Abstract
Using real-time PCR and immunohistochemistry, we have examined the expression of carbonic anhydrase isozymes (CA) I, II, III, IV, IX, XII, XIII and XIV in the brain, kidney, stomach and colon of the wild-type, CA II-deficient (Car2−/−), and CA IX deficient (Car9−/−) mice. The expression of Car4, Car12, Car13 and Car14 mRNAs did not show any significant deviations between the three groups of mice, whereas both groups of CA deficient mice showed decreased expression levels of Car1 in the colon and Car3 in the kidney. The Car2 mRNA level was greatly reduced but not completely abolished in all four tissues from the Car2−/− mice in which no CA II protein was expressed. Sequencing the Car2 cDNA isolated from C57BL6 Car2−/− mice revealed two nucleotide differences from the wild-type C57BL6 mice. One is a silent polymorphism found in Car2 mRNA from wild-type DBA mice, which is the strain that provided the original mutagenized chromosome. The second change is a mutation that causes prematurely terminated translation at codon 155 (Gln155X). Car9 mRNA and CA IX protein expression levels were up-regulated about 2.5- and 3.6-fold, respectively, in the stomach of the Car2−/− mice. These results suggest that the loss of function of cytosolic CA II in the stomach of Car2−/− mice leads to up-regulation of an extracellular CA, namely CA IX, which is expressed on the cell surface of the gastric epithelium.
Carbonic anhydrases (CAs) are zinc-containing metalloenzymes that catalyse the reversible hydration of carbon dioxide in the reaction CO2+ H2O ⇌ HCO3−+ H+, and participate in various biological processes, including CO2 transport, regulation of pH homeostasis, bone resorption, ureagenesis, gluconeogenesis, production of body fluids, and fertilization (Sly & Hu, 1995; Parkkila, 2000; Lehtonen et al. 2004; Kivela et al. 2005a). The mammalian α-CA family is comprised of 16 different isoforms, among which 13 (CA I, II, III, IV, VA, VB, VI, VII, IX, XII, XIII, XIV and XV) are enzymatically active, whereas the other three (CA-RP VIII, X and XI) appear to lack CA activity because of substitutions in one or more of the functionally important histidine residues (Sly & Hu, 1995; Tashian et al. 2000; Lehtonen et al. 2004; Hilvo et al. 2005; Kivela et al. 2005a). In addition, the receptor-type protein-tyrosine phosphatases (RPTP) β and γ have also been reported to contain ‘CA-like’ domains. The 13 active CA isozymes differ in their subcellular localizations: CA I, II, III, VII and XIII are cytosolic, CA IV, IX, XII, XIV and XV are membrane associated, CA VA and VB are mitochondrial, and CA VI is secreted.
The expression of CAs in mammalian tissues has been intensively studied during the past three decades by means of reverse transcription polymerase chain reaction (RT-PCR), Northern blot, Western blot and immunohistochemical staining (Kumpulainen & Korhonen, 1982; Fleming et al. 1995; Ivanov et al. 1998; Saarnio et al. 1998; Parkkila, 2000; Ivanov et al. 2001; Wykoff et al. 2001; Leppilampi et al. 2003; Kivela et al. 2005b). These studies have provided important background information to understanding the roles of different isozymes in each type of tissue. The most distinctive features of CA expression are: (1) CA II appears to be the most widely expressed isozyme which is present in all major mammalian organs. As a high-activity isozyme, it may represent the most important isozyme for several fundamental biological processes (Sly & Hu, 1995; Parkkila & Parkkila, 1996); (2) two CA isozymes, CA IX and XII, are called cancer-associated isozymes due to their overexpression in certain carcinomas (Pastorekova et al. 2004; Pastorekova & Zavada, 2004); (3) multiple isozymes are often expressed in a particular mammalian tissue or organ, e.g. the murine kidney expresses at least CA II, IV, XII, XIII, XIV and XV (Brion et al. 1997; Kaunisto et al. 2002; Kyllonen et al. 2003; Lehtonen et al. 2004; Hilvo et al. 2005). The co-expression of several isozymes in different cell types and in different subcellular localizations in one organ suggests different functions for the different isozymes, which may or may not be redundant in a given tissue. Loss of one may lead to a compensatory up-regulation of another. Such compensatory mechanisms have never been studied thoroughly. In this study, we used quantitative real-time RT-PCR and immunohistochemical staining to assess the expression levels of CA isozymes I, II, III, IV, IX, XII, XIII and XIV in the brain, kidney, stomach and colon of the wild-type, CA II-deficient (Car2−/−), and CA IX-deficient (Car9−/−) mice.
