 37°C. In L broth, total and viable cell numbers per millilitre at defined optical density points were comparable at 37°C, and their cell and nucleoid morphologies were indistinguishable (data not shown).
The differences between circular and linear genomes would be expected to follow topological constraints. Thus, we examined whether there was a difference in growth between strains with circular or linear genomes in DNA gyrase (gyrB) (see Methods) and topoIV (parC and ParE) temperature-sensitive mutants (see Methods). We found no differences in colony-forming capacity at different temperatures (data not shown). The bacterial genome is organized into closed domains, which are regions of DNA that are topologically constrained at their ends and behave independently of the rest of the chromosome (Postow et al, 2001; Espeli & Marians, 2004). This might explain gyrB-dependent growth of the linear genome strain. Conversely, catenations are thought to accumulate at the replication termination region owing to incomplete resolution of positive supercoils and conversion from precatenanes (Postow et al, 2001; Espeli & Marians, 2004), which are most likely formed between the replicated duplexes behind the replication fork. However, the above results are hard to reconcile with this interpretation. Catenanes might accumulate at multiple genomic regions, in addition to the replication termination region. For example, precatenanes might remain at multiple chromosomal regions in the topoIV mutant, or multiple replication origins, as in eukaryotic genomes. The latter possibility, however, seems unlikely because the linear genome strain showed dnaA+-dependent growth and the replication origin of this strain seemed restricted to the oriC region, according to gene dosage analysis using DNA microarrays (K.-i.K. & T.H., unpublished data). Whole-genome sequence data of bacteria with linear genomes showed the presence of genes coding for both DNA gyrase and topoIV, suggesting that both are required for the growth of these organisms (Fraser et al, 1997; Bentley et al, 2002; Ikeda et al, 2003). However, these previous data could not exclude the possibility that these enzymes were required for replication and segregation of circular plasmids, which, now or previously, coexisted in these linear genome bacteria. Our present data, however, eliminate this possibility.
As topological constraints affect transcription, we examined genome-wide gene expression patterns of the linear and circular genome strains grown in LB broth at 37°C. The expression of a total of 4,300 genes was compared and only three genes were found to have significantly different expression (>fourfold difference). The expression of two of these, avtA (alanine–valine transaminase gene: 80 min) and sulA (cell division inhibitor gene: 22 min), was increased 6.49- and 5.55-fold in the linear genome strain, respectively, whereas the other was decreased (ygdB_b2824 at 64 min: 0.22-fold). Thus, gene expression remains largely unchanged between linear and circular genome forms. Gene chip data were deposited in Arrayexpress under accession number E-MEXP-910.
Another structural difference to be expected is that the circular, but not linear, genome can form a dimer. With circular chromosomes, a dimer is formed through odd numbers of recombination events between the sister chromosomes (Kuempel et al, 1991), but with linear genomes no dimers can be produced even if sister-chromosomal recombination occurs. The resolution of the dimer requires dif site-specific recombination catalysed by cer-specific recombinase C and D (XerCD) and FtsK proteins. Thus, on wild-type backgrounds, dif or xerCD mutants show slower growth, produce more elongated cells and partially induce an SOS response (Kuempel et al, 1991; Cornet et al, 1996). To examine whether these phenotypes are also generated in organisms with a linear genome, we compared the growth and cell morphology of Δdif mutants with circular or linear genomes. The three Δdif circular genome derivatives (MG1655Δdif, MGΔdif(N15) and MGΔdif(tos)) grew more slowly and produced more elongated cells than the parental strain, as shown in Fig 3A,C–E. By contrast, growth and cell morphology of the linear genome strain MGΔdif(tos)(N15) was similar to that of the parental strain. Thus, normal growth of a strain with a linear genome does not require the dif site-specific recombination system. This is consistent with sequence data from naturally occurring bacteria with linear genomes (Fraser et al, 1997; Recchia & Sherratt 1999; Bentley et al, 2002; Ikeda et al, 2003), which lack genes homologous to xerCD.
Figure 3.

Effect of dif deficiency on the linear genome strains. Four different dif-defective MG1655 derivatives were constructed and their growth rates and cell morphologies were compared. (A) Growth curves were produced as in Fig 2C. Cell morphologies of the parental and the four derivative strains are shown: (B) MG1655, (C) MGΔdif, (D) MGΔdif(tos), (E) MGΔdif(N15) and (F) MGΔdif(tos)(N15). OD, optical density.
