Abstract
MicroRNAs (miRNAs) are a recently identified class of epigenetic elements consisting of small noncoding RNAs that bind to the 3′ untranslated region of mRNAs and down-regulate their translation to protein. miRNAs play critical roles in many different cellular processes including metabolism, apoptosis, differentiation, and development. We found 33 miRNAs expressed in CD34+ hematopoietic stem-progenitor cells (HSPCs) from normal human bone marrow and mobilized human peripheral blood stem cell harvests. We then combined these data with human HSPC mRNA expression data and with miRNA-mRNA target predictions, into a previously undescribed miRNA:mRNA interaction database called the Transcriptome Interaction Database. The in silico predictions from the Transcriptome Interaction Database pointed to miRNA control of hematopoietic differentiation through translational control of mRNAs critical to hematopoiesis. From these predictions, we formulated a model for miRNA control of stages of hematopoiesis in which many of the genes specifying hematopoietic differentiation are expressed by HSPCs, but are held in check by miRNAs until differentiation occurs. We validated miRNA control of several of these target mRNAs by demonstrating that their translation in fact is decreased by miRNAs. Finally, we chose miRNA-155 for functional characterization in hematopoiesis, because we predicted that it would control both myelopoiesis and erythropoiesis. As predicted, miRNA-155 transduction greatly reduced both myeloid and erythroid colony formation of normal human HSPCs.
Keywords: expression analysis, hematopoiesis, stem cell
MicroRNAs (miRNAs) consist of a class of epigenetic elements that inhibit translation of mRNA to protein and also may result in gene silencing through chromatin remodeling (1). miRNAs have been identified in many organisms, including plants, Drosophila, rats, mice, and humans, where they have been shown to control major cellular processes, including metabolism, apoptosis, differentiation, and development (2). miRNAs have been implicated in the differentiation of mammalian blood cell lineages. miRNA-181 seems to bias mouse lymphoid progenitors toward B lymphoid development, and miRNA-146 and -223 bias toward T lymphopoiesis (3). In accordance with the model we propose herein, miRNA-221 and -222 are critically involved in the negative control of human erythropoiesis (4) and miRNA-223 in the down-regulation of mouse granulopoiesis (5). Given that miRNAs regulate normal cellular functions, such as apoptosis, it is not surprising that miRNAs also are involved in carcinogenesis. miRNAs have been identified at deleted or amplified genomic regions or translocation breakpoints in human cancers (6); e.g., miRNA-15 and -16 are the critical genetic elements deleted from 13q14 chromosomal region in a subset of chronic lymphocytic leukemia cases (7).
To better understand the role of miRNAs in normal human hematopoiesis, we determined the miRNA expression profile of human hematopoietic stem-progenitor cells (HSPCs). We then bioinformaticly combined (i) these miRNA expression data, (ii) mRNA expression data obtained for human CD34+ cells that we had determined in a previous study (8), and (iii) the predicted mRNA targets of all known miRNAs (9–11). Combining these five data sets into one database allowed us to examine, in silico, the interactions of HSPC-expressed miRNAs (HE-miRNAs) and mRNAs. Based on pairing HE-miRNAs with their putative HSPC-expressed mRNA targets, along with annotation implicating certain of these targets as associated with hematopoietic differentiation, we made predictions as to which HE-miRNAs might control hematopoietic differentiation.
We found that 33 miRNAs are expressed in CD34+ HSPCs obtained from both bone marrow (BM) and mobilized blood peripheral blood stem cell (PBSC) harvests, and we demonstrate that translation of several mRNAs associated with hematopoietic differentiation is controlled by these HE-miRNAs. Each of these HE-miRNAs targets multiple hematopoietic differentiation-associated mRNAs, suggesting that a relatively small subset of HE-miRNAs negatively regulates hematopoiesis. Taken together, these observations led us to propose a model in which many of the protein-coding genes that specify hematopoietic differentiation are expressed at an early time point by undifferentiated HSPCs; yet, these factors appear to be held in check by HE-miRNA-mediated repression of protein translation.
Results
miRNA Expression in CD34+ Cells.
We measured the expression of 228 human miRNAs in CD34+ HSPCs obtained from normal donor PBSC or BM harvests [supporting information (SI) Table 3]. Mean background was 47,458 relative fluorescence units (RFU) for PBSC and 34,676 RFU for BM CD34+ cells. After normalization of the data with GeneSpring software, the minimum cutoff for an expressed miRNA was calculated to be 96,900 RFU for PBSC and 74,509 RFU for BM CD34+ cells. Filtering for expression, in combination with a Student's t test significance threshold of 0.05, yielded 35 miRNAs expressed in PBSC and 78 miRNAs in BM CD34+ cells (SI Table 4). Intersection analysis (8) of these data sets (Table 1) identified 31 mature miRNAs and 2 precursor miRNAs, 33 miRNAs in total, expressed by CD34+ cells from both PBSC and BM, which we designated hematopoietic-expressed-miRNAs.