Methods
The experimental mice
The brain, stomach, colon and kidney samples were collected from five wild-type (WT), three Car2−/−, and four Car9−/− mice. The C57BL/6 Car2−/− mice were originally generated by chemical mutagenesis (Lewis et al. 1988; Spicer et al. 1989). The Car9−/− mice were recently produced by targeted disruption of Car9 gene as described in Ortova Gut et al. (2002). These mice were introduced into the animal facility of Oulu University by embryo transfer, backcrossed 10 generations (F10) to the C57BL/6J strain, and then heterozygous mice were intercrossed to produce mice homozygous for the targeted gene. The mice were fed with standard feed (R36 for mice and rats, Lactamin, Stockholm, Sweden), and housed in pathogen-free conditions. The study protocols were approved by the Animal Care Committee of the Oulu University.
Determination of Car9 genotype
DNA was isolated from mice ear markings using NucleoSpin Tissue (Macherey-Nagel, Düren, Germany) DNA extraction kit. Car9 gene was amplified with PCR using Reddy Mix PCR Master Mix (ABgene, Epsom, UK). The template DNA for each reaction was 150 ng. The primers were: 5′-CCAGTCAGCTGCATGGCC-3′ and 3′-AGGAGCCTCGGGAGTCGA-5′ for the WT allele, and 5′-AGGAGCAAAGCTGCTATTGG-3′ and 3′-AGGAGCCTCGGGAGTCGA-5′ for the targeted Car9 allele. The primers were chosen from the first exon that was disrupted in the knockout mice (Ortova Gut et al. 2002). The PCR program was: 96°C for 5 min, 35 cycles of: 96°C for 30 s, 56°C for 60 s, 72°C for 60 s. The PCR products were characterized in 1.2% agarose (LE, analytical grade, Promega Madison, WI, USA) gel containing 0.005% nucleic acid gel stain GelStar (BMA, Rockland, ME, USA) and visualized by UV light.
Determination of Car2 genotype
The Car2 genotypes were determined by phenotypic analysis of the mice. CA activity was assayed from the blood sample (treated with EDTA, diluted 1 : 5000) with the imidazole–tris technique (Brion et al. 1988). Since CA II constitutes the major fraction of CA activity of the blood, the test is considered reliable in monitoring CA II activity, and therefore, the Car2 genotype.
RNA extraction and first strand cDNA synthesis
All tissue samples used for quantitative real-time PCR were stabilized in RNAlater (Ambion, Austin, TX, USA) and stored at −80°C until use. Total RNA was isolated from tissue samples using RNeasy Mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. RNA concentration and purity were determined spectrophotometrically at 260 and 280 nm. RNAs were pooled for each group (wild-type, Car2−/− and Car9−/−) to eliminate the potential individual variation. Three micrograms of total RNA was converted into first strand cDNA using First Strand cDNA Synthesis kit (Fermentas, Burlington, Canada), utilizing random hexamers, according to the protocol recommended by the manufacturer.
Quantitative real-time PCR
The amount of mouse Car1, Car2, Car3, Car4, Car9, Car12, Car13 and Car14 transcripts in different tissues was assessed by real-time RT-PCR using the Lightcycler detection system (Roche, Rotkreuz, Switzerland). Real-time PCR primers were designed based on the complete cDNA sequences deposited in GenBank (accession numbers: NM_009799 for Car1, NM_009801 for Car2, NM_007606 for Car3, NM_007607 for Car4, NM_139305 for Car9, NM_178396 for Car12, NM_024495 for Car13, and NM_011797 for Car14). To avoid amplification of contaminating genomic DNA, the two primers of all primer sets were placed in different exons. β-Actin was used as an internal RNA control to normalize the RNA samples for differences (Chtanova et al. 2001). The primer sequences are shown in Table 1.
Table 1.
Primer sequences for real-time RT-PCR (Tm: annealing temperature)
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | PCR product size (bp) | Tm (°C) |
|---|---|---|---|---|
| β-actin | AGAGGGAAATCGTGCGTGAC | CAATAGTGATGACCTGGCCGT | 138 | 57 |
| Car1 | TTGATGACAGTAGCAACC | CCAGTGAACTAAGTGAAG | 161 | 51 |
| Car2 | CAAGCACAACGGACCAGA | ATGAGCAGAGGCTGTAGG | 122 | 56 |
| Car3 | GCTCTGCTAAGACCATCC | ATTGGCGAAGTCGGTAGG | 160 | 54 |
| Car4 | CTC CTT CTT GCT CTG CTG | GAC TGC TGA TTC TCC TTA | 145 | 55 |
| Car9 | CTGAAGACAGGATGGAGAAG | GCAGAGTGCGGCAGAATG | 221 | 57 |
| Car12 | CCTATGTTGGTCCTGCTG | CGTTGTAACCTTGGAACTG | 143 | 53 |
| Car13 | AATACGACTCCTCACTCC | TGCCGCAACCTGTAGTTC | 116 | 52 |
| Car14 | TGTTGTTCTTCGCTCTCCTG | CACTGTCTGTCTGGATATTG | 161 | 53 |
Every PCR was performed in a total reaction volume of 20 µl containing 0.5 µl of first strand cDNA, 1 × QuantiTect SYBR Green PCR Master Mix (Qiagen), and 0.5 µm of each primer. Amplification and detection were carried out as follows. After an initial activation step of 15 min at 95°C, amplification was performed in a three-step cycling procedure: denaturation at 95°C, 15 s, ramp rate 20°C s−1; annealing at Tm, 20 s, ramp rate 20°C s−1; and elongation at 72°C, 20 s, ramp rate 20°C s−1 for 45 cycles and a final cooling step. The melting curve analysis was always performed after the amplification to check PCR specificity. To quantify the concentration of β-actin and the eight CA transcripts in the four tissues of this study, a standard curve for each gene was established using 5-fold serial dilutions of known concentrations of purified PCR products generated from the same primer sets. Each cDNA sample was tested in duplicate and the crossing point (Cp) value obtained allowed the determination of the amount of the starting message using the specific standard curve. The final real-time PCR result was indicated as the copies of target gene per 1000 copies of β-actin.
