Abstract
The αB-crystallin and HspB2 genes are located ≈0.9 kb apart in a head-to-head arrangement in mammals. Previous experiments have shown that a truncated −668/+45 αB-crystallin enhancer/promoter fragment from blind mole rats (Spalax ehrenbergi), which have nonfunctional lenses, lacks lens activity and has enhanced muscle activity in transgenic mice. Here we show that the full-length mole rat αB-crystallin intergenic region behaves similarly in transgenic mice. A two-nucleotide mutation (−273CA→G) in the mouse αB-crystallin enhancer/promoter fragment mimicking the wild-type mole rat sequence functionally converted the mouse promoter fragment to that of the wild-type mole rat promoter when tested in transgenic mice. The reciprocal mutation in the mole rat promoter fragment (−272G→CA) did not affect its activity. Oligonucleotides from the wild-type mouse and mole rat αB-crystallin promoter region under study formed distinct complexes with nuclear proteins from cultured cells. The mouse mutant sequence lost binding ability, whereas the mutated mole rat sequence gained the ability to form a complex similar in size to that of the wild-type mouse oligonucleotide. Our data support the idea that blind mole rats' αB-crystallin promoter activity was modified during the evolution of subterranean life and shows that tissue-specific promoter activity can be modulated by changing as few as two apparently neutral nucleotides in the mouse αB-crystallin enhancer region, implying the importance of the context of regulatory sequences for promoter activity.
Keywords: evolution, gene expression, lens, muscle
The α-crystallins (αA and αB) are among the major soluble proteins of mammalian lenses (1). αB-crystallin is a stress-inducible, small heat-shock protein that is constitutively expressed in the lens and to a lesser extent in many other tissues, especially heart and skeletal muscle (2, 3). An upstream, muscle-preferred enhancer (−426/−259) of the αB-crystallin gene is essential for expression in non-lens tissues and boosts expression in the lens (4), whereas a proximal promoter fragment (−164/+44) without the enhancer is sufficient for lens-specific promoter activity in transgenic mice (5, 6). The enhancer contains at least five cis-elements and is orientation-dependent in its influence on the αB-crystallin promoter in transgenic mice (7). The proximal promoter fragment (−164/+44) without the enhancer has lens-specific activity but not muscle activity in transgenic mice (5, 6) and has regulatory motifs that bind Pax 6, large Maf proteins, and retinoic acid receptors (8, 9). In mammals, the αB-crystallin gene is situated in a head-to-head arrangement with the Mkbp/HspB2 (myotonic dystrophy protein kinase-binding protein/ heat shock protein B2) gene, another member of the small heat-shock protein family, with an intergenic region of ≈0.9 kb (10, 11). Unlike the αB-crystallin gene, the Mkbp/HspB2 gene is not expressed in the mouse lens and is not stress-inducible (11).
The adult subterranean blind mole rat (Spalax ehrenbergi) has degenerate s.c. eyes with lenses containing disorganized, vacuolated cells (12, 13). A low level of α-crystallin gene expression has been detected in the eye and lens of the blind mole rat (12, 14, 15). When tested in transgenic mice, the αB-crystallin-truncated enhancer/promoter fragment (−668/+45) of blind mole rats has little if any activity in the lens but an excess of activity in skeletal muscle relative to the corresponding (−661/+43) mouse promoter fragment (15). However, a later report indicated that a mole rat αB-crystallin promoter fragment spanning the entire intergenic region between the αB-crystallin and the Mkbp/HspB2 genes can drive a reporter gene in the lens of transgenic Xenopus larvae (16). This finding raises the possibility that the control elements for lens-specific promoter activity are present upstream of the truncated mole rat promoter fragment that we used in the transgenic mouse experiments (15). Here we consider that the truncated αB-crystallin enhancer/promoter fragment (−668/+45) of blind mole rats lacks one or more critical DNA regulatory elements needed for lens expression. Indeed, DNA control elements might be situated at considerable distances from the crystallin gene that they regulate (17). Moreover, similar cis-control elements are often configured with distinct spatial arrangements in different crystallin genes (18–23). We showed that the entire mole rat intergenic region between the αB-crystallin and Mkbp/HspB2 genes has low lens-promoter activity and elevated muscle activity in adult transgenic mice relative to that of the comparable mouse αB-crystallin enhancer/promoter fragment. We thus explored whether specific sequence changes could be found that might contribute to the low lens activities and elevated muscle activities of the blind mole rat αB-crystallin enhancer/promoter fragment in transgenic mice. We found, surprisingly, that a 2-nt mutation 3′ to the known cis-regulatory motifs of the mouse αB-crystallin enhancer mimicking the mole rat-like sequence in that region (which is variable among mammals) abolishes lens promoter activity and elevates muscle promoter activity in transgenic mice.
Results
Mouse and Mole Rat Promoter Activities in Transgenic Mice.