Table 1.
Levels of the HE-miRNAs expressed in CD34+ cells from PBSC and BM
| miRNA* | PBSC RFU | BM, RFU |
|---|---|---|
| hsa-let-7b | 163,441 | 202,826 |
| hsa-mir-10a | 123,147 | 104,124 |
| hsa-mir-16a | 123,486 | 86,828 |
| hsa-mir-17 | 169,601 | 839,361 |
| hsa-mir-20 | 168,040 | 657,934 |
| hsa-mir-23a | 445,017 | 896,002 |
| hsa-mir-23b | 450,110 | 947,757 |
| hsa-mir-24-1 | 96,912 | 105,926 |
| hsa-mir-25 | 211,423 | 3,574,408 |
| hsa-mir-26a | 364,607 | 867,620 |
| hsa-mir-26b | 368,293 | 2,851,706 |
| hsa-mir-30b | 136,572 | 91,862 |
| hsa-mir-30d | 225,309 | 241,004 |
| hsa-mir-92-1 | 4,115,379 | 5,793,765 |
| hsa-mir-92-2-p | 3,477,367 | 4,760,875 |
| hsa-mir-93-1 | 127,149 | 483,003 |
| hsa-mir-103-2 | 103,209 | 686,012 |
| hsa-mir-103-1 | 158,154 | 885,352 |
| hsa-mir-106a | 160,635 | 729,776 |
| hsa-mir-107 | 136,376 | 832,992 |
| hsa-mir-128a | 319,848 | 183,657 |
| hsa-mir-130a | 186,958 | 420,271 |
| hsa-mir-146 | 626,547 | 444,647 |
| hsa-mir-155 | 355,623 | 639,819 |
| hsa-mir-181a | 110,744 | 598,830 |
| hsa-mir-191 | 247,692 | 3,563,677 |
| hsa-mir-193 | 104,803 | 481,103 |
| hsa-mir-197 | 182,695 | 231,027 |
| hsa-mir-213 | 272,646 | 1,088,714 |
| hsa-mir-221 | 354,047 | 549,559 |
| hsa-mir-222 | 354,343 | 200,347 |
| hsa-mir-222-p | 186,393 | 138,190 |
| hsa-mir-223 | 2,038,358 | 504,773 |
RFU, relative florescence units.
*All miRNAs listed are the mature form, except those suffixed with “-p” which are the precursor forms (RFU values shown in italics).
HE-miRNAs Are Predicted to Regulate Several mRNAs Expressed in CD34+ Cells.
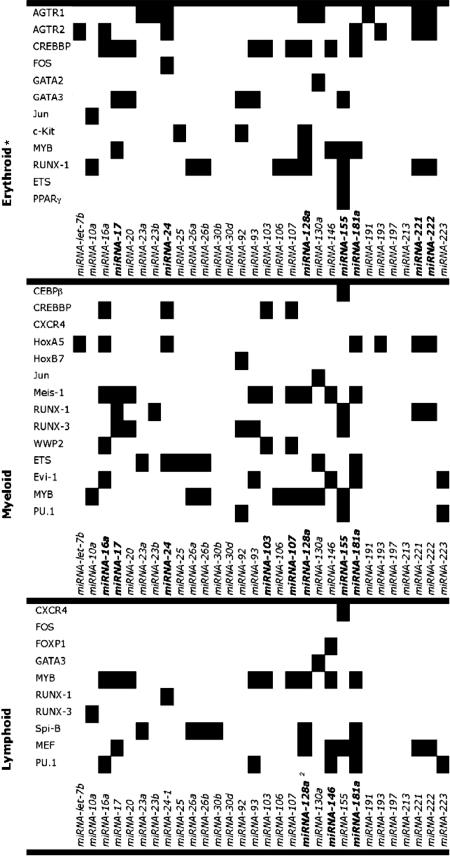
Combining our prior HSPC mRNA expression data sets (8) with the miRNA expression data generated in the current study, along with the miRNA target predictions (9–11), we mapped each of the 33 HE-miRNAs to its multiple predicted target mRNA transcripts expressed in CD34+ cells (from both PBSC and BM). mRNAs (16.7%) expressed by CD34+ HSPCs (HE-mRNAs) from both PBSC and BM are predicted to be regulated by at least one HE-miRNA (i.e., 1,230 of the 7354 mRNAs expressed by CD34+ cells potentially were HE-miRNA-regulated; SI Table 5 and 6), including several mRNAs thought to be associated with hematopoietic differentiation (Table 2). These in silico data allowed us to formulate a bioinformatic model predicting which HE-miRNAs may control hematopoietic differentiation and at which hematopoietic stages they may act (Fig. 1).
Table 2.
HSPC-expressed mRNAs associated with hematopoietic differentiation and predicted to be regulated by HE-miRNAs
miRNAs in bold are those that, based on the number of predicted target mRNAs they hit as well as on the strength of association to each category based on our annotation, we predict will be the major regulators of each particular category of differentiation. mRNAs are grouped into categories reflecting their roles in lymphoid, myeloid, and/or erythroid differentiation.