Sequencing of Car2 full-length cDNA
Primers were designed to amplify the full-length cDNA from stomach samples of wild-type and CA II-deficient mice according to the sequence of Car2 cDNA (accession numbers: NM_009801). The primer sequences were as follows: F: 5′-CAAGCACAACGGACCAGA-3′, R2: 5′-TGTGGTATGTTATCTGAAGG-3′. Amplification was carried out in a 50 µl reaction containing 1 × Extensor Hi-Fidelity PCR Master Mix (Abgene), 0.2 µm of each primer, and 1 µl of cDNA. The PCR profile was: 94°C denature for 2 min followed by 35 cycles of denaturation at 94°C for 10 s, annealing at 57°C for 30 s, and extension at 68°C for 1 min, followed by a final extension at 68°C for 2 min. The 1386 bp PCR fragment was purified with the GFX PCR DNA and Gel Band purification Kit (Amersham Biosciences, Buckinghamshire, UK) and cloned into the pGEM-T vector (Promega) according to the instructions of the manufacturer. The plasmid DNA was purified using QIAprep Spin Miniprep kit (Qiagen) and then used as a template in the sequencing PCR. T7 5′-TAATACGACTCACTATAGGCG-3′), Sp6 5′-ATTTAGGTGACACTATAGA-3′) and Car2-specific primers (F1: 5′-TGAAGAAGCGATGGTGGA-3′, R1: 5′-TACAGAGAGGCGGTCACA-3′) were used for the sequencing PCR. The sequencing PCR reaction contained 4 µl of ABI PRISM BigDyeTM Terminator Cycle Sequencing Ready Reaction Mix v. 3.1 (Applied Biosystems, Foster City, CA, USA), 0.16 µm of primer, and 200–500 ng of plasmid DNA, and the PCR profile was 25 cycles of denaturation at 94°C for 10 s, annealing at 50°C for 5 s, and extension at 68°C for 10 min.
Immunohistochemistry and digital image analysis
The tissue specimens from the wild-type, Car2−/− and Car9−/− mice were fixed in 4% neutral-buffered formaldehyde at +4°C for 8–27 days. The samples were then dehydrated in an alcohol series, treated with xylene, embedded in paraffin wax, and 4 µm sections were cut and placed on Superfrost microscope slides.
The immunohistochemical staining was performed for CA II, IV, IX, XII and XIV, but only CA II, IX and XII antigens were preserved well in the fixation conditions used. The rabbit polyclonal antibodies for each isozyme have been described and used previously in the following studies: anti-CA II (Lehtonen et al. 2004), anti-CA IV (Brion et al. 1997), anti-CA IX (Ortova Gut et al. 2002), anti-CA XII (Kyllonen et al. 2003), and anti-CA XIV (Parkkila et al. 2001).
After removal of paraffin, the sections were immunohistochemically stained using automated Laboratory Vision Autostainer 480 and Power Vision+ Poly HRP IHC Kit (both from ImmunoVision Technologies, Co., Brisbane, CA, USA) reagents according to the following procedure: (1) rinsing with 1 × tris-buffered saline (TBS) containing 0.05% Tween; (2) blocking with 3% H2O2 for 5 min and rinsing with 1 × TBS containing 0.05% Tween; (3) incubation in 5 times diluted cow colostral whey for 30 min and rinsing with 1 × TBS containing 0.05% Tween; (4) incubation with primary antibody diluted 1: 2000 for 30 min and rinsing with 1 × TBS containing 0.05% Tween for 3 × 5 min; (5) incubation with Poly HRP anti-mouse/rabbit IgG polymer for 30 min and rinsing with 1 × TBS containing 0.05% Tween for 3 × 5 min; (6) treatment with 1 × 3,3′-diaminobenzidine tetrahydrochloride (DAB) solution for 6 min and rinsing with distilled water. The slides were then examined and photographed using a Zeiss Axioskop 40 microscope (Carl Zeiss, Göttingen, Germany). The images photographed with 200 × magnification were subjected to digital image analysis. The staining extent was analysed with analySIS software (Soft Imaging System GmbH, Münster, Germany). From the stomach, eight and six rectangular regions were scored for the wild-type and Car2−/− mice, respectively, using colour threshold values of 145. Each analysed region (700 µm × 550 µm) covered the whole thickness of the gastric mucosa from the surface to the base of the glands. The relative area value obtained indicates the mean percentage of stained area within the analysed regions. An unpaired Student t test was used to evaluate the significance of differences in protein expression in different groups of mice detected by immunohistochemical staining and digital image analysis.