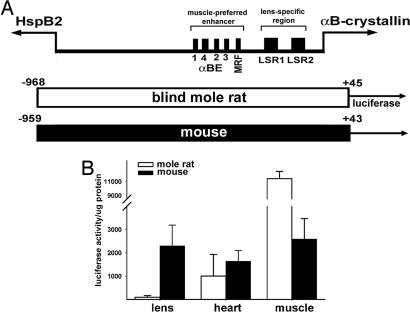
The blind mole rat αB-crystallin and Mkbp/HspB2 genes are situated head-to-head along the chromosome with an intergenic distance of 906 bp. We isolated the −968/+45 fragment of the blind mole rat αB-crystallin gene, which spans the whole intergenic region (Fig. 1A). The −968/+45 blind mole rat sequence is 78% identical to the corresponding −959/+43 mouse sequence, with 89% identity in the muscle-preferred enhancer region (−426/−259) and 93% identity in the lens-specific promoter (−164/+44) region.
Fig. 1.
Activity of the full-length (entire intergenic region) blind mole rat and mouse αB-crystallin promoters. (A) Schematic representation of the bidirectional organization of the αB-crystallin and HspB2 genes. Individual regulatory elements identified in the αB-crystallin enhancer (αBE) and proximal promoter are shown. Luciferase reporter gene constructs from the mole rat (white bar) and mouse (black bar) are shown. The αB-crystallin and HspB2 genes are indicated by the labeled arrows. All numbers are relative to the transcription initiation site (+1) of the αB-crystallin gene of the respective species. The translational initiation sites at positions +48 (mole rat) and +46 (mouse) were not included in the reporter gene constructs. The HspB2 genes begin at positions −863 (mouse) and −905 (mole rat), respectively. MRF, myogenic regulatory factor binding site; LSR, lens-specific regulatory element. (B) Blind mole rat and mouse αB-crystallin promoter activity in transgenic mice. White bars represent average values for four founder lines of transgenic mice with the mole rat promoter:luciferase transgene; black bars represent average values for two founder lines of transgenic mice with the mouse promoter:luciferase transgene. Three to six siblings were analyzed per founder.
The blind mole rat αB-crystallin −968/+45 gene sequence was subcloned upstream of the luciferase reporter gene, and its promoter activity was compared with the corresponding mouse αB-crystallin −959/+43 gene sequence (7). Preliminary transfection experiments were conducted with mouse αTN4 lens cells and C2C12 mouse myoblasts that differentiate in culture into myotubes. Although both promoter fragments showed preferential activity in the muscle cells, the ratio of mole rat to mouse promoter activity was 0.3 in the αTN4 lens cells and 1.8 in the differentiated C2C12 myotubes (data not shown).
We next generated transgenic mice with the mole rat −968/+45 promoter:luciferase transgene. Consistent with the transfection results, the −968/+45 mole rat promoter fragment showed low lens activity but high skeletal muscle activity in four lines of transgenic mice (Fig. 1B). The average luciferase activity generated by the mole rat intergenic promoter fragment was ≈100-fold less in the lens than in skeletal muscle. The lens activity of the mouse intergenic promoter fragment, by contrast, was comparable to the skeletal muscle activity in the transgenic mice. The activity of the mole rat intergenic promoter was 25-fold less than that of the mouse intergenic promoter fragment in lens and 4-fold more than that of the mouse promoter in skeletal muscle in the transgenic mice. The mole rat and mouse intergenic promoter fragment activities were comparable in the heart of the transgenic mice. The low lens activity and high muscle activity of the −968/+45 mole rat promoter fragment in the transgenic mice are consistent with our previous observations made by using a truncated −668/+45 blind mole rat promoter fragment (15) and establish that the tissue-specific activity of the blind mole rat αB-crystallin intergenic promoter fragment in transgenic mice differs from that of the mouse.
Functional Switch of the Mouse Promoter Activity by a 2-nt Mutation.
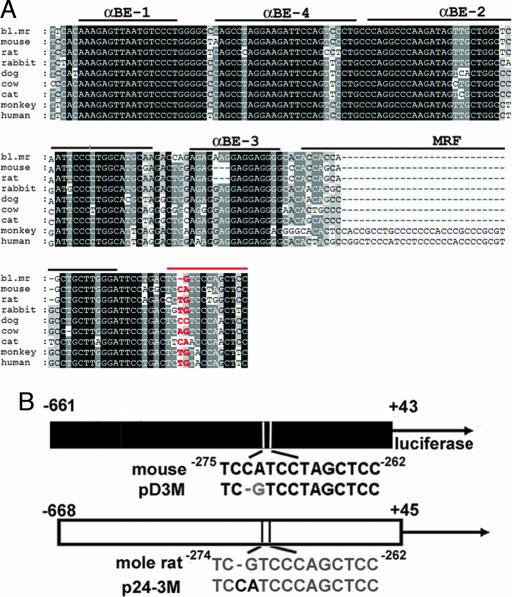
As an initial step in identifying a candidate sequence contributing to the functional differences between their tissue-specific activities, we compared the mole rat and mouse sequences in the lens-specific proximal promoter and the muscle-preferred enhancer regions (21, 24). We noted a potential regulatory sequence at position −274 to −262 in the blind mole rat immediately downstream of the muscle regulatory factor-binding element of the αB-crystallin enhancer (Fig. 2A). The −274/−262 mole rat sequence, but not its mouse counterpart, resembles the DNA recognition sequences for the Pax3 paired domain (see figure 3B of ref. 25). This resemblance kindled our interest inasmuch as Pax3 contributes to gene expression in muscle (26), where the mole rat αB-crystallin promoter is very active in the transgenic mice. The −274/−262 mole rat sequence differs from its mouse counterpart by three nucleotides, the first two of which appear critical for Pax-3 binding and bear the least sequence conservation among different species (see Fig. 2A). Indeed, this region is more variable than enhancer regulatory elements αBE-1, αBE-4, and αBE-2 and is 3′ to all of the enhancer elements that have been identified to date (Fig. 2A). Because the mole rat αB-crystallin promoter is preferentially active in muscle, we explored the possible functional significance of the −274/−262 sequence further.
