Abstract
Motor proteins are essential in life processes because they convert the free energy of ATP hydrolysis to mechanical work. However, the fundamental question on how they work when different amounts of free energy are released after ATP hydrolysis remains unanswered. To answer this question, it is essential to clarify how the stepping motion of a motor protein reflects the concentrations of ATP, ADP, and Pi in its individual actions at a single molecule level. The F1 portion of ATP synthase, also called F1-ATPase, is a rotary molecular motor in which the central γ-subunit rotates against the α3β3 cylinder. The motor exhibits clear step motion at low ATP concentrations. The rotary action of this motor is processive and generates a high torque. These features are ideal for exploring the relationship between free energy input and mechanical work output, but there is a serious problem in that this motor is severely inhibited by ADP. In this study, we overcame this problem of ADP inhibition by introducing several mutations while retaining high enzymatic activity. Using a probe of attached beads, stepping rotation against viscous load was examined at a wide range of free energy values by changing the ADP concentration. The results showed that the apparent work of each individual step motion was not affected by the free energy of ATP hydrolysis, but the frequency of each individual step motion depended on the free energy. This is the first study that examined the stepping motion of a molecular motor at a single molecule level with simultaneous systematic control of ΔGATP. The results imply that microscopically defined work at a single molecule level cannot be directly compared with macroscopically defined free energy input.
INTRODUCTION
In many biological ATP-driven motor proteins, stepping motion (linear or rotary) is observed at a single molecule level. In these cases, the free energy liberated by ATP hydrolysis (ΔGATP) is the energy input, and the mechanical step motion is the apparent output. This gives rise to an intriguing question: how does the change in ΔGATP affect the stepping motion? For example, how do molecular motors behave at a very low ΔGATP? As free energy is a macroscopically defined quantity and stepping motion of a single molecule is a microscopic feature, the above question is asking how to relate the law of the macroscopic world to observation in the microscopic world.
At first, the following may be considered as the possible behaviors:
The stepping velocity depends on ΔGATP; therefore, there is a change in the mechanical work done by the individual step motion.
The motor generates a step motion of a fixed size and velocity, irrespective of the ΔGATP input. However, when the chemical energy input (ΔGATP) becomes lower than the mechanical energy output in one step, it simply stops all step motions except those required for thermal fluctuation.
It might be possible that when one ATP is hydrolyzed, constant mechanical work is derived irrespective of the ΔGATP. However, the frequency of the forward step motion decreases as ΔGATP decreases and the back step increases due to the reverse reaction.
However, each of the three possibilities stated above has its limitations. In the case of possibility 1 or 2, a single motor molecule has to detect ΔGATP, which is a function of [ATP], [ADP] and [Pi] in a bulk medium. In addition, the mechanism of changing or stopping the stepping motion in response to ΔGATP remains unclear. Alternatively, in the case of possibility 3, it might be possible that the constant mechanical work exceeds the ΔGATP input; this seems to violate the law of thermodynamics.
The simple abovementioned question on the action of molecular motors has remained unanswered due to many experimental difficulties. For example, myosin II easily detaches from an actin filament, preventing the continuous observation of a single molecule under different conditions. Myosin V or kinesin's step motions yield a relatively low mechanical energy. Moreover, in the presence of a high concentration of ADP or Pi, which is required for controlling ΔGATP, carrying out a single molecule motility assay for these linear motors is quite difficult. This study aims to experimentally answer the above question by using a rotary molecular motor, namely, F1-ATPase.