Results
Expression of CA transcripts in tissues of wild-type mice
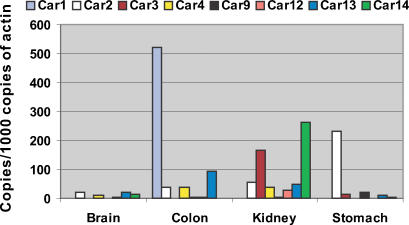
In this report, the expression levels of Car1, Car2, Car3, Car4, Car9, Car12, Car13 and Car14 mRNAs were examined by quantitative real-time RT-PCR. As shown in Fig. 1, each of the CA isozymes exhibited a distinct expression profile between different tissues of the wild-type mice. Car2 mRNA was expressed in all four tissues studied and represented the most abundant CA mRNA in the brain and stomach. This similar broad expression pattern was found for the Car13 mRNA. It was expressed in all four tissues (high levels in the colon and less in the brain, kidney and stomach). Car4 also showed a broad expression profile. Its mRNA was detected in the brain, colon and kidney (higher levels in the colon and kidney). Car1, Car3 and Car9 mRNAs exhibited clearly different expression patterns. Car1 mRNA was detected only in the colon where it was actually the highest CA transcript. Car3 mRNA was highly expressed in the kidney and less in the stomach. Car9 mRNA showed the strongest signal in the stomach and was negligible in the brain, kidney and colon. Positive expression of Car12 mRNA was detected in three tissues. The signal was highest in the kidney and very low in the brain and colon. Car14 mRNA showed a very high signal in the kidney, and in fact, it was the most abundant CA mRNA in this particular organ. Car14 mRNA was also expressed in the brain and stomach, although the expression levels were markedly lower in these tissues than in the kidney.
Figure 1. Expression of Car1, 2, 3, 4, 9, 12, 13 and 14 mRNAs in the wild-type mice detected by real-time RT-PCR.
Note: Car1 mRNA in colon is expressed as copies/100 copies of actin.
Expression of CA transcripts in tissues of Car2−/− and Car9−/− mice
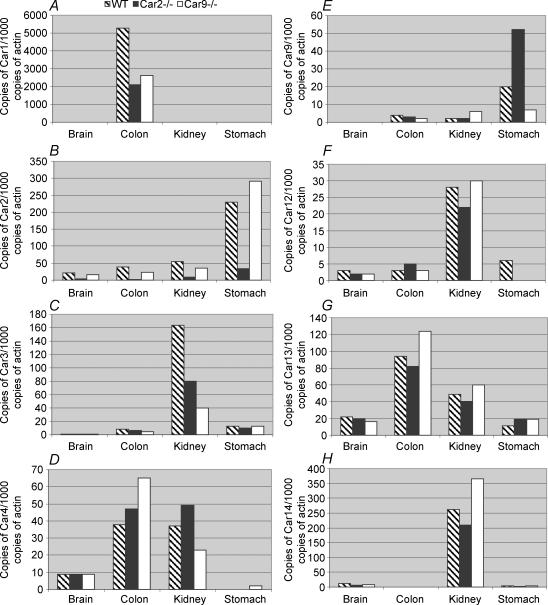
The expression of Car1, Car2, Car3, Car4, Car9, Car12, Car13 and Car14 mRNAs in tissues of Car2−/− and Car9−/− mice in comparison with wild-type mice is shown in Fig. 2. Interestingly, the Car1 (Fig. 2A) and Car3 (Fig. 2C) mRNA levels were down-regulated in the colon and kidney, respectively, of Car2−/− and Car9−/− mice. It is noteworthy that all the transcripts showed some variability between different groups of mice analysed. However, the Car2 mRNA was very clearly decreased in all tissues of Car2−/− mice as compared with the wild-type and Car9−/− mice (Fig. 2B). It is also notable that the expression of Car9 mRNA was markedly up-regulated in the stomach of Car2−/− mice (Fig. 2E), whereas this effect was not observed in the brain, colon or kidney.
Figure 2. Expression of Car1, 2, 3, 4, 9, 12, 13 and 14 mRNAs in the wild-type, Car2−/− and Car9−/− mouse tissues.