Fig. 2.
Sequence comparisons and mutagenesis of the αB-crystallin enhancer/promoter fragments. (A) The blind mole rat (bl. mr) enhancer sequence (−422/−262) is compared with those from mouse, rat, rabbit, dog, cow, cat, monkey, and human. The conserved sequences are shaded based on the identity. The cis-regulatory elements identified by DNase I footprinting (43) are indicated above the sequences. The red line indicates the putative Pax3 binding site; the two nucleotides selected for mutation in the mole rat and mouse are indicated in red letters. (B) Site-directed mutagenesis creating a mouse −661/+43 mutant (pD3M) in which the −273CA was converted into G and a blind mole rat −668/+45 mutant (p24-3M) in which the −272G was converted into CA. Gray letters indicate the blind mole rat sequences; black letters indicate the mouse sequences.
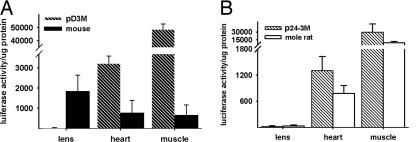
Fig. 3.
Promoter activities of the pD3M and p24-3M mutants in transgenic mice. (A) Promoter activities of the mouse wild-type (black bars) and mutant (pD3M; hatched bars) αB-crystallin promoter constructs in transgenic mice. Black bars represent averages of four founder lines of transgenic mice with the mouse promoter:luciferase transgene. Patterned bars represent averages of four founder lines of transgenic mice with the pD3M mutant:luciferase transgene. (B) Promoter activities of the mole rat wild-type (white bars) and mutant (p24-3M; hatched bars) αB-crystallin promoter constructs in transgenic mice. White bars represent averages of four founder lines of transgenic mice with the mole rat promoter:luciferase transgene. Patterned bars represent averages of two founder lines of transgenic mice with the p24-3M mutant promoter:luciferase transgene. Three to six siblings were analyzed for each founder.
The −273CA nucleotides in the mouse αB-crystallin −661/+43 promoter were changed to a single G, which occurs in the mole rat promoter, and the −272G nucleotide in the mole rat αB-crystallin −668/+45 promoter was mutated to CA, which occurs in the mouse promoter in that region (Fig. 2B). We named the mutant mouse promoter fragment pD3M and the mutant mole rat promoter fragment p24-3M (Fig. 2B). The promoter activities of the mutant fragments were compared with their wild-type counterparts. Transfection experiments showed that pD3M promoter activity was similar to that of the wild-type −661/+43 mouse promoter in lens cells but higher than that of the wild-type promoter in myoblasts (data not shown).
Transgenic mice were made by using a pD3M:luciferase transgene, and the luciferase activity in the lens, heart, and skeletal muscle of those mice was compared with that in the corresponding tissues in 8-week-old transgenic mice carrying the wild-type −661/+43 mouse promoter:luciferase transgene. As expected, the wild-type mouse promoter exhibited its highest activity in the lens (15). The average wild-type mouse promoter activities in heart and skeletal muscle were ≈41% and 35% of the lens activity, respectively (Fig. 3A). Surprisingly, pD3M promoter activity in the transgenic mouse lens was <1% of that in skeletal muscle. The pD3M mutant promoter activity was at least 100-fold less in the lens and 74-fold greater in the muscle than the wild-type promoter activity in the transgenic mice (Fig. 3A). The pD3M mutant promoter activity was ≈4-fold higher in heart than the wild-type mouse promoter activity. In view of the unexpected nature of this result, the pD3M:luciferase transgene was isolated by PCR from a transgenic mouse, and the mutated region was confirmed by sequencing (data not shown). Hence, the mutation of −273CA→G converted the mouse promoter into a wild-type mole rat promoter with respect to having little lens activity and enhanced muscle activity.
We next attempted to functionally convert the mole rat promoter into a mouse promoter by producing a corresponding site-specific mutation in the same region. A reciprocal mutant of pD3M, termed p24-3M, was generated in which the single −272G nucleotide of the mole rat αB-crystallin −668/+45 promoter was modified into −273CA nucleotides similar to those existing in the analogous region of the mouse promoter (Fig. 2B). The mutant p24-3M and wild-type mole rat promoter activities were comparable in the transfected αTN4-1 lens cells and C2C12 myoblasts (data not shown). In transgenic mice, the p24-3M:luciferase transgene was expressed in the lens at <1% of that expressed in skeletal muscle, which is similar to the muscle-preferred activity of the wild-type mole rat promoter (Fig. 3B). The fact that the mole rat p24-3M mutant αB-crystallin promoter did not adopt a mouse-like functional activity level indicates that the −273CA (mouse)/−272G (mole rat) nucleotide difference is not sufficient to account for the functional differences in αB-crystallin promoter activity between these two rodent species.