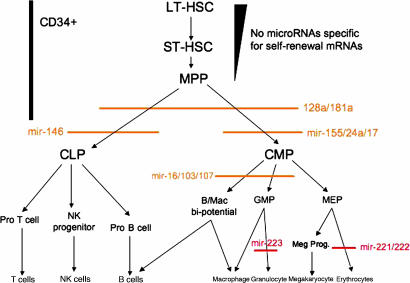
Fig. 1.
A model for HE-miRNA-mediated control of human hematopoiesis. Determination and electronic annotation of the predicted target HSPC-expressed mRNAs for the HE-miRNAs yielded predictions of the differentiation stages where HE-miRNAs might block hematopoietic differentiation. Those HE-miRNAa shown in red are predicted to regulate hematopoiesis by our study and supported by other reports; those in orange are our additional predictions. The hierarchy of hematopoietic differentiation is that proposed by Kondo et al. (17). LT-HSC, long-term-repopulating HSC; ST-SHC, short-term-repopulating HSC; MPP, multipotent progenitor cell; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocyte-erythrocyte progenitor.
HE-miRNAs Regulate Hematopoietic Differentiation-Associated mRNAs in CD34+ cells.
Of the HSPC-expressed mRNAs putatively targeted by HE-miRNAs, we selected 18 mRNAs, because they were either well established and/or annotated to have a role in regulation of hematopoietic differentiation. For each of these selected “hematopoietic differentiation-associated” mRNAs, the 3′ UTR was cloned into the 3.1-Luc expression vector. K562 cells were transduced with each chimeric luc-3′ UTR mRNA. Luc activity in the transduced K562 cells for the following mRNAs was reduced by 52–92% as compared with the luc-positive control: BCL2, C/EBPβ, CXCR4, GATA2, GATA3, HES1, HOXA9, HOXA10, HOXB5, HOXB7, KLF2, MEIS1, MYB, AML1/RUNX1, and RUNX3 (Fig. 2). However, luc activity of the SOCS2 and WWP2 constructs was increased compared with the luc-positive control; these two activities were identical to that of a chimeric mRNA (Luc-nmiR) containing a 3′UTR devoid of predicted miRNA-binding sites, which (for unknown reasons) resulted in almost twice the control luc activity. Thus, our predictions of targets of the HE-miRNA data resulted in a false-positive rate of 10.5%, considerably lower than the 25–40% false-positive rate postulated by Bentwich (12).
Fig. 2.
Translation of 16 of 18 tested hematopoietic differentiation-associated mRNAs is controlled by HE-miRNAs. Chimeric mRNAs, consisting of a luc reporter followed by the 3′ UTR from the mRNAs shown, were expressed in K562 cells from a CMV-driven plasmid construct. All values are the percent of luc expression as compared with the control plasmid construct containing luc only. Results are combined from four separate replicate experiments.
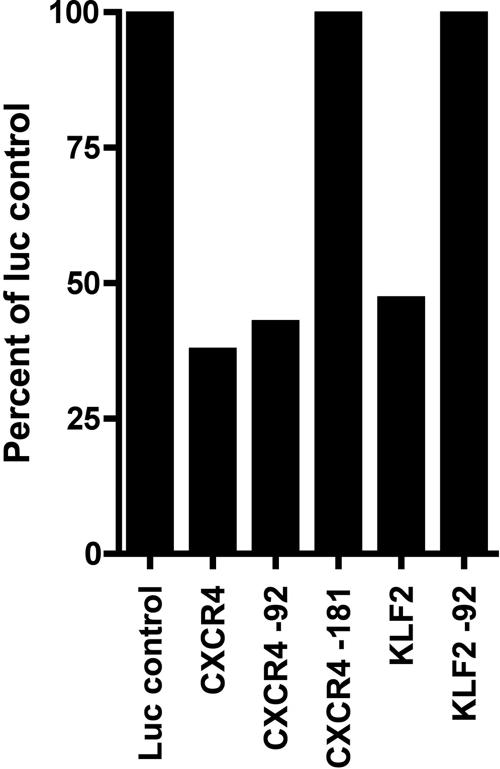
We further characterized HE-miRNA translational control of two of the hematopoietic differentiation-associated mRNAs, by deleting the 21- to 25-nt miRNA binding sites from the 3′ UTRs in the chimeric luc reporter constructs (Fig. 3). Deletion of the miRNA-92-binding site from the KLF2 3′ UTR restored luc expression, demonstrating that miRNA-92 regulates KLF2 in hematopoietic cells. Similarly, deletion of the binding site for miRNA-181 from the CXCR4 3′ UTR resulted in restoration of luc activity, whereas deletion of the miRNA-92-binding site from the CXCR4 3′UTR did not significantly change luc activity, indicating that CXCR4 is specifically modulated by miRNA-181.
Fig. 3.