Car2 mRNA in Car2−/− mice
CA II-deficient mice were originally produced by random mutagenesis induced by N-ethyl-N-nitrosourea treatment of male DBA mice (Lewis et al. 1988). CA II-deficient mice produced by this method have been shown to lack CA II activity and protein. However, the only report of the mutational basis of this deficiency is a review citing unpublished data (Sly & Hu, 1995). In the present study, real-time RT-PCR analysis revealed a significantly decreased Car2 mRNA expression in the brain, kidney, stomach and colon of the Car2−/− mice as compared with the wild-type and Car9−/− mice (Fig. 2B). Immunohistochemical staining confirmed the absence of CA II protein in all the tissues of Car2−/− mice observed in the present study (data not shown).
As Car2 mRNA expression was not completely absent in the CA II-deficient mice, full-length cDNAs could be amplified from the mRNAs obtained from the stomach samples of the wild-type and Car2−/− mice. The cDNAs were cloned and the resultant plasmids sequenced. Comparing the sequences revealed two nucleotide variations in the Car2−/− mice from the sequence obtained from Car2 mRNA from C57BL6 mice. The first was at nucleotide position 145 (G→C), which caused no change in amino acid sequence. Sequencing the mRNA from the DBA mouse strain revealed that this is a polymorphism present in the DBA mouse and not an induced mutation. The second difference was at position 508 (C→T), which produced a prematurely terminated protein at codon 155 (Gln155X). Figure 3 shows the alignment result of the amino acid sequences translated from the two groups.
Figure 3. Alignment of the wild-type and mutant mouse CA II sequences.
WT, wild-type mice; M, CA II-deficient mice. Asterisks indicate untranslated amino acids.
Car9 mRNA and CA IX protein content is up-regulated in the stomach of Car2−/− mice
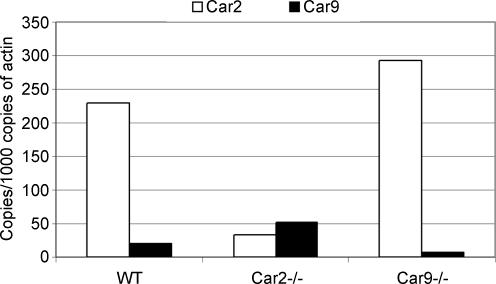
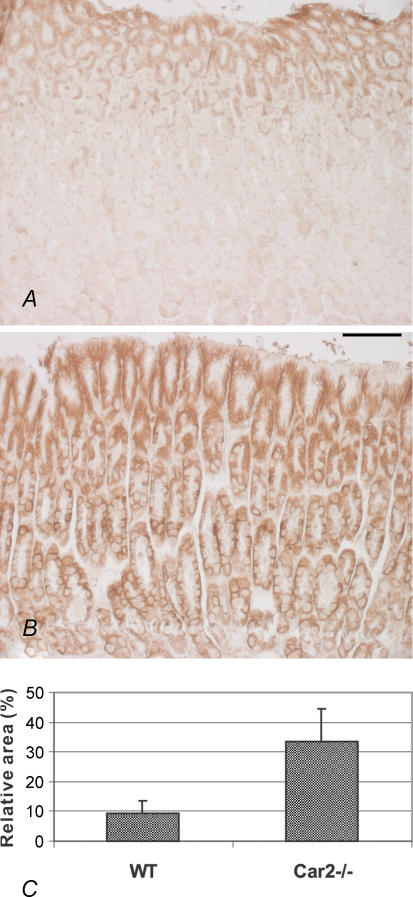
The results summarized in Fig. 4 indicate that the expression of Car9 mRNA was increased about 2.5-fold in the stomach of Car2−/− mice in comparison with wild-type. To determine whether the elevated mRNA expression resulted in an increased expression of CA IX protein, immunohistochemical staining was carried out for the same tissue specimens. The results presented in Fig. 5A and B show that CA IX protein was, indeed, more widely expressed in the Car2−/− mice compared with the wild-type mice. In the Car2−/− mice, the strongly positive area covered the whole mucosa from the deep gastric glands to the most superficial part. In the wild-type mice, the expression was high in the superficial part of the mucosa involving the mucus-producing epithelial cells, while weaker reactions were detected in the gastric glands. Figure 5C shows the digital image analysis results based on the immunohistochemical staining of CA IX. These results confirmed that the extent of CA IX protein staining was significantly (P = 0.0022) increased (about 3.6-fold) in the stomach of the Car2−/− mice as compared with the wild-type mice.
Figure 4. Expression of Car2 and Car9 mRNAs in the stomach of the wild-type, Car2−/− and Car9−/− mice.
Car9 mRNA expression was increased in the stomach of the Car2−/− mice. Also the signal for Car2 mRNA was slightly elevated in the Car9−/− mice.
Figure 5. Immunohistochemical staining of CA IX protein in the stomach of the wild-type and CA II-deficient mice.