EMSAs.
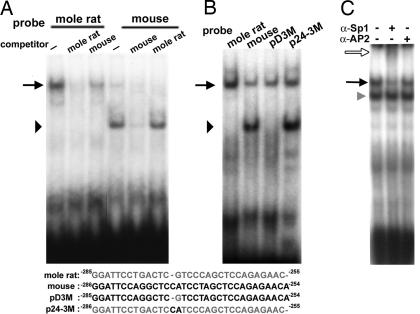
We tested whether oligonucleotides derived from the wild-type mole rat and mouse promoter sequences bind similar nuclear proteins when EMSAs are performed and whether the introduced mutations affect such binding. EMSAs were performed by using radiolabeled oligonucleotides spanning the mole rat αB-crystallin −285/−255 region, and nuclear extracts were used from αTN4-1 mouse lens cells, C2C12 mouse myoblasts, and HeLa cells. The results were similar with each extract and are shown for the C2C12 cells (Fig. 4A). Incubation of the labeled mole rat probe with C2C12 nuclear extract produced one major complex (Fig. 4A, black arrow). A 100-fold molar excess of unlabeled mole rat probe abolished complex formation with the labeled probe; comparable incubation with 100-fold molar excess of the analogous unlabeled mouse oligonucleotide reduced but did not abolish the radioactive EMSA band. Use of the mouse radioactively labeled αB-crystallin −286/−254 probe produced a weak complex that comigrated with that produced by the mole rat oligonucleotide and a major, more rapidly migrating complex (Fig. 4A; black arrowhead). Both mouse radioactive complexes were virtually abolished in the presence of a 100-fold molar excess of unlabeled mouse probe; by contrast, a 100-fold molar excess of the analogous mole rat unlabeled probe abolished the larger complex but did not affect the formation of the smaller complex. Thus, the mouse −286/−254 promoter sequence complexes with a distinct factor(s) that does not bind the corresponding mole rat sequence, and the mole rat −285/−255 promoter sequence complexes strongly with a nuclear protein(s) that is bound weakly by the corresponding mouse sequence.
Fig. 4.
Nuclear protein–DNA complexes using the mole rat −285/−255 and mouse −286/−254 αB-crystallin oligonucleotides. (A) EMSAs were carried out by using C2C12 nuclear extracts and radiolabeled oligonucleotides spanning either the −285/−255 sequence of the mole rat promoter or the −286/−254 sequence of the mouse αB-crystallin promoter. A 100-fold molar excess of unlabeled competitor oligonucleotides was included as indicated. The arrow indicates the mole rat complex, and the arrowhead indicates the mouse complex. (B) EMSAs were carried out by using C2C12 nuclear extracts and radiolabeled oligonucleotides derived from wild-type mole rat and mouse and mutant mole rat (p24-3M) and mouse (pD3M) promoter fragments. Oligonucleotide sequences of the radiolabeled probes are shown below the gel images. Gray letters represent the blind mole rat nucleotides; black letters represent the mouse nucleotides. The arrowhead indicates the major complex formed with the mouse oligonucleotides, and the arrow indicates the complex formed with the mole rat oligonucleotides. (C) The radiolabeled mole rat probe was incubated with a HeLa nuclear extract and specific Sp1 or AP2 antibodies. The black arrow indicates the major complex formed by using the mole rat oligonucleotide; the white arrow indicates the supershifted complex containing anti-Sp1 antibody; and the gray arrowhead indicates a nonspecific complex that could not be competed with the unlabeled mole rat probe (data not shown).
We next performed EMSAs with the mutant mouse pD3M and mole rat p24-3M oligonucleotides. The pD3M oligonucleotide did not form the small complex that is characteristic of the wild-type mouse oligonucleotide (Fig. 4B) and did not compete for complex formation with the wild-type mouse probe (data not shown). The pD3M probe did form a large complex that comigrated with that produced by the analogous −285/−255 wild-type mole rat oligonucleotide (Fig. 4B). The unlabeled wild-type mole rat oligonucleotide competed with the radiolabeled pD3M probe for formation of the large complex, consistent with the pD3M and p24-3M large complexes comprising the same proteins (data not shown). The mole rat p24-3M oligonucleotide formed a complex that comigrated with the large complex of the wild-type mole rat oligonucleotide and a complex that comigrated with the small complex of the wild-type mouse oligonucleotide (Fig. 4B).