Deletion of specific predicted miRNA-binding sites restored translation of hematopoietic differentiation-associated mRNAs. Individual miRNA-binding sites were deleted from the luc-CXCR4 and luc-KLF2 chimeric mRNA constructs and then transfected into K562 cells. Labels show the mRNA name followed by the miRNA-binding site deleted from that 3′ UTR. All values are the percent of luc expression as compared with the plasmid construct containing luc only. Results are combined from four separate replicate experiments.
miRNA-155 Controls Myeloid and Erythroid Differentiation.
miRNA-155 was predicted to block hematopoietic differentiation along more than one lineage (Fig. 1), because it was predicted to block protein expression of CREBBP, CXCR4, JUN, MEIS1, PU.1, AGTRI, AGTRII, FOS, and GATA3. Consistent with this prediction, <1% of miRNA-155-transduced K562 cells, as compared with ≈65% of control cells, were treated with hemin differentiated toward erythrocytes (data not shown). In addition, 57% ± 3% of miRNA-155-transduced K562 cells expressed CD41 (mean fluorescence intensity (MFI) = 11.5 ± 1.2), in comparison with 78 ±5% of control K562 cells (MFI = 32.8 ± 3.5) treated with IDB-expressed CD41 (data not shown). Both the decrease in percentage of cells positive for CD41, as well as the ≈3-fold decrease in MFI, are indicative of a large decrease in K562 cell megakaryocytic differentiation (13–15). K562 cells transduced with miRNA-155 proliferated at a similar rate to those transduced with control vector (data not shown), indicating that the observed lack of differentiation was not due to cell cycle arrest or decreased cell growth rate.
We next transduced normal human PBSC CD34+ cells with FUGW-miRNA-155, and we assayed hematopoietic colony formation. miRNA-155-transduced cells generated 5-fold fewer myeloid and 3-fold fewer erythroid colonies. Furthermore, the colonies generated by miRNA-155-transduced cells were much smaller than controls (Fig. 4). These data demonstrate that miRNA-155 can control both myeloid and erythroid differentiation of human HSPCs.
Fig. 4.
mir-155 inhibited generation of myeloid and erythroid colonies by normal primary human CD34+ cells. Normal PBSC CD34+ cells were transduced with FUGW (empty) or mir-155 lentivector. (A and B) Myeloid (A) and erythroid (B) colonies per 1,000 cells plated. (C) Representative colonies generated by control and mir-155-transduced cells. Results are representative of three separate experiments.
Discussion
We used bioinformatic techniques to combine human HSPC miRNA expression with mRNA expression to begin to define the roles that HE-miRNAs play in human hematopoiesis. Our bioinformatic and functional analyses of HE-miRNA action indicate that multiple miRNAs, including miRNA-17, -24, -146, -155, -128, and -181, may hold early hematopoietic cells at an early stem-progenitor stage, blocking their differentiation to more mature cells. miRNA-16, -103, and -107 may block differentiation of later progenitor cells; miRNA-221, -222, and -223 most likely act to control terminal stages of hematopoietic differentiation (Fig. 1). As an initial validation of the in silico predictions, we demonstrate that miRNA-155 is a potent inhibitor of HSPC differentiation.
In our prior mRNA expression study (8), we were perplexed by the finding that many genes known to be associated with differentiation of HSPCs (including RUNX1, MEIS1, ETS1, and PU.1) are equivalently expressed at the mRNA level in both the HSC-enriched and the more differentiated HPC-enriched populations. Our Transcriptome Interaction Database predicts that ≈16.7% of the mRNAs expressed by HSPCs may be regulated at the translational level by the 33 HE-miRNAs. Taking into account the consensus that such predictions likely contains 25% false-positives (12), we estimate that 10.5% of the HSPC-expressed mRNAs are controlled by HE-miRNAs. This is in concordance with a recent study (16), which compared a limited proteome of HSPCs to various transcriptome studies (including our own study; ref. 8) and found that the expression of ≈15% of proteins were underestimated by their mRNA expression levels.
Based on their predicted mRNA targets, we found that the HE-miRNAs annotated as having hematopoietic function fall into three categories (Fig. 1): (i) those which might inhibit differentiation of all hematopoietic lineages (including B/T/NK lymphoid) by regulating molecules critical to very early steps in hematopoiesis (miRNA-181 and -128); (ii) those that might act in differentiation of (a) lymphoid (miRNA-146); (b) granulocytic/monocytic/erythroid/megakaryocytic/B-lymphoid (miRNA-155, -24, and -17), or (c) granulocytic/monocytic/B-lymphoid (miRNA-16, -103, and -107); and (iii) in combination with other published studies (3–5), those that act only on one specific lineage of blood cells (miRNA-221 and -222 in erythroid development and miRNA-223 in granulocytic development).