F1-ATPase, a water-soluble portion of ATP synthase (1), is a rotary molecular motor in which the central γ-subunit rotates against the α3β3 cylinder, hydrolyzing ATP (2). The rotation is stepwise. When one ATP molecule is hydrolyzed, the γ-subunit makes a 120° step (anticlockwise, viewed from the Fo side) (3). The structure of this motor is highly stable, giving rise to a continuous, processive stepping rotation. In addition, the torque generated is considerably high (2–6). These features are ideal to explore the relationship between free energy input and mechanical work output. The only drawback is that this motor protein is severely inhibited by ADP. This ADP inhibition, also known as ADP-Mg inhibition, is not a simple product inhibition. Extensive kinetic analyses of ADP-Mg inhibition have lead to the conclusion that when ADP-Mg is formed during ATP hydrolysis or if it binds from the external medium, then the enzyme enters an inhibited state, which barely releases the bound ADP-Mg (7–9). Due to this specific inhibition, it has been practically impossible to observe continuous rotations while controlling ΔGATP by adding high concentrations of ADP. To overcome this problem, we introduced several mutations in α3β3γ, the minimal subcomplex of F1 that shows ATP-catalyzed rotation (Fig. 1). Using the mutant, termed as GT mutant, we investigated the relationship between ΔGATP and the stepping motion against viscous load. This is the first report in which the stepping motion of a molecular motor was examined at a single molecule level while ΔGATP was systematically controlled.
FIGURE 1.
Mutations specifically introduced in the β-subunit of the GT mutant. Mutations common to the previous mutants used in the rotation assay are not shown.
MATERIALS AND METHODS
Protein
The mutant of the α3β3γ subcomplex resistant to ADP-Mg inhibition was derived from F1-ATPase of a thermophilic Bacillus PS3 by introducing the following mutations: β-G181A (10) and β-T165S (11) for minimizing ADP-Mg inhibition; α-C193S, γ-S107C, and γ-I210C for specific biotinylation of the γ-subunit (12); β-His10 at the amino terminus (2); α-W463F and β-Y341W (13) (Fig. 1). The mutant was named as GT mutant. In this manuscript, we call α-W463F / β-Y341W mutant as wild-type F1. The protein was expressed and purified by the conventional method described in Dou et al. (14). The purified enzyme hardly retained bound nucleotides (<0.05 mol/mol of enzyme).
ATP hydrolysis was measured as a decrease in the absorbance of NADH at 340 nm at 25°C by using an ATP-regenerating system. Since the GT mutant showed a lag phase in its ATPase activity, the maximum rate of ATP hydrolysis was determined several minutes after the start of the reaction.
Rotation assay
Carboxy-modified polystyrene beads (0.35 μm in diameter; Polysciences, Warrington, PA) were modified with 0.5 mg·ml−1 biotin-X cadaverine (Molecular Probes, Eugene, OR) and 2.5 mg·ml−1 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, Pierce, Rockford, IL) at room temperature for 2 h in dark; unreacted biotin was removed by centrifugation. Next, 1 mg ml−1 NeutrAvidin or streptavidin (Sigma, St. Louis, MO) was added, and the excess avidin was washed off. Biotin (BiotinPEAC5maleimide; Dojindo, Kumamoto, Japan) was coupled to Cys residues in the γ-subunit of F1 by a 30-min incubation at room temperature at 1∼8 μM of F1 at F1/biotin = 1:2. To observe the rotation, ∼100 pM F1 in buffer A was infused into a flow chamber that comprised of two glass coverslips. The volume of the chamber was ∼15 μl.
F1 tended to attach to the glass surface in a nonspecific manner. After 2 min, the chamber was washed with 20 mM MOPS-KOH, 10 mM MgCl2, 10 mg·ml−1 BSA, 0.1 M potassium phosphate at pH 7, and the avidin-coated beads at 0.1% concentration were then infused. Avidin on the beads specifically attached to the γ-subunit in a single F1, while α3β3 was immobilized on the surface. Stepping between α3β3 and γ was visualized as bead displacement. The buffer system used for the rotation assay contained 20 mM MOPS-KOH, 10 mM MgCl2, 10 mg·ml−1 BSA, 0.1 M potassium phosphate at pH 7, and 2 or 10 μM ATP. To change the ΔGATP, ∼40 or 60 μl of the buffer containing different concentrations of ADP was infused at least twice.
In this study, [ATP] was fixed at 2 or 10 μM, [Pi] was fixed at 0.1 M, and [ADP] was varied from 0.2 to 1000 μM to change ΔGATP. The reason that we chose only [ADP] as a variable to control ΔGATP was that we wanted to maintain ionic strength (nearly) constant by keeping high [Pi] constant. Change in the ionic strength may cause change in friction between beads probe and glass surface. Addition of other salts to adjust the ionic strength might change enzymatic activity of F1-ATPase. The stock solution of ADP was treated with hexokinase and glucose to remove any contaminating ATP. The level of ATP contamination after treatment was measured using a luciferase assay and was found to be <0.05%. ΔGATP was calculated by the following equation according to Nicholls (15).