Finally, Pax3 does not appear to interact with the mole rat probe as was originally considered. A Pax3 consensus sequence (27) did not compete with the complex formed between mole rat probe and C2C12 nuclear extracts, and the Pax3 consensus sequence complex migrated faster than that formed by using the mole rat probe incubated in a C2C12 nuclear extract (data not shown). Initial experiments performed with HeLa nuclear extract showed that Sp-1 antibody was able to supershift the complex with the wild-type mole rat probe (Fig. 4C). In addition, a 10-fold molar excess of the unlabeled Sp1 consensus sequence (28) abolished complex formed between the mole rat probe and C2C12 nuclear extract (data not shown). These data suggest that the mole rat oligonucleotide probe interacts with a Sp1-like protein and not Pax3.
Discussion
Blind mole rat eyes regress during embryogenesis, resulting in s.c. degenerate eyes that are compatible with the subterranean lifestyle of the mature animals (13, 29–31). Although visually nonfunctional, the atrophied blind mole rat eye appears to have a role in controlling circadian rhythms (32, 33). In the present study we show that, in contrast to the mouse intergenic promoter fragment, the entire intergenic region of the blind mole rat αB-crystallin gene shows little lens-promoter activity and has appreciable skeletal muscle activity in transgenic mice. Thus, it does not appear that cis-regulatory elements required for lens activity in transgenic mice are situated between the αB-crystallin and HspB2 genes upstream of the truncated enhancer/promoter fragment of the mole rat αB-crystallin gene that we investigated earlier (15), a possibility that was raised by transgenic Xenopus experiments (16). Because the relative amount of mole rat promoter activity was not quantitated in the transgenic Xenopus larvae (16), it remains possible that the mole rat promoter activity is considerably reduced in the transgenic Xenopus larval lens, as it is in the transgenic mouse lens. Alternatively, the mole rat promoter might function more effectively in the lens of transgenic Xenopus larvae than that of transgenic mice.
The present data suggest but do not prove that the loss of lens functioning and gain of muscle activity caused by the mole rat αB-crystallin promoter in transgenic mice represent adaptive changes in gene expression associated with evolution to adjust to subterranean life. Moreover, the previous finding (15) that the mole rat enhancer/promoter fragment functions in the transgenic mouse lens during early development correlates well with the regression of the mole rat eye during development. It remains to be shown, however, whether the tissue-specific expression pattern of the endogenous mole rat αB-crystallin gene is the same as the activity of its promoter in transgenic mice.
It is well known that selection on gene expression regulated by enhancers and promoters has important roles in evolution (34–40). There have been examples showing that even a few changes in transcriptional factor binding sites can affect gene expression levels (40, 41). Even a single nucleotide polymorphism in the promoter of the IL-4 gene, which influences the balance of cytokine signaling in the immune system, is favored in human species as a result of positive selection (42). We show here that a 2-nt mutation (−273CA→G) in the mouse αB-crystallin enhancer/promoter fragment changes its promoter activity from that characteristic of the mouse (high lens activity and moderate skeletal muscle activity) to that characteristic of its blind mole rat orthologue (little lens activity and high skeletal muscle activity) when tested in transgenic mice. We do not know whether different nucleotides used in this location would cause comparable changes in mouse promoter activity or whether the lowering of lens activity and elevation of muscle activity are mechanistically linked. This functional conversion of promoter activity is unexpected because the mutation in the mouse promoter fragment involved two nonconserved nucleotides 3′ to the known regulatory motifs of the mouse enhancer (8, 9, 16, 21, 43, 44). Moreover, the mouse proximal promoter alone without the enhancer can drive modest lens-specific gene expression in transgenic mice, indicating that the mutated region is not necessary for promoter activity in the lens (5, 6). In general, then, our results indicate that even a 2-nt change in a variable sequence stretch that is not absolutely required for tissue-specific promoter activity can, under certain circumstances, have a profound, tissue-specific effect on promoter activity. Apparently, lack of strict sequence conservation within a gene control region does not mean a priori that the region is functionally neutral and raises interesting questions as to the neutrality of any polymorphism or very short nonconserved sequence within a larger regulatory region.
In contrast to the striking conversion of the mouse αB-crystallin promoter to the blind mole rat pattern of tissue-specific activity by a 2-nt mutation, the reciprocal mutation changing the mole rat promoter fragment to a mouse sequence in this region (−272G→CA) does not affect the tissue-specific pattern of mole rat promoter activity in the transgenic mice. However, it is likely that additional nucleotide sequence and/or transfactor differences between mole rat and mouse are responsible for the loss of lens activity and gain of skeletal muscle promoter activity of the mole rat αB-crystallin intergenic fragment in our transgenic mouse experiments. Indeed, there are numerous stretches of sequence disparities between the mouse and mole rat enhancers (see Fig. 2A, αBE-3 and MRF, for example) as well as in other areas of the fragments. Because the mole rat mutant p24-3M oligonucleotide forms the small, fast migrating complex (Fig. 4B, black arrowhead) characteristic of that of the wild-type mouse oligonucleotide but the p24-3M promoter neither activates lens activity nor decreases muscle activity, it is unlikely that absence of the small complex contributes significantly to the low lens activity or high muscle activity of the wild-type mole rat promoter. Because the mouse mutant pD3M oligonucleotide forms the large complex (Fig. 4B, black arrow), it remains possible that the large complex contributes to the reduction of lens promoter activity and/or activation of muscle promoter activity, although it is not sufficient to account for the differences in tissue-specific activities of the mouse and mole rat promoter fragments. Further studies are necessary to extend our preliminary investigation suggesting that the complex with the mole rat probe involves binding of a Sp1-like protein, a finding consistent with reports that Sp1 and Sp3 contribute to the regulation of the lens-preferred δ-crystallin (45, 46) and MIP (major intrinsic protein) genes (47).