We wished to validate our model and specific in silico predictions by testing HE-miRNA translational repression of selected predicted target mRNAs. Of the 18 hematopoietic differentiation-associated mRNAs tested in our model system, we found that protein expression from 16 were down-regulated greatly in K562 cells expressing the implicated HE-miRNAs. The translational repression we observed was generally quantitatively greater than that of other past miRNA functional studies, where a 40–60% miRNA-mediated decrease in protein expression has proven to be biologically relevant (4, 5). We feel that these other studies, although correct, may greatly underestimate the translational repression mediated by HE-miRNAs, because of several factors. First, we extensively optimized our assays to quantify differences in protein expression. Because the CMV promoters used in these plasmids are such strong initiators of transcription, we strove for a “low-level” transfection in an attempt to minimize the number of plasmids entering individual cells. Further, we added the additional control of a chimeric-mRNA containing a 3′ UTR having no predicted miRNA-binding sites. This control vector resulted in twice the amount of luc expression compared with the control vector containing no 3′UTR sequence. It may be that using only a luc alone leads to a major underestimation of miRNA action, and the luc-nmiR may be a much more biologically relevant control, because it mimics the natural structure of mRNAs. To test more directly which miRNAs mediate the observed decrease in protein expression, for two targeted mRNAs, we produced chimeric mRNAs with 3′ UTRs that had specific deletions of only the predicted miRNA-binding site. In both cases, deletion of miRNA-binding sites completely restored protein expression.
To begin to confirm the biological effects of HE-miRNAs, miRNA-155 was studied because it was predicted to target multiple hematopoietic differentiation-associated molecules, including C/EBPβ, CREBBP, JUN MEIS1, PU.1, AGTR1, AGTR2, and FOS. In addition, it has been reported that miRNA-155 is overexpressed in undifferentiated CD34+/CD38− acute myeloid leukemia (AML) stem cells and that ectopic over-expression of miRNA-155 in AML cells blocks differentiation‖. In our studies, transduction of cells with miRNA-155 followed by treatment with each of two chemical inducers of differentiation showed that miRNA-155 decreased both erythroid and megakaryocytic differentiation in K562 cell line models of human hematopoiesis. More important, miRNA-155-transduced normal human CD34+ cells generated far fewer (and smaller) myeloid and erythroid colonies than controls. Thus, miRNA-155 negatively regulates normal myelopoiesis and erthryopoiesis
Most of the cells within the CD34+ population we studied have differing degrees of self-renewal capacity. In that light, perhaps it is not surprising that we did not identify any HE-miRNAs that targeted mRNAs and are thought to be involved in self-renewal, such as HOX-B4 (18) and BMI-1 (19, 20).
Three other studies have begun to examine the functional role of miRNAs in the differentiation of specific aspects of hematopoiesis. Fazi et al. (5) showed that murine mmu-miRNA-223 up-regulates mouse granulopoiesis by blocking protein expression of NFI-A, a factor that represses granulopoiesis. The myeloid regulator C/EBPα drives miRNA-223 expression, thereby removing NFI-A mediated differentiation repression. Our model of human HE-miRNA regulation of hematopoiesis agrees with this finding in that the Transcriptome Interaction Database predicts that human hsa-mir-223 regulates human NFI-A translation.
Felli et al. (4) showed that human miRNA-221 and -222 block erythropoiesis via translational repression of c-KIT. Our findings strongly support that miRNA-221 and -222 block erythropoiesis. Not only do our in silico studies predict that miRNA-221 and -222 block c-KIT translation, but they also predict that they may block CREBBP, FOS, and PPARγ (additional molecules highly associated with erythroid differentiation) (21–24). miRNA-221 and -222 also may be more generalized inhibitors of hematopoietic differentiation, because our data (Table 2) predict that they also may regulate myeloid differentiation-associated molecules (Fig. 2; MEIS1 and ETS1, both proteins shown to be involved in myeloid development (25, 26), and FOS, which has been implicated in lymphopoiesis; ref. 27 and unpublished data). Consistent with this, Felli et al. (4) found that overexpression of miRNA-221 and -222 inhibited engraftment of human CD34+ cells in NOD-scid mice, pointing to a more generalized role in differentiation.
Chen et al. (3) studied the role of mmu-mir-142, -181, and -223 in mouse hematopoiesis and found that ectopic overexpression of miRNA-181 in mouse HSPCs resulted in an increase in B-lymphopoiesis, and miRNA-142 and -223 resulted in small but significant increases in T-lymphopoiesis. Unfortunately, the relevant target mRNAs were not determined in that early study. It is important to note that although miRNA-181 is strongly expressed in human CD34+ cells, in the murine system, it seems to be expressed selectively in B lymphocytes. In fact, in human BM, miRNA-181 is expressed more weakly than miRNA-146, an miRNA that Chen et al. (3) finds expressed strongly in all mouse hematopoietic tissues. In addition, we found that miRNA-142 is not expressed in human HSPCs. Thus, these HE-miRNAs may be expressed and act differently in human hematopoiesis than in mouse hematopoiesis; this interpretation is substantiated by the finding that the same types of human and mouse hematopoietic cells show large differences in miRNA expression (28). Although our predictive data are consistent with a possible outcome that miRNA-181 may affect B lymphoid development, our in silico model indicates that miRNA-181 and miRNA-146 might block differentiation very early in human lymphopoiesis.