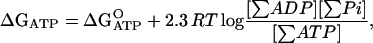 |
at pH 7.0, 10 mM Mg2+.
Here, R is the gas constant (8.3 J·mol−1·K−1), and T is the absolute temperature (300 K). In the calculation of the mass-action ratio, the sums of different ionic species of ATP, ADP, and Pi are used and apparent equilibrium constant (K′) of 105 is applied. Therefore,  here. The rotation of the same F1 molecule was compared at different ΔGATP levels defined above. Observations were recorded at room temperature (25 ± 2 °C).
here. The rotation of the same F1 molecule was compared at different ΔGATP levels defined above. Observations were recorded at room temperature (25 ± 2 °C).
RESULTS
Enzymatic activity and resistance to ADP-Mg inhibition of the GT mutant
The GT mutant was made by introducing several mutations to get resistant to ADP-Mg inhibition as described in Materials and Methods (Fig. 1). It is known that initial rate of ATP hydrolysis by wild-type F1 is severely inhibited by preincubation with ADP due to the ADP-Mg inhibition. But in case of the GT mutant, which has a lag phase in the initial rate of ATP hydrolysis, it was activated by ADP preincubation rather than inhibited (Fig. 2). The initial rate and steady-state rate after ADP preincubation are plotted against ADP concentration in Fig. 3. The causes of the lag phase in ATP hydrolysis of the GT mutant and elimination of it with ADP preincubation (Fig. 2) are not clear. We suspect nucleotide binding to noncatalytic sites is necessary for full activity of ATP hydrolysis for the GT mutant, but further investigation is required on this point.
FIGURE 2.
Effect of ADP preincubation on the GT mutant and wild-type F1. Wild-type F1 and GT mutant were preincubated with or without 100 μM ADP for 10 min and aliquots were injected into an ATPase assay mixture containing ATP regenerating system at the time indicated by the arrow. The hydrolysis of ATP was monitored as the oxidation of NADH (decrease in the absorbance at 340 nm). ATP concentration was 4 mM. (Left panel, wild-type; right panel, GT mutant).
FIGURE 3.
Effect of ADP preincubation on the initial and steady-state rate of the ATPase activity of GT mutant GT mutant was preincubated with various concentrations of ADP and ATPase activity was measured as in Fig. 2. Initial rate (open circle, determined from the slope between 3 and 8 s after injection) and steady-state rate (solid square, determined several minutes after injection) were plotted against ADP concentration during preincubation.
The Vmax was estimated at 250 ± 20 (SE) s−1 and the Km was estimated at 37 ± 10 μM at 25°C; these values are comparable to 247 s−1 and 19 μM, which are the values for wild-type F1 in the presence of lauryl dimethylamine oxide (LDAO). LDAO is known to reduce ADP-Mg inhibition in wild-type F1. However, the GT mutant did not require LDAO for hydrolysis measurement because of the absence of ADP-Mg inhibition.
NaN3, a potent inhibitor of F1-ATPase, which inhibits F1 by stabilizing ADP-Mg inhibited state had significantly less effect on the GT mutant than the wild-type (Fig. 4). These results together indicate that the GT mutant is highly resistant to ADP-Mg inhibition. Thus, it was possible to observe rotation with ADP at a concentration 500 times higher than ATP in this mutant.
FIGURE 4.
Inhibitory effect of NaN3 on wild-type F1 and GT mutant. Wild-type F1 and GT mutant were injected into an ATPase assay mixture containing 4 mM ATP at the indicated arrow. Then 10 mM NaN3 was injected at the indicated arrow. (Upper line, wild-type; lower line, GT mutant).
Rotation assay at different ΔGATP levels
After the mutant subcomplex was attached to a glass surface, a duplex of beads (0.35 μm in diameter) coated with streptavidin was attached to the γ-subunit that had been biotinylated at two engineered Cys residues. Relatively large beads were used so that the viscous friction against the beads was the major rate-limiting factor during step motions. ΔGATP was controlled by perfusing different media and the rotation of the same bead duplex (the same F1 molecule) was compared at different ΔGATP levels.