In summary, although our results do not establish which regulatory sequences are responsible for the differences between the tissue-specific activities of the blind mole rat and mouse αB-crystallin enhancer/promoter fragments in transgenic mice, they do demonstrate that a 2-nt mutation in the mouse fragment results in a functional conversion to that of the blind mole rat promoter fragment when tested in transgenic mice. Most surprising is our finding that this tissue-specific conversion of the mouse αB-crystallin promoter activity occurs as the result of sequence modification of two apparently neutral nucleotides that are not absolutely required for lens specificity of the mouse wild-type promoter fragment (5, 6). This result emphasizes the importance of context within functional enhancers and promoters: Tissue-specific changes in promoter activity can occur without the modification of nucleotides comprising the known conserved cis-control elements. The ability of sequence alterations in short variable stretches of promoter regions to affect tissue-specific promoter activity increases the opportunities for independent evolutionary changes in gene expression among different animals and would be expected to contribute to diversity. Such considerations are relevant to the convergent evolution involved in recruiting the various taxon-specific lens crystallins in different species (48–51). Similar considerations of the meaning of neutrality extend to amino acid sequences of proteins, which also are separated into conserved regions with known functional significance (such as active sites of enzymes) and variable regions that are not known to be associated with functional importance. However, mutations within these variable regions no doubt often cause small changes in the ability of proteins to interact with other proteins, resulting in the development of additional functions by a gene-sharing mechanism that may be selected during evolution (see ref. 52).
Materials and Methods
Intergenic Regions.
The −968/+45 fragment of the blind mole rat (S. galili) αB-crystallin gene was isolated from the genome by using PCR and Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA). A 5′ primer (5′-TGCAGCCCCAACAAGCTCAGTACG-3′) derived from the Nannospalax ehrenbergi αB-crystallin gene (GenBank accession no. AJ617819) and a 3′ primer (5-GTAGGGGGTCAGCTGGCTGGTCAG-3′) derived from the blind mole rat αB-crystallin gene (GenBank accession no. AJ293658) were used to amplify the intergenic promoter fragment. The PCR product was cloned into the pRLFL-Null vector (7) and sequenced. Intergenic sequences of human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), monkey (Macaca mulatta), dog (Canis familiaris), cow (Bos taurus), and rabbit (Oryctolagus cuniculus) were retrieved from the assembled genomic sequences (www.ensembl.org/index.html). Intergenic sequences of cat (Felis catus; GenBank accession no. AJ617823) were retrieved from GenBank database.
Mutagenesis.
Site-specific mutations (pD3M and p24-3M) were introduced by using QuikChange II site-directed mutagenesis kits (Stratagene, La Jolla, CA). The mutated oligonucleotide for creation of pD3M contained sequence TCGTCCTAGCTCC, and the mutated oligonucleotide for p24-3M contained TCCATCCCAGCTCC; the italic letters denote substituted nucleotides.
Transgenic Mice.
Transgenic mice were produced by the National Eye Institute's Transgenic Mouse Facility. The DNA fragment containing αB-crystallin:luciferase was isolated from pRLFL-αB-crystallin by using BamHI digestion or from pGL-αB-crystallin by using KpnI and SalI digestion. The DNA fragments were purified by using agarose gel electrophoresis, eluted by using the Geneclean kit (MP Biomedicals, Aurora, OH), purified further by using chloroform extraction and ethanol precipitation, and injected into the pronuclei of fertilized oocytes of FVB/N mice. Genomic DNAs from founder mice were screened, and positive founders were mated with wild-type FVB/N mice to obtain F1 offspring. F1 transgenic mice were killed at 8 wk of age, and tissues were homogenized in passive lysis buffer. Tissue homogenates were centrifuged at ≈10,000 × g; protein concentrations of the supernatant fractions were determined by using a bicinchoninic acid protein assay (BCA; Pierce, Rockford, IL). Supernatant fractions from different tissues containing ≈10 μg of protein were assayed for luciferase activity.
EMSAs.
The following are the sequences of the oligonucleotides used in EMSAs. The italic and underlined letters indicate the mutations; the bold letters indicate additional nucleotides comprising 5′ overhangs used for labeling. Mole rat: GATCGGATTCCTGACTCGTCCCAGCTCCAGAGAAC; mouse: GATCGGATTCCAGGCTCCATCCTAGCTCCAGAGAACA; pD3M: GATCGGATTCCAGGCTCGTCCTAGCTCCAGAGAACA; p24-3M: GATCGGATTCCTGACTCCATCCCAGCTCCAGAGAAC. Radiolabeled DNA probes were synthesized by annealing complementary strands, followed by filling-in single-stranded overhangs with dATP, dGTP, TTP, and 3,000 Ci/mmol (1 Ci = 37 GBq) [α-32P]dCTP (MP Biomedicals) by using the Klenow fragment of DNA polymerase I.