In Streptomyces cells, each end of the linear genome was found to be associated with the other (Yang & Losick, 2001), probably through the two terminal proteins bound covalently to the 5′ end of each terminus. To identify the position of each terminus of the E. coli linear genome in cells, we followed the method developed by Sherratt's group (Wang et al, 2005; see Methods). We investigated the position of fluorescence foci of cyan fluorescent protein and yellow fluorescent protein derivatives of the LacI and TetR repressors binding to their operator arrays inserted into sites close to the two termini (designated ter1 and ter2 sites: −20 and +20 kb from dif). Two control strains were used for associated and separated foci: one is the isogenic circular genome strain and the other is the strain carrying one array located at the site close to oriC and the other at ter1. Microscopy using samples from cultures grown in log phase in M9-succinate minimal medium (generation time about 120 min) showed that, as expected, the two foci at oriC and ter1 were spatially separated in most of the cells (87.5%; Fig 4A,D), whereas almost all foci at ter1 and ter2 on the circular genome were close to one another or were overlapping (97.5%; Fig 4B,D) throughout the cell cycle. By contrast, the two foci in linear genome cells showed two different patterns (Fig 4C,D). One was similar to the typical ter1–ter2 patterns of circular genome strains, where the two foci are associated at the middle of the cell (37.6%). However, the other was never seen in circular genome samples; each focus was separated at each end of the cell (49.7%). This pattern was also found most commonly (52.3%) in the presence of both inducers, isopropyl-β-D-thiogalactoside (5 μM) and anhydrotetracycline (40 ng/ml; supplementary Fig 2 online; see Methods). In the experiments reported by Wang et al (2005), microscopy showed that ter1 (−20 kb) and ter2 (+20 kb) on the linear genome behave in a strikingly similar manner to ter4 (−210 kb) and ter2 (+210 kb; this is different from our ter2 site) on the circular genome. As the oriC site in the linear genome seems to behave in a similar manner to the same site in the circular genome (data not shown), most (>90%) of the chromosome (ter4-oriC-ter2 region, according to Sheratt's nomenclature in Wang et al (2005)) might behave in a similar manner irrespective of whether the genome is circular or linear. In any case, and this is different from Streptomyces, there is a stage when the two termini separate and locate in defined side regions. This pattern is shown as a model in supplementary Fig 3 online, together with the pattern of a circular genomic cell for comparison.
Figure 4.
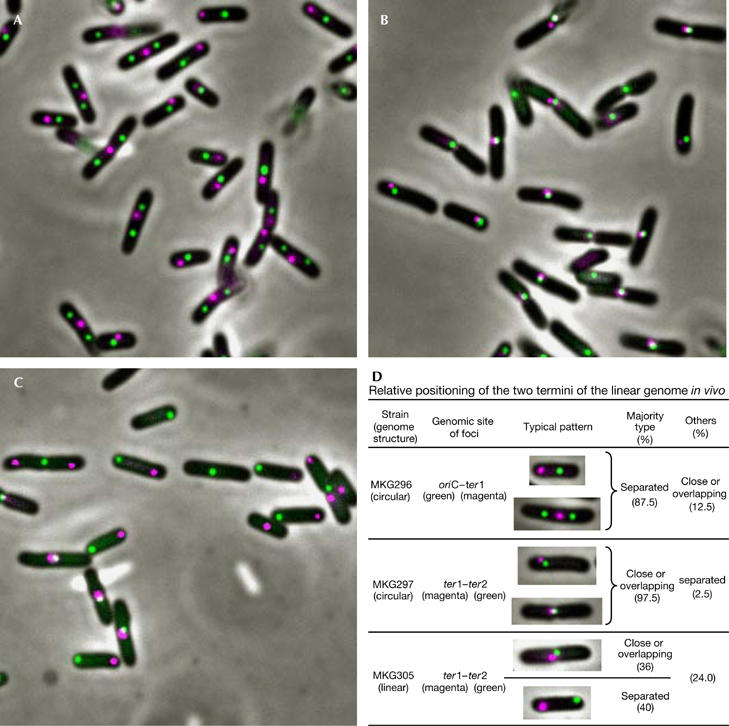
Relative positioning of the two termini of the Escherichia coli linear genome in vivo. For details on the strains and experiments, see text and Methods. (A) Strain MKG296 (circular genome). Relative positioning of oriC (green) and ter1 (red, −20 kb from dif) can be seen. (B) Strain MK297 (circular genome). Relative positioning of ter1 (red) and ter2 (green, +20 kb from dif) can be seen. (C) Strain MK305 (linear genome). Relative positioning of ter1 (red) and ter2 (green) can be seen. (D) Summary table of relative positions of the two foci in each strain. All cells (126 cells of MKG296; 234 cells of MKG297; 189 cells of MKG305 were used) were classified into the following three patterns: two foci are (i) separated, (ii) close or overlapping, or (iii) have other patterns. The percentage of each pattern and the typical principal patterns are shown in the table. oriC, replication origin.