In the wild-type mouse, the staining is mainly located in the mucus-producing epithelial cells of the outer part of the mucosa (A). In the Car2−/− mouse, the signal was widely spread, covering intensively the whole thickness of the gastric mucosa (B). Original magnifications × 200. C shows digital image analysis results, comparing the staining extent of CA IX protein in the stomach of the wild-type and Car2−/− mice (P = 0.0022). Scale bar, 100 µm, and applies to A and B.
Discussion
A number of studies have examined the expression of different CA isozymes in mammalian tissues (Fleming et al. 1995; Kivela et al. 2005a; Purkerson & Schwartz, 2005). These investigations provided important background information for later functional studies using CA inhibitors and various cell culture and animal models. Previous studies made clear that several isozymes could be identified in the same mammalian tissue or organ. In many cases, the different isozymes were expressed in different cell types in these tissues and/or in different subcellular localizations. Examples of tissues which express multiple CA isozymes include the four tissues selected for the present investigation.
The purpose of this study was to determine whether loss of function of CA II or CA IX in the respective knockout mouse models leads to compensatory changes in other CAs. The mRNA expression of Car4, Car12, Car13 and Car14 did not show any significant changes in the brain, colon, stomach and kidney of either the Car2−/− or Car9−/− mice. The unchanged level of CA XII protein expression in these tissues as examined by immunohistochemical staining confirmed the lack of compensatory change in this isozyme (data not shown). In a previous study, CA IV protein levels were reported to be up-regulated in the central nervous system of the Car2−/− mice (Brion et al. 1994). However, no such alteration in the expression of Car4 mRNA in the brain of Car2−/− mice was detected in the present study. We were unable to document the protein levels of CA IV by immunohistochemistry since the antigenicity for CA IV was lost in these samples due to fixation.
The expression of Car1 mRNA in the colon of wild-type mice was extremely high in comparison with other isozymes. Car3 mRNA level was also high in the kidney of wild-type mice. Interestingly, both groups of CA-deficient mice showed a down-regulation of Car1 and Car3 mRNAs in the colon and kidney, respectively. The mechanism for this change is unclear, although it may simply reflect a disturbed pH regulation in these organs due to CA II or CA IX deficiency.
A somewhat surprising finding was the high basal expression of Car14 mRNA in the kidney relative to the mRNAs for the other CAs in the wild-type mice. The relative copy number of Car14 mRNA was approximately 5 or 10 times higher than those of Car2 or Car12, respectively. This finding suggests an important role of CA XIV in mouse kidney function. Recent studies have also indicated that CA XIV has even higher CA activity than murine CA II (Whittington et al. 2004). The same report indicated that CA XIV is nearly five times as active as murine CA IV. These results add further weight to the idea that CA XIV is probably a very important enzyme for renal physiology in the mouse.
Comparing the full-length Car2 cDNAs from the wild-type C57BL and Car2−/− mice revealed two nucleotide changes in the cDNA from the Car2−/− mice. One is the chemically induced mutation that causes an early stop codon in the Car2 transcript. This nonsense mutation at codon 155 (Gln155X) would account for the absence of CA II protein in the Car2−/− mice. It is also likely that the reduced level of mRNA in the Car2−/− mice is explained by the nonsense-mediated decay mechanism, which has been well characterized (Conti & Izaurralde, 2005). The original report of the Car2−/− mouse detected no difference in the expression of Car2 mRNA between the Car2−/− and wild-type mice (Lewis et al. 1988). However, the real-time RT-PCR method used in the present study is more sensitive to quantitative differences.
CA II has been considered the major isozyme in the stomach, where it participates in the production of gastric acid by proton secretion from the parietal cells, and on the other hand, protects the epithelium from acid digestion by bicarbonate secretion from the mucus-producing epithelial cells. To date, no gastrointestinal symptoms have been reported in CA II-deficient humans or mice (Ohlsson et al. 1986; Lewis et al. 1988). CA IX is another isozyme expressed in the gastric epithelium. The absence of functional CA IX enzyme has been linked to abnormal morphogenesis of the gastric mucosa (Ortova Gut et al. 2002). In the present study, the expression of Car9 mRNA and CA IX protein in the Car2−/− mice was found to be significantly increased compared with levels in the wild-type mice. This result is interesting in that it apparently reflects a compensatory up-regulation of a CA expressed on the cell surface of the gastric mucosal cells in response to the loss of the cytosolic isozyme CA II. Comparison of the gastric function in Car2−/− and Car9−/− knockout mice may provide clear insights into the relative contribution of each to gastric acidification and indicate whether the apparent compensatory increase in CA IX in CA II deficiency is effective.