This in silico bioinformatic study attempts to combine global miRNA and mRNA expression data into a model for exploring the action of miRNAs within a cellular system. We validated the in silico predictions in two ways: (i) We show that 16 of 18 “hematopoietic” mRNAs are controlled by HE-miRNAs and (ii) we functionally validated miRNA-155 as potent controller of normal human myelopoiesis and erythropoieis. We hope that this study will serve to guide, and will be greatly refined by, the work of other researchers examining the role of miRNAs in hematopoiesis. Specifically, we look forward to results of experiments testing our model in which many of the genetic components associated with hematopoietic differentiation are expressed at an early time point by undifferentiated HSPCs but are controlled by HE-miRNA-mediated repression of protein translation until differentiation is initiated.
Methods
Cell Samples.
Human CD34+ cells.
Cryopreserved cadavaric BM and PBSC CD34+ cells from normal adult donors were obtained from the National Heart, Lung, and Blood Institute Program of Excellence in Gene Therapy, Hematopoietic Cell Processing Core (Fred Hutchison Cancer Center). Each BM sample was a pool of cells from two donors. Each PBSC sample was a pool from three to five donors. CD34+ cells were thawed and washed twice with PBS (8).
Cell lines.
HEK 293T, Hal-01, HL60, K562, KG1, KG1a, MV-4-1, REH, TF1, and TF1a cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA). A different K562 cell subline, used in the cellular differentiation studies, was provided by Michael McDevitt (The Johns Hopkins University) (29). All cells were cultured by following instructions provided by the ATCC.
miRNA Expression Microarrays.
CD34+ cells were disrupted with TRIzol Reagent (15596-018; Invitrogen. Carlsbad, CA). Five micrograms of total RNA obtained from the TRIzol preparations were analyzed with miRNA chips (30). Standard microarray methods were used for normalization and statistical analysis of the miRNA expression data. Statistical analyses were carried out in GeneSpring 7.0 (Silicon Genetics/Agilent Technologies, Palo Alto, CA) to determine the RFU value for a 95% confidence cutoff for miRNA expression. Significantly expressed miRNAs were identified by using the GeneSpring Cross-Gene Error Model (8). Thus, for each set of samples (PBSC or BM,) values from individual samples were normalized and averaged, then expressed miRNAs were identified as those with a P < 0.05 of expression over background (i.e., the weighted averaged values of all chip measurements). Then, to perform intersection analysis, we determined those microRNAs expressed by both PBSC and BM CD34+ samples at the P < 0.05 significance level (Student's t test).
Bioinformatic Integration of mRNA and miRNA Expression Data.
miRNA expression data, mRNA expression data (determined in ref. 8), and miRNA target prediction data (from Lewis et al., refs. 10 and 11; www.targetscan.org), Sloan–Kettering Cancer Center Human MicroRNA Targets Database (www.microRNA.org; ref. 9) and miRBase Targets (http://microrna.sanger.ac.uk/targets/v3) were combined in a Filemaker Pro 7.0 (Filemaker Corp., Santa Clara, CA) relational database, which we call the “Transcriptome Interaction Database.” Individual data tables consisted of miRNA expression from each individual sample, PBSC CD34+ cell mRNA expression data, BM CD34+ cell mRNA expression data, and miRNA targets predicted by each of the three databases. This integrated database allows us to interrogate directly the predicted interactions of HE-miRNAs with HSPC-expressed mRNAs. Regulated mRNAs were annotated for possible hematopoietic function through both manual and electronic annotation. Manual annotation consisted of the authors scanning the lists of mRNA targets for those with known hematopoietic function. Electronic annotation consisted of querying the PubMed (www.ncbi.nlm.nih.gov/entrez/query.fcgi?holding = jhumlib_fft_ndi) and Gene Ontology (www.geneontology.org) databases with search terms of the “gene common name” and terms including “hematopoiesis,” “myeloid,” “erythroid,” “blood,” etc. After compiling the list of candidate hematopoietic differentiation-associated mRNAs, PubMed searches were performed, and peer-reviewed publications were reviewed to assure that these mRNAs actually had been implicated in hematopoietic differentiation. At least two articles, each directly demonstrating either that the mRNA of interest is involved in differentiation or critical to a molecular pathway of a known hematopoietic regulator, were required for the mRNA to be scored as hematopoietic differentiation-associated. In addition, we also classified certain genes expressed in the HSC-enriched population (8) as hematopoietic differentiation-associated.
Functional Analysis of Expressed HE-miRNAs.
Plasmids and constructs: Luciferase(luc)-3′ UTR reporter plasmids.