First of all, we attempted to examine the rotation at a relatively high ATP concentration (10 μM) with 100 mM Pi and various concentrations of ADP (1 μM to 1000 μM); this corresponds to ΔGATP of −64∼−37 pN·nm/ATP. Fig. 5 a shows the time course of the rotation. Under these conditions, clear step rotation could not be resolved, and the average rate of rotation was plotted as a function of ΔGATP in Fig. 5 b. Due to data scattering, we are not sure if the relation is linear or not. Nevertheless, Fig. 5 b clearly indicates a correlation between the average rotational rate and ΔGATP. If we regard the average rotational rate ×120° as proportional to the mechanical work done by the hydrolysis of one ATP molecule, it indicates that the mechanical work done by one ATP molecule appeared to change as a function of ΔGATP when our time resolution was low.
FIGURE 5.
Rotation of the GT mutant at 10 μM ATP and various ΔGATP levels. (a) Rotation at 10 μM ATP, 100 mM Pi and 10, 100, or 1000 μM ADP. Nominally calculated ΔGATP values are −37, −46, and −55 pN·nm/ATP, respectively. (b) Average rotation speed was plotted against −ΔGATP. In this case, ΔGATP initially increased from −64 pN·nm/ATP to −37 pN·nm/ATP and then decreased again to −55 pN·nm/ATP. The same F1 molecule (beads duplex) was observed throughout the experiment. The beads flowed away before the last observation at −64 pN·nm/ATP. The straight line was drawn by least square fitting.
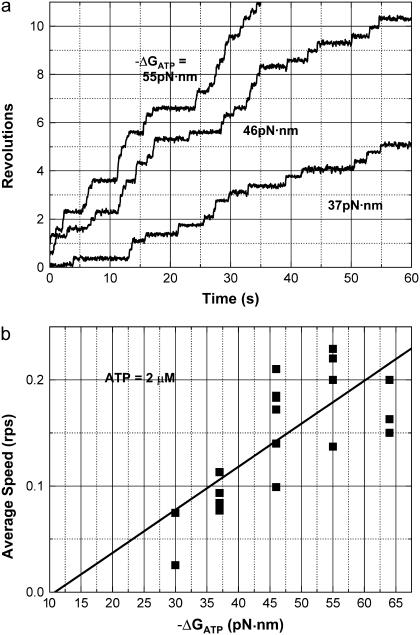
Next, we attempted to reduce the ATP concentration to resolve the individual step motions. At 2 μM ATP, we observed the step motions as shown in Fig. 6 a. The average rotational rate showed a similar tendency observed at a concentration of 10 μM ATP (Fig. 6 b). Fig. 7 shows the analysis of the rate of rise of individual steps and the dwell time between them. Fig. 7, b–e, indicates that the dwell time increases as ΔGATP increases. In contrast, Fig. 7, f–i, shows that the rate of rise in individual steps barely depended on ΔGATP. The decrease in the average velocity was attributed to the increase in dwell time. Due to the difficulties in repeated perfusion of different media for the same specimen, we could examine an ADP concentration range of 0.2–1000 μM (nominal ΔGATP was −64 to −30 pN·nm/ATP.) in only six independent experiments. ADP concentration ranges of 0.2–200 μM and 2–1000 μM were examined in one experiment. In all the experiments, the rate of rise in individual steps did not demonstrate significant ΔGATP dependency. The rate of rise at −30 pN·nm/ATP of ΔGATP ([ADP] = 1000 μM) was 92% ± 9.9% (average ± SD) of the rate at −64 pN·nm/ATP of ΔGATP ([ADP] = 0.2 μM) for the six experiments. The apparent “work” per 120° step motion was calculated using the viscosity of bulk water and rate of rise in individual steps for each experiment. The values varied with each experiment, probably due to the difficulty in determining the precise rotation radius. At −30 pN·nm/ATP of ΔGATP, the experimentally obtained minimum and maximum value was 19.3 pN·nm/step and 36.1 pN·nm/step, respectively. The average (±SD) was 27.1 (±6.0) pN·nm/step. As described in Discussion, the “work” associated with the step motion, estimated by using the viscosity of bulk water, is the minimum limit of its true value. Based on these results, we conclude that the rate of rise in each step motion and hence the apparent work per 120° step motion is essentially independent of the free energy input, i.e., ΔGATP.