Ring chromosomes have been found in various organisms, including humans. Budding and fission yeast cells containing circular chromosomes are unstable, both mitotically and meiotically (Haber et al, 1984; Fan et al, 1992). A fission yeast with three circularized chromosomes grew slowly and contained cells showing anaphase bridges and aneuploidy in mitotic division; they were also infertile (Naito et al, 1998). Thus, eukaryotic circular chromosomes behave in the same way as the circular chromosome in the dif− mutant, whereas E. coli linear chromosomes behave like normal mitotic eukaryotic chromosomes with telomeres.
The termini of the linear genome we constructed here are much closer to dif (−3 kb from dif; Fig 1). If the ends are farther away from dif, does any phenotypic change occur? To examine this, the tos site was moved to five different positions on the chromosome, as shown in Fig 5A. E. coli cells containing a linear chromosome could be generated, except when the tos site was close to the region of replication origin (94 min). When the linearization occurred close to the terminus region, no appreciable differences in growth rate were detected between strains MG1655(−3 kb∷tos)(N15) (the original linear genome strain), MG(−200 kb∷tos)(N15) or MG(+200 kb∷tos)(N15) (Fig 5B). The further away the linearization of the chromosome occurred from the terminus region, the stronger the growth defect was relative to wild-type cells. In samples of +200 kb and −200 kb linear genome strains, microscopy showed that cell size was normal but filamentous cells were produced, although in small number. Conversely, there was an apparent increase in the cell size of 7 and 20 min linear genome strains, and also in the proportion of filamentous cells compared with the non-filamentous cells. This correlation might be caused by unbalanced replication of a pair of chromosome arms with different lengths—a disadvantage of polar sequence-dependent translocation activity of the FtsK protein (Lesterlin et al, 2004), and/or collision between replication and transcription. In any case, our linear genome system will provide a very useful tool to address many issues related to chromosome biology in prokaryotes.
Figure 5.
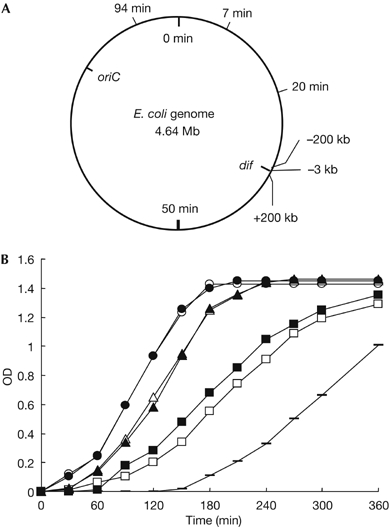
Terminus location-dependent growth of strains with a linear genome. (A) The circle represents a 0–100 min chromosome map, on which two cis sites, dif and oriC, are shown inside. The other six tos insertion sites, 94 min, 7 min, 20 min, −200 kb, −3 kb (the original linear genome strain) and +200 kb from the dif site, are also shown outside. (B) We inserted the tos-Km fragment into different sites of the chromosome of MG1655 (or MG) shown in (A), isolated their N15 lysogens and measured their growth as follows: 100 μl of overnight culture of each strain was inoculated into 10 ml of fresh L broth, shaken at 37°C and growth measured spectroscopically at OD600 nm. MG1655(wild type) (open circles); MG(−3 kb∷tos)(N15) (filled circles); MG(−200 kb∷tos)(N15) (open triangles); MG(+200 kb∷tos)(N15) (open squares); MG(+200 kb∷tos)(tus−)(N15) (filled triangles); MG(7 min∷tos)(tus−)(N15) (horizontal bars); MG(20 min∷tos)(tus−) (N15) (filled squares). OD, optical density; oriC, replication origin.