Acknowledgments
We thank Dr Marta Ortova Gut and Professor Gerolf Gros for providing the founders of Car9−/− and Car2−/− mice and Dr Daniela Ungureanu and Dr Markus Sjöblom for technical assistance. This work was supported by grants from the Sigrid Juselius Foundation, Academy of Finland, Bayer Corporation, Slovak Grant Agencies VEGA (2/3055) and APVT (51-005802), and National Institutes of Health (GM34182, DK40163).
References
- Brion LP, Cammer W, Satlin LM, Suarez C, Zavilowitz BJ, Schuster VL. Expression of carbonic anhydrase IV in carbonic anhydrase II-deficient mice. Am J Physiol. 1997;273:F234–F245. doi: 10.1152/ajprenal.1997.273.2.F234. [DOI] [PubMed] [Google Scholar]
- Brion LP, Schwartz JH, Zavilowitz BJ, Schwartz GJ. Micro-method for the measurement of carbonic anhydrase activity in cellular homogenates. Anal Biochem. 1988;175:289–297. doi: 10.1016/0003-2697(88)90391-0. [DOI] [PubMed] [Google Scholar]
- Brion LP, Suarez C, Zhang H, Cammer W. Up-regulation of carbonic anhydrase isozyme IV in CNS myelin of mice genetically deficient in carbonic anhydrase II. J Neurochem. 1994;63:360–366. doi: 10.1046/j.1471-4159.1994.63010360.x. [DOI] [PubMed] [Google Scholar]
- Chtanova T, Kemp RA, Sutherland APR, Ronchese F, Mackay CR. Gene microarrays reveal extensive differential gene expression in both CD4+ and CD8+ and type 2 T cells. J Immunol. 2001;167:3057–3063. doi: 10.4049/jimmunol.167.6.3057. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Fleming RE, Parkkila S, Parkkila AK, Rajaniemi H, Waheed A, Sly WS. Carbonic anhydrase IV expression in rat and human gastrointestinal tract regional, cellular, and subcellular localization. J Clin Invest. 1995;96:2907–2913. doi: 10.1172/JCI118362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilvo M, Tolvanen M, Clark A, Shen B, Shah GN, Waheed A, Halmi P, Hänninen M, Hämäläinen JM, Vihinen M, Sly WS, Parkkila S. Characterization of CA XV, a new GPI-anchored form of carbonic anhydrase. Biochem J. 2005;392:83–92. doi: 10.1042/BJ20051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J, Zavada J, Waheed A, Sly W, Lerman MI, Stanbridge EJ. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905–919. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov SV, Kuzmin I, Wei MH, Pack S, Geil L, Johnson BE, Stanbridge EJ, Lerman MI. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc Natl Acad Sci U S A. 1998;95:12596–12601. doi: 10.1073/pnas.95.21.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaunisto K, Parkkila S, Rajaniemi H, Waheed A, Grubb J, Sly WS. Carbonic anhydrase XIV: luminal expression suggests key role in renal acidification. Kidney Int. 2002;61:2111–2118. doi: 10.1046/j.1523-1755.2002.00371.x. [DOI] [PubMed] [Google Scholar]
- Kivela AJ, Kivela J, Saarnio J, Parkkila S. Carbonic anhydrases in normal gastrointestinal tract and gastrointestinal tumours. World J Gastroenterol. 2005a;11:155–163. doi: 10.3748/wjg.v11.i2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivela AJ, Parkkila S, Saarnio J, Karttunen TJ, Kivela J, Parkkila AK, Bartosova M, Mucha V, Novak M, Waheed A, Sly WS, Rajaniemi H, Pastorekova S, Pastorek J. Expression of von Hippel-Lindau tumor suppressor and tumor-associated carbonic anhydrases IX and XII in normal and neoplastic colorectal mucosa. World J Gastroenterol. 2005b;11:2616–2625. doi: 10.3748/wjg.v11.i17.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpulainen T, Korhonen LK. Immunohistochemical localization of carbonic anhydrase isoenzyme C in the central and peripheral nervous system of the mouse. J Histochem Cytochem. 1982;30:283–292. doi: 10.1177/30.4.6801110. [DOI] [PubMed] [Google Scholar]
- Kyllonen MS, Parkkila S, Rajaniemi H, Waheed A, Grubb JH, Shah GN, Sly WS, Kaunisto K. Localization of carbonic anhydrase XII to the basolateral membrane of H+-secreting cells of mouse and rat kidney. J Histochem Cytochem. 2003;51:1217–1224. doi: 10.1177/002215540305100912. [DOI] [PubMed] [Google Scholar]
- Lehtonen J, Shen B, Vihinen M, Casini A, Scozzafava A, Supuran CT, Parkkila AK, Saarnio J, Kivela AJ, Waheed A, Sly WS, Parkkila S. Characterization of CA XIII, a novel member of the carbonic anhydrase isozyme family. J Biol Chem. 2004;279:2719–2727. doi: 10.1074/jbc.M308984200. [DOI] [PubMed] [Google Scholar]