The luc-coding sequence and 3′ multiple-cloning-site (MCS) from the promoterless pSP-luc+NF fusion vector (E4471; Promega, Madison, WI) were subcloned into the NheI and XhoI sites of the pcDNA 3.1(+) expression vector (V79020; Invitrogen). The resulting plasmid (Luc-3′ UTR) construct contains a strong CMV promoter driving a luc-expression cassette, followed by a 3′ MCS. The 3′ UTR sequences from several hematopoietic differentiation-associated mRNAs were isolated by RT-PCR with primers containing restriction enzyme linkers (http://www.pnas.org/cgi/content/full/0610983104/DC1SI Methods). These 3′ UTR PCR fragments were cloned into the 3′ MCS of our Luc-3′ UTR reporter construct. The resulting chimeric mRNAs, containing luc and the 3′ UTR of the mRNA of interest, allowed for rapid functional analysis of miRNA translational control of mRNAs of interest (SI Fig. 5).
In addition, for certain of the luc-3′ UTR reporter plasmids, we produced variant constructs, in which specific miRNA-binding sites were deleted from the 3′ UTR (Luc-3′ UTR-DL constructs). Approximately 60-mer PCR primers, consisting of the 20-nucleotide (nt) sequence on either side of the miRNA-binding sites plus an intervening 25-bp spacer containing a PacI site, were generated (http://www.pnas.org/cgi/content/full/0610983104/DC1SI Methods) and then used in full plasmid PCR (QuikChange XL Kit 200516; Stratagene, La Jolla, CA). Briefly, these primers span the deletion site but replace the sequence to be deleted with a unique restriction enzyme site that allows for rapid screening of clones with the deletion. The entire mutant plasmid then is produced via PCR, lacking the miRNA-binding site, as described in the manufacturer's protocol and http://www.pnas.org/cgi/content/full/0610983104/DC1SI Methods.
miRNA expression constructs.
The hsa-mir-155 gene was cloned from K562 cell genomic DNA via PCR by using primers containing restriction enzyme linkers (http://www.pnas.org/cgi/content/full/0610983104/DC1SI Methods) that would allow cloning into either the pcDNA3.1(+) expression vector (V79020; Invitrogen) or the FUGW lentiviral expression vector (31). We used vectors containing strong PolII promoters, because miRNA expression and processing by Drosha, Exportin V, and Dicer seem to be much more reliable with longer 5′-capped-mRNA sequences produced by PolII, rather than shorter RNAs produced by PolIII promoters (32). The fragments produced by PCR contained the miRNA precursor (70–120 nt) flanked on both the 5′ and 3′ sides by 250 nt of genomic sequence, resulting in inserts of 570–620 nt.
Expression of miRNA precursor and mature sequences were tested by Lipofectamine (11668-019; Invitrogen) transfection of 293T cells with these expression constructs. Small RNAs were isolated from the transfected cells (MirVana miRNA Isolation Kit 1560; Ambion, Austin, TX). Precursor and mature miRNA expression was determined by miRNA RT-PCR (mirVana qRT-PCR miRNA Detection Kit 1158; Ambion; using specific mirVana qRT-PCR Primer Sets; Ambion) and/or by Northern blotting with 32P-radiolabeled probes specific for the mature miRNA sequence (mirVana miRNA Probe Construction Kit 1550; Ambion).
HE-miRNA Translational Control of Hematopoietic Differentiation-Associated mRNAs Expressed in HSPCs.
We determined the miRNA expression of 10 hematopoietic cell lines and found that the K562 cell line expresses many HE-miRNAs (SI Table 7). Luc-3′ UTR and Luc-3′ UTR-DL constructs were electroporated into K562 cells (Nucleofector V Kit VCA-1003; Amaxa, Cologne, Germany; Nucleofector II Device AAD-1001; Amaxa). Control plasmids of backbone Luc-3.1 and Luc-nmiR (containing a 3′ UTR with no predicted miRNA-binding sites) controls also were used. Each electroporation included 100 ng of β-galactosidase (β-gal)-3.1 plasmid to allow normalization of luc activity. Three days after transfection, cells were lysed (Passive Lysis Buffer E194A; Promega), and luminescence was measured to quantify luc protein expression (Model 1500; Analytical Luminescence Laboratories, San Diego, CA; Luciferase Assay System E1501; Promega) and β-gal expression (Galactolight Plus Kit; Tropix, Bedford, MA). Relative β-gal units were used to generate individual normalization factors to standardize the Relative Luc Units for each sample to the Luc-nmiR-positive control.
Cellular Differentiation Assays.
K562 cells were transduced with FUGW-miRNA-155 or FUGW-parental control (i.e., empty vector) lentivirus at a multiplicity of infection of 5. Each of these vectors contains a GFP cassette driven from a second promoter (31). Cells were cultured for 2 days and then checked for GFP expression, indicating lentiviral transduction. Cells then were washed with PBS and cultured in complete growth media containing ingenol 3,20-dibenzoate (IDB, 1 μg/ml; Sigma) or hemin (50 μM; Sigma). On day 4, IDB-treated cells were assayed by monoclonal antibody staining and flow cytometry for CD41 (Becton Dickinson, Franklin Lake, NJ) expression, indicating megakaryocytic differentiation (13–15). Hemin-treated cells were assayed similarly on day 6 for benzidine staining, indicating erythroid differentiation (33).