FIGURE 6.
Rotation of the GT mutant at 2 μM ATP and at various ΔGATP levels. (a) Rotation at 2 μM ATP, 100 mM Pi, and 2, 20, or 200 μM ADP. Nominally calculated ΔGATP values are −37, −46, and −55 pN·nm/ATP, respectively. (b) Average rotation speed was plotted against −ΔGATP. In this case, ΔGATP initially increased from −64 to −30 pN·nm/ATP by perfusing different solutions containing different concentrations of ADP. The same F1 molecule (beads duplex) was observed throughout the experiment and average speed was defined in the time courses where very long pauses were not included. Therefore, we plotted more than one point at the same ΔGATP in panel d. The straight line was drawn by least-square fitting.
FIGURE 7.
Statistical analyses of the rotation of the GT mutant at 2 μM ATP and at various ΔGATP. (a) Rates of rise and dwell time that were analyzed are shown along the time course of rotation. (b–e) Analyses of dwell time between steps. The time constant was deduced by fitting with a single exponential curve to the dwell time histogram (b–d) and plotted against the −ΔGATP in panel e. (f–i) Analyses of the rate of rise at the step. The rate of rise was estimated for every step motion, and the histograms were constructed (f–h). The mean and standard errors were plotted against −ΔGATP in panel i. In panel i, 3.0 rps corresponds to the mechanical “work” of 27 pN·nm/step without the correction of the viscosity increase near the glass surface. See text for details.
DISCUSSION
The results in Figs. 6 and 7 clearly demonstrated that the apparent “work” in a single step motion, which was defined as the rate of rise (=angular velocity) × frictional drag coefficient × step angle (=2/3 π), was almost constant, irrespective of the free energy input (ΔGATP). Therefore, the possibility 1 proposed in the Introduction is disproved. The apparent “work” was almost the same or occasionally higher than the lowest value of −ΔGATP that was examined (30 pN·nm/ATP). But the value of the apparent “work” seems to be underestimated for the following reasons: In the above calculation, the frictional drag coefficient of bulk water was used. When the increase in viscosity near the glass surface is considered, the value of the frictional coefficient should be approximately twice of that in bulk water (16). Alternatively, when the actin filament attached to the γ-subunit was used as a spring balance, the torque was estimated to be 50 ± 6 pN·nm (17) rather than 40 pN·nm estimated by simple hydrodynamic calculation using a viscosity of the bulk water (3,18). The result indicates that the true value of the torque should be at least 1.2 times larger than that estimated above. Thus, the true value of the apparent “work” plus ΔGATP easily becomes positive with either of these corrections. Therefore, possibility 2 also appears unlikely. Possibility 3 predicted that constant mechanical work is derived in a one-step motion even with an increase in the ADP concentration, and that partial product inhibition or back reaction by ADP and Pi occurs. In this study, the ΔGATP-independent rate of rise in Fig. 7 B agrees with this prediction. Further, the increase in the dwell time observed in Fig. 7 A agrees with the predicted product inhibition; however, a clear increase in the back step was not observed. But the apparent absence of full back steps (120° back step) was anticipated because when considering the equilibrium constant of 105 M for ATP hydrolysis, the ratio of the forward reaction to the backward reaction is in the order of 1000 even at the lowest −ΔGATP examined in this study (2 μM ATP, 1000 μM ADP, and 100 mM Pi).