Nuclear extracts from cultured αTN4-1 lens cells, HeLa cells, and C2C12 myoblasts, and differentiated C2C12 myotubes were prepared by using a nuclear extract kit (Active Motif, Carlsbad, CA). Extracts containing ≈5 μg of nuclear proteins were incubated with 0.05 pmol of radiolabeled double-stranded oligonucleotides in a 20-μl reaction mixture of 20 mM Hepes (pH7.9)/5% glycerol/50 mM KCl/5 mM MgCl2/100 μg/μl BSA/1 mM DTT/2 μg of poly(dI·dC). For competition experiments, nuclear extracts were first incubated with a 10-, 30-, 100-, or 300-fold molar excess of nonradioactive probe for 20 min at room temperature and then mixed with 0.02 pmol of radiolabeled probes for 30 min at room temperature. For supershift assays, the anti-Sp1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was incubated with the nuclear extracts for 30 min on ice, followed by the addition of the radiolabeled mole rat probes. The protein-DNA complexes were resolved in 6% acrylamide gel in 0.5× TBE buffer (1× TBE = 89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3) and visualized by using autoradiography.
Acknowledgments
We thank Zbynek Kozmik, Michael Spencer, and Vasilis Vasiliou for constructive discussions and Shivalingappa Swamynathan, Janine Davis, and Barbara Norman for valuable advice concerning experimental procedures.
Footnotes
The authors declare no conflict of interest.
References
- 1.de Jong WW, Leunissen JA, Voorter CE. Mol Biol Evol. 1993;10:103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- 2.Bhat SP, Nagineni CN. Biochem Biophys Res Commun. 1989;158:319–325. doi: 10.1016/s0006-291x(89)80215-3. [DOI] [PubMed] [Google Scholar]
- 3.Dubin RA, Wawrousek EF, Piatigorsky J. Mol Cell Biol. 1989;9:1083–1091. doi: 10.1128/mcb.9.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubin RA, Gopal-Srivastava R, Wawrousek EF, Piatigorsky J. Mol Cell Biol. 1991;11:4340–4349. doi: 10.1128/mcb.11.9.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopal-Srivastava R, Piatigorsky J. Nucleic Acids Res. 1994;22:1281–1286. doi: 10.1093/nar/22.7.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopal-Srivastava R, Kays WT, Piatigorsky J. Mech Dev. 2000;92:125–134. doi: 10.1016/s0925-4773(99)00341-x. [DOI] [PubMed] [Google Scholar]
- 7.Swamynathan SK, Piatigorsky J. J Biol Chem. 2002;277:49700–49706. doi: 10.1074/jbc.M209700200. [DOI] [PubMed] [Google Scholar]
- 8.Gopal-Srivastava R, Cvekl A, Piatigorsky J. J Biol Chem. 1996;271:23029–23036. doi: 10.1074/jbc.271.38.23029. [DOI] [PubMed] [Google Scholar]
- 9.Gopal-Srivastava R, Cvekl A, Piatigorsky J. J Biol Chem. 1998;273:17954–17961. doi: 10.1074/jbc.273.28.17954. [DOI] [PubMed] [Google Scholar]
- 10.Iwaki A, Nagano T, Nakagawa M, Iwaki T, Fukumaki Y. Genomics. 1997;45:386–394. doi: 10.1006/geno.1997.4956. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki A, Sugiyama Y, Hayashi Y, Nyu-i N, Yoshida M, Nonaka I, Ishiura S, Arahata K, Ohno S. J Cell Biol. 1998;140:1113–1124. doi: 10.1083/jcb.140.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quax-Jeuken Y, Bruisten S, Bloemendal H, de Jong WW, Nevo E. Mol Biol Evol. 1985;2:279–288. doi: 10.1093/oxfordjournals.molbev.a040351. [DOI] [PubMed] [Google Scholar]
- 13.Sanyal S, Jansen HG, de Grip WJ, Nevo E, de Jong WW. Invest Ophthalmol Vis Sci. 1990;31:1398–1404. [PubMed] [Google Scholar]
- 14.Avivi A, Joel A, Nevo E. Gene. 2001;264:45–49. doi: 10.1016/s0378-1119(00)00603-x. [DOI] [PubMed] [Google Scholar]
- 15.Hough RB, Avivi A, Davis J, Joel A, Nevo E, Piatigorsky J. Proc Natl Acad Sci USA. 2002;99:8145–8150. doi: 10.1073/pnas.122231099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerwald L, van Rheede T, Dirks RP, Madsen O, Rexwinkel R, van Genesen ST, Martens GJ, de Jong WW, Lubsen NH. J Mol Evol. 2004;59:674–686. doi: 10.1007/s00239-004-2659-y. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, Chauhan BK, Gao CY, Cveklova K, Duncan MK, et al. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi S, Goto K, Okada TS, Kondoh H. Genes Dev. 1987;1:818–828. doi: 10.1101/gad.1.8.818. [DOI] [PubMed] [Google Scholar]
- 19.Klement JF, Wawrousek EF, Piatigorsky J. J Biol Chem. 1989;264:19837–19844. [PubMed] [Google Scholar]
- 20.Sax CM, Piatigorsky J. Adv Enzymol Relat Areas Mol Biol. 1994;69:155–201. doi: 10.1002/9780470123157.ch5. [DOI] [PubMed] [Google Scholar]