Methods
Strains, phage, plasmid, media and routine methods. The E. coli strains, plasmids and phages used are described in the supplementary information online. Media and E. coli genetic methods were as described previously (Miller, 1992). DNA manipulations, such as extraction, DNA digestion, cloning and sequencing, were carried out as described previously (Sambrook et al, 1989). Agarose and pulse-field gel electrophoresis are described in the supplementary information online.
Insertion of the tos-Km fragment into E. coli chromosome. Essentially, the linear DNA transformation method (Datsenko & Wanner, 2000) was used and is described in the supplementary information online.
Construction of lacO and tetO arrays. We followed the method developed by Dr Sherratt's group (Lau et al, 2003; see the supplementary information online for details).
Microscopy, PCR assay for genome linearity and Affymetrix DNA microarray analysis. These were carried out as described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr N. Ravin (Russian Academy of Sciences, Moscow, Russia), Dr D. Sherratt (University of Oxford), Dr J. Kato (Tokyo Metropolitan University), Dr T. Baba (Nara Institute of Science and Technology), Dr A. Nishimura (National Institute of Genetics) and Dr S. Yasuda (National Institute of Genetics) for strains, phage and plasmid. We thank Dr A. Kikuchi (Nagoya University) and G. Austen (National Institute for Basic Biology) for discussion and critical reading of the manuscript. This work was supported in part by grant 13141205 from the Ministry of Education, Science and Culture, Japan. T.C. was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship.
References
- Bentley SD et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147 [DOI] [PubMed] [Google Scholar]
- Cornet F, Louarn J, Patte J, Louarn JM (1996) Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev 27: 1152–1161 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneke J, Ziegelin G, Lurz R, Lanka E (2000) The protelomerase of temperate Escherichia coli phage N15 has cleaving–joining activity. Proc Natl Acad Sci USA 97: 7721–7726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Marians KJ (2004) Untangling intracellular DNA topology. Mol Microbiol 52: 925–931 [DOI] [PubMed] [Google Scholar]
- Fan JB, Rochet C, Gaillardin C, Smith CL (1992) Detection and characterization of a ring chromosome in the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res 20: 5943–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdows MS, Barbour AG (1989) Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc Natl Acad Sci USA 86: 5969–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586 [DOI] [PubMed] [Google Scholar]
- Haber JE, Thorburn PC, Rogers D (1984) Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics 106: 185–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21: 526–531 [DOI] [PubMed] [Google Scholar]
- Kuempel PL, Henson JM, Dircks L, Tecklenburg M, Lim DF (1991) dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol 3: 799–811 [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okastad O-A, Barre F-X, Sherratt DJ (2003) Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol 49: 731–743 [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Barre FX, Cornet F (2004) Genetic recombination and the cell cycle: what we have learned from chromosome dimmers. Mol Microbiol 54: 1151–1160 [DOI] [PubMed] [Google Scholar]
- Miller JH (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Naito T, Matsuura A, Ishikawa F (1998) Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet 20: 203–206 [DOI] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzaelli NR (2001) Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci USA 98: 8219–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV (2003) Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol Lett 221: 1–6 [DOI] [PubMed] [Google Scholar]
- Recchia GD, Sherratt DJ (1999) Conservation of xer site-specific recombination genes in bacteria. Mol Microbiol 34: 1146–1148 [DOI] [PubMed] [Google Scholar]
- Rybchin VN, Svarchevsky AN (1999) The plasmid prophage N15: a linear DNA with covalently closed ends. Mol Microbiol 33: 895–903 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning; A Laboratory Manual, 2nd ed. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Stuart D, Ellison K, Graham K, McFadden G (1992) In vitro resolution of poxvirus replicative intermediates into linear mini-chromosomes with hairpin termini by a virally induced. J Virol 66: 1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Altenbuchner J (2000) A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol Lett 186: 143–150 [DOI] [PubMed] [Google Scholar]
- Wang X, Possoz C, Sherratt D (2005) Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev 19: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MC, Losick R (2001) Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J Bacteriol 183: 5180–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