From the viewpoint of enzyme kinetics, this result would be expected because increases in [ADP] will cause product inhibition of the forward reaction. Previous single molecule work has established that the individual ATP-triggered steps occur rapidly. If ADP dissociation was required before ATP could bind, then high ADP would increase the dwell time, but not influence the rise time once it was gone and ATP had bound. This would be true even if ADP dissociated from a different subunit than the one to which ATP bound. However, if we think of the balance between input free energy (ΔGATP) and output mechanical work, there is still a problem to be clarified. In fact, if we define the energy transduction efficiency as the ratio of the apparent work of individual steps to −ΔGATP, these results indicate this apparent efficiency will exceed 100%. From this result, one may conclude that the biological molecular motor can violate the law of thermodynamics. This conclusion, of course misdirected, is based on a misinterpretation of the law of thermodynamics. The problem resides in the definition of “work”. A simple reasoning may be explained as follows: the reason that the maximum work is limited by the free energy input is that if it exceeds the free energy, it would result in an entropy decrease of the whole system. A prerequisite for this statement is that the thermodynamically defined work does not contribute to entropy production in any way (19). In this situation, however, the apparent “work” is done against viscous load and hence dissipated as heat. Therefore, it eventually contributes to the increase in entropy. Thus, the apparent “work” against viscous load is different from thermodynamically defined work. Therefore, even if the apparent “work” defined for the individual rate of rise exceeds the magnitude of input free energy (−ΔGATP), it does not violate the law of thermodynamics. This outcome reflects statistical nature of thermodynamics.
To compare the work done by a molecular stepping motor with thermodynamic parameters such as ΔGATP, Wang and Oster theoretically proposed that it is necessary to average the rotation speed over a long time including interstep dwells (20). The efficiency calculated using the averaged speed (Stokes efficiency) is limited to 100% even if the work is done against frictional load (20). In this case, averaging the speed is a type of statistical treatment by which work and chemical potential can be compared.
In a sense, it can be said that thermodynamic parameters are defined macroscopically with low spatial and temporal resolutions. The low resolution data in Fig. 5 in which we cannot discriminate individual steps matches the chemical potential rather than the data in Fig. 6. In Fig. 5, our temporal resolution was not high enough to resolve individual steps at 10 μM ATP and we could only get information on the average rotational velocity. But using this rotational velocity, we could calculate apparent work that showed clear correlation with ΔGATP (Fig. 5 b). At 2 μM ATP (Figs. 6 and 7), the temporal resolution was relatively higher than at 10 μM ATP and we could resolve each step motion. Then, the apparent “work” calculated from the rate of rise of individual steps lost correlation with ΔGATP. Instead, time-averaged velocity that was obtained indirectly after statistical treatment, showed correlation with ΔGATP (Fig. 6 b). As our temporal and spatial resolution get higher, we can observe single molecule events microscopically. At such a high resolution, what we observe is a single molecular motor exchanging thermal energy to and from the environment to achieve its function. Then the results of our observation can be related to macroscopically defined thermodynamic parameters after appropriate statistical treatment. At the high resolution, the field of microscopic vision shows us a world where ambient noise cannot be reduced, and what is directly seen is not directly compared with macroscopically defined thermodynamics (21).
Acknowledgments
We thank to Dr. Fumio Oosawa, Dr. Shigeru Hayashi, Dr. Kazuhiko Kinosita, and Dr. Ken Sekimoto for their helpful advice and encouragement. We also thank Dr. Kengo Adachi for his insightful discussions and for providing us with a useful plug-in for Image J that was used in the image analyses. We thank Dr. Takayuki Ariga and Dr. Katsuya Shimabukuro, and our laboratory member for their helpful advice and technical assistance.