- 21.Cvekl A, Piatigorsky J. BioEssays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 22.Ilagan JG, Cvekl A, Kantorow M, Piatigorsky J, Sax CM. J Biol Chem. 1999;274:19973–19978. doi: 10.1074/jbc.274.28.19973. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Chauhan BK, Cveklova K, Cvekl A. J Mol Biol. 2004;344:351–368. doi: 10.1016/j.jmb.2004.07.102. [DOI] [PubMed] [Google Scholar]
- 24.Gopal-Srivastava R, Piatigorsky J. Mol Cell Biol. 1993;13:7144–7152. doi: 10.1128/mcb.13.11.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalepakis G, Gruss P. Gene. 1995;162:267–270. doi: 10.1016/0378-1119(95)00345-7. [DOI] [PubMed] [Google Scholar]
- 26.Goulding M, Lumsden A, Paquette AJ. Development (Cambridge, UK) 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- 27.Fortin AS, Underhill DA, Gros P. Nucleic Acids Res. 1998;26:4574–4581. doi: 10.1093/nar/26.20.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadonaga JT, Courey AJ, Ladika J, Tjian R. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 29.Quax-Jeuken Y, Quax W, van Rens G, Khan PM, Bloemendal H. Proc Natl Acad Sci USA. 1985;82:5819–5823. doi: 10.1073/pnas.82.17.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendriks W, Leunissen J, Nevo E, Bloemendal H, de Jong WW. Proc Natl Acad Sci USA. 1987;84:5320–5324. doi: 10.1073/pnas.84.15.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong WW, Hendriks W, Sanyal S, Nevo E. Prog Clin Biol Res. 1990;335:383–395. [PubMed] [Google Scholar]
- 32.Avivi A, Oster H, Joel A, Beiles A, Albrecht U, Nevo E. Proc Natl Acad Sci USA. 2002;99:11718–11723. doi: 10.1073/pnas.182423299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avivi A, Oster H, Joel A, Beiles A, Albrecht U, Nevo E. J Biol Rhythms. 2004;19:22–34. doi: 10.1177/0748730403260622. [DOI] [PubMed] [Google Scholar]
- 34.Wang RL, Stec A, Hey J, Lukens L, Doebley J. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 35.Wittkopp PJ. Cell Mol Life Sci. 2005;62:1779–1783. doi: 10.1007/s00018-005-5064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittkopp PJ. Heredity. 2006;97:139–147. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- 37.Wittkopp PJ, Haerum BK, Clark AG. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 38.Wilkins AS. The Evolution of Developmental Pathways. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 39.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 40.Schulte PM, Glemet HC, Fiebig AA, Powers DA. Proc Natl Acad Sci USA. 2000;97:6597–6602. doi: 10.1073/pnas.97.12.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Meaux J, Goebel U, Pop A, Mitchell-Olds T. Plant Cell. 2005;17:676–690. doi: 10.1105/tpc.104.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockman MV, Hahn MW, Soranzo N, Goldstein DB, Wray GA. Curr Biol. 2003;13:2118–2123. doi: 10.1016/j.cub.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 43.Gopal-Srivastava R, Haynes JI, II, Piatigorsky J. Mol Cell Biol. 1995;15:7081–7090. doi: 10.1128/mcb.15.12.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan MK, Cvekl A, Kantorow M, Piatigorsky J. In: Development of the Ocular Lens. Robinson ML, Lovicu FJ, editors. New York: Cambridge Univ Press; 2004. pp. 119–150. [Google Scholar]
- 45.Das GC, Piatigorsky J. Proc Natl Acad Sci USA. 1986;83:3131–3135. doi: 10.1073/pnas.83.10.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayashi S, Kondoh H. Mol Cell Biol. 1986;6:4130–4132. doi: 10.1128/mcb.6.11.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S, Ge H, Ohtaka-Maruyama C, Chepelinsky AB. Mol Vis. 1999;5:12–21. [PubMed] [Google Scholar]
- 48.de Jong WW, Hendriks W, Mulders JW, Bloemendal H. Trends Biochem Sci. 1989;14:365–368. doi: 10.1016/0968-0004(89)90009-1. [DOI] [PubMed] [Google Scholar]
- 49.Piatigorsky J. J Biol Chem. 1992;267:4277–4280. [PubMed] [Google Scholar]
- 50.Tomarev SI, Piatigorsky J. Eur J Biochem. 1996;235:449–465. doi: 10.1111/j.1432-1033.1996.00449.x. [DOI] [PubMed] [Google Scholar]
- 51.Wistow GJ, Piatigorsky J. Annu Rev Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 52.Piatigorsky J. Gene Sharing and Evolution: The Diversity of Protein Functions. Cambridge, MA: Harvard Univ Press; 2007. [Google Scholar]