- Leppilampi M, Saarnio J, Karttunen TJ, Kivela J, Pastorekova S, Pastorek J, Waheed A, Sly WS, Parkkila S. Carbonic anhydrase isozymes IX and XII in gastric tumors. World J Gastroenterol. 2003;9:1398–1403. doi: 10.3748/wjg.v9.i7.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE, Erickson RP, Barnett LB, Venta PJ, Tashian RE. N-ethyl-N-nitrosourea-induced null mutation at the mouse Car-2 locus: an animal model for human carbonic anhydrase II deficiency syndrome. Proc Natl Acad Sci U S A. 1988;85:1962–1966. doi: 10.1073/pnas.85.6.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson A, Cumming WA, Paul A, Sly WS. Carbonic anhydrase II deficiency syndrome: recessive osteopetrosis with renal tubular acidosis and cerebral calcification. Pediatrics. 1986;77:371–381. [PubMed] [Google Scholar]
- Ortova Gut MO, Parkkila S, Vernerova Z, Rohde E, Zavada J, Hocker M, Pastorek J, Karttunen T, Gibadulinova A, Zavadova Z, Knobeloch KP, Wiedenmann B, Svoboda J, Horak I, Pastorekova S. Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology. 2002;123:1889–1903. doi: 10.1053/gast.2002.37052. [DOI] [PubMed] [Google Scholar]
- Parkkila S. An overview of the distribution and function of carbonic anhydrase in mammals. Exs. 2000;90:79–93. doi: 10.1007/978-3-0348-8446-4_4. [DOI] [PubMed] [Google Scholar]
- Parkkila S, Parkkila AK. Carbonic anhydrase in the alimentary tract. Roles of the different isozymes and salivary factors in the maintenance of optimal conditions in the gastrointestinal canal. Scand J Gastroenterol. 1996;31:305–317. doi: 10.3109/00365529609006403. [DOI] [PubMed] [Google Scholar]
- Parkkila S, Parkkila AK, Rajaniemi H, Shah GN, Grubb JH, Waheed A, Sly WS. Expression of membrane-associated carbonic anhydrase XIV on neurons and axons in mouse and human brain. Proc Natl Acad Sci U S A. 2001;98:1918–1923. doi: 10.1073/pnas.98.4.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorekova S, Parkkila S, Pastorek J, Supuran CT. Carbonic anhydrases: current state of the art, therapeutic applications and future prospects. J Enzyme Inhib Med Chem. 2004;19:199–229. doi: 10.1080/14756360410001689540. [DOI] [PubMed] [Google Scholar]
- Pastorekova S, Zavada J. Carbonic anhydrase IX (CA IX) as a potential target for cancer therapy. Cancer Ther. 2004;2:245–262. [Google Scholar]
- Purkerson JM, Schwartz GJ. Expression of membrane-associated carbonic anhydrase isoforms IV, IX, XII, and XIV in the rabbit: induction of CA IV and IX during maturation. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1256–R1263. doi: 10.1152/ajpregu.00735.2004. [DOI] [PubMed] [Google Scholar]
- Saarnio J, Parkkila S, Parkkila AK, Waheed A, Casey MC, Zhou XY, Pastorekova S, Pastorek J, Karttunen T, Haukipuro K, Kairaluoma MI, Sly WS. Immunohistochemistry of carbonic anhydrase isozyme IX (MN/CA IX) in human gut reveals polarized expression in the epithelial cells with the highest proliferative capacity. J Histochem Cytochem. 1998;46:497–504. doi: 10.1177/002215549804600409. [DOI] [PubMed] [Google Scholar]
- Sly WS, Hu PY. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- Spicer SS, Lewis SE, Tashian RE, Schulte BA. Mice carrying a CAR-2 null allele lack carbonic anhydrase II immunohistochemically and show vascular calcification. Am J Pathol. 1989;134:947–954. [PMC free article] [PubMed] [Google Scholar]
- Tashian RE, Hewett-Emmett D, Carter N, Bergenhem NC. Carbonic anhydrase (CA) -related proteins (CA-RPs), and transmembrane proteins with CA or CA-RP domains. Exs. 2000;90:105–120. doi: 10.1007/978-3-0348-8446-4_6. [DOI] [PubMed] [Google Scholar]
- Whittington DA, Grubb JH, Waheed A, Shah GN, Sly WS, Christianson DW. Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes. J Biol Chem. 2004;279:7223–7228. doi: 10.1074/jbc.M310809200. [DOI] [PubMed] [Google Scholar]
- Wykoff CC, Beasley N, Watson PH, Campo L, Chia SK, English R, Pastorek J, Sly WS, Ratcliffe P, Harris AL. Expression of the hypoxia-inducible and tumor-associated carbonic anhydrases in ductal carcinoma in situ of the breast. Am J Pathol. 2001;158:1011–1019. doi: 10.1016/S0002-9440(10)64048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]