Human PBSC CD34+ cells were transduced with miRNA-155 or control lentivirus and assayed by standard colony-forming unit (cfu) assays for myeloid and erythroid differentiation (34).
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA70970 and CA60441 (to C.I.C.), a Fellow Award from the National Foundation for Cancer Research (to C.I.C.), a Kimmel Foundation for Cancer Research award (to G.A.C.), and Program Project grants from the National Cancer Institute (to C.M.C.).
Abbreviations
- BM
bone marrow
- HE-miRNA
hematopoietic-expressed microRNA
- HSPCs
hematopoietic stem-progenitor cells
- miRNAs
microRNAs
- PBSC
peripheral blood stem cell
- RFU
relative fluorescence units.
Footnotes
Conflict of interest statement: The Johns Hopkins University holds patents on CD34 monoclonal antibodies and inventions related to stem cells. C.I.C. is entitled to a share of the sales royalty received by the university under licensing agreements between the university, Becton Dickinson Corp., and Baxter HealthCare Corp. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610983104/DC1.
Lechman, E. R., Hope, K. J, Saiz, F. J. S., Takenaka, K., Croece, C. M., Minden, M. D., Dick, J. E. (2005) Blood Vol. 106, p. 140a (abstr.).
References
- 1.Finnegan EJ, Matzke MA. J Cell Sci. 2003;116:4689–4693. doi: 10.1242/jcs.00838. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen CZ, Li L, Lodish HF, Bartel DP. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 4.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgantas RW, III, Tanadve V, Malehorn M, Heimfeld S, Chen C, Carr L, Martinez-Murillo F, Riggins G, Kowalski J, Civin CI. Cancer Res. 2004;64:4434–4441. doi: 10.1158/0008-5472.CAN-03-3247. [DOI] [PubMed] [Google Scholar]
- 9.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 12.Bentwich I. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 13.Elagib KE, Xiao M, Hussaini IM, Delehanty LL, Palmer LA, Racke FK, Birrer MJ, Shanmugasundaram G, McDevitt MA, Goldfarb AN. Mol Cell Biol. 2004;24:7779–7794. doi: 10.1128/MCB.24.17.7779-7794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldfarb AN, Delehanty LL, Wang D, Racke FK, Hussaini IM. J Biol Chem. 2001;276:29526–29530. doi: 10.1074/jbc.M103825200. [DOI] [PubMed] [Google Scholar]
- 15.Xie P, Chan FS, Ip NY, Leung MF. Leuk Res. 1999;23:1113–1119. doi: 10.1016/s0145-2126(99)00141-1. [DOI] [PubMed] [Google Scholar]
- 16.Unwin RD, Smith DL, Blinco D, Wilson CL, Miller CJ, Evans CA, Jaworska E, Baldwin SA, Barnes K, Pierce A, et al. Blood. 2006;107:4687–4694. doi: 10.1182/blood-2005-12-4995. [DOI] [PubMed] [Google Scholar]
- 17.Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 18.Sauvageau G, Iscove NN, Humphries RK. Oncogene. 2004;23:7223–7232. doi: 10.1038/sj.onc.1207942. [DOI] [PubMed] [Google Scholar]
- 19.Lessard J, Sauvageau G. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 20.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 21.Blobel GA. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 22.Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, Brindle PK. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- 23.Nagasawa E, Abe Y, Nishimura J, Yanase T, Nawata H, Muta K. Exp Hematol. 2005;33:857–864. doi: 10.1016/j.exphem.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Bieker JJ. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azcoitia V, Aracil M, Martinez AC, Torres M. Dev Biol. 2005;280:307–320. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L, et al. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumura T, Kametani Y, Ando K, Hirano Y, Katano I, Ito R, Shiina M, Tsukamoto H, Saito Y, Tokuda Y, et al. Exp Hematol. 2003;31:789–797. doi: 10.1016/s0301-472x(03)00193-0. [DOI] [PubMed] [Google Scholar]
- 28.Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP, Jr, Sloand EM, Kajigaya S, Young NS. Leuk Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Racke FK, Wang D, Zaidi Z, Kelley J, Visvader J, Soh JW, Goldfarb AN. J Biol Chem. 2001;276:522–528. doi: 10.1074/jbc.M005236200. [DOI] [PubMed] [Google Scholar]
- 30.Liu C-G, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Zhan X, D'Costa J, Tanavde VM, Ye Z, Peng T, Malehorn MT, Yang X, Civin CI, Cheng L. Mol Ther. 2003;7:827–838. doi: 10.1016/s1525-0016(03)00104-7. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Alder JK, Chun JH, Friedman AD, Heimfeld S, Cheng L, Civin CI. Stem Cells. 2006;24:876–888. doi: 10.1634/stemcells.2005-0598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.