References
- 1.Boyer, P. D. 1993. The binding change mechanism for ATP synthase: some probabilities and possibilities. Biochim. Biophys. Acta. 1140:215–250. [DOI] [PubMed] [Google Scholar]
- 2.Noji, H., R. Yasuda, M. Yoshida, and K. Kinosita Jr. 1997. Direct observation of the rotation of F1-ATPase. Nature. 386:299–302. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda, R., H. Noji, K. Kinosita Jr., and M. Yoshida. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120o steps. Cell. 93:1117–1124. [DOI] [PubMed] [Google Scholar]
- 4.Muneyuki, E., H. Noji, T. Amano, T. Masaike, and M. Yoshida. 2000. FoF1-ATP synthase: general structural features of ‘ATP-engine’ and a problem on free energy transduction. Biochim. Biophys. Acta. 1458:467–481. [DOI] [PubMed] [Google Scholar]
- 5.Noji, H., and M. Yoshida. 2001. The rotary machine in the cell, ATP synthase. J. Biol. Chem. 276:1665–1668. [DOI] [PubMed] [Google Scholar]
- 6.Kinosita, K., Jr., K. Adachi, and H. Itoh. 2004. Rotation Of F1-ATPASE: how an ATP-driven molecular machine may work. Annu. Rev. Biophys. Biomol. Struct. 33:245–268. [DOI] [PubMed] [Google Scholar]
- 7.Vasilyeva, E. A., I. B. Minkov, A. F. Fitin, and A. D. Vinogradov. 1982. Kinetic mechanism of mitochondrial adenosine triphosphatase—ADP-specific inhibition as revealed by the steady state kinetics. Biochem. J. 202:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milgrom, Y. M., and P. D. Boyer. 1990. The ADP that binds tightly to nucleotide-depleted mitochondrial F1-ATPase and inhibits catalysis is bound at a catalytic site. Biochim. Biophys. Acta. 1020:43–48. [DOI] [PubMed] [Google Scholar]
- 9.Jault, J. M., and W. S. Allison. 1993. Slow binding of ATP to noncatalytic nucleotide binding sites which accelerate catalysis is responsible for apparent negative cooperativity exhibited by the bovine mitochondrial F1-ATPase. J. Biol. Chem. 268:1558–1566. [PubMed] [Google Scholar]
- 10.Masaike, T., H. Noji, E. Muneyuki, R. Yasuda, K. Kinosita, Jr., and M. Yoshida. 2000. Rotation of F1-ATPase and the hinge residues of the β subunit. J. Exp. Biol. 203:1–8. [DOI] [PubMed] [Google Scholar]
- 11.Jault, J. M., C. Dou, N. B. Grodsky, T. Matsui, M. Yoshida, and W. S. Allison. 1996. The α3β3γ subcomplex of the F1-ATPase from the thermophilic bacillus PS3 with the βT165S substitution does not entrap inhibitory MgADP in a catalytic site during turnover. J. Biol. Chem. 271:28818–28824. [DOI] [PubMed] [Google Scholar]
- 12.Yasuda, R., H. Noji, M. Yoshida, K. Kinosita Jr., and H. Itoh. 2001. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 410:898–904. [DOI] [PubMed] [Google Scholar]
- 13.Dou, C., P. A. G. Fortes, and W. S. Allison. 1998. The α3(βY341W)3 γ subcomplex of the F1-ATPase from the thermophilic Bacillus PS3 fails to dissociate ADP when MgATP is hydrolyzed at a single catalytic site and attains maximal velocity when three catalytic sites are saturated with MgATP. Biochemistry. 37:16757–16764. [DOI] [PubMed] [Google Scholar]
- 14.Dou, C., N. B. Grodsky, T. Matsui, M. Yoshida, and W. S. Allison. 1997. ADP-fluoroaluminate complexes are formed cooperatively at two catalytic sites of wild-type and mutant α3β3γ subcomplexes of the F1-ATPase from the thermophilic Bacillus PS3. Biochemistry. 36:3719–3727. [DOI] [PubMed] [Google Scholar]
- 15.Nicholls, D. G. 1982. Bioenergetics: An Introduction to the Chemiosmotic Theory. Academic Press, New York.
- 16.Svoboda, K., and S. M. Block. 1994. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 23:247–285. [DOI] [PubMed] [Google Scholar]
- 17.Panke, O., D. A. Cherepanov, K. Gumbiowski, S. Engelbrecht, and W. Junge. 2001. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 81:1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noji, H., D. Bald, R. Yasuda, H. Itoh, M. Yoshida, and K. Kinosita, Jr. 2001. Purine, but not pyrimidine, nucleotides support rotation of F1-ATPase. J. Biol. Chem. 276:25480–25486. [DOI] [PubMed] [Google Scholar]
- 19.Kittel, C., and H. Kroemer. 1980. Thermal Physics. W. H. Freeman and Company, San Francisco, CA and London, UK.
- 20.Wang, H., and G. Oster. 2002. The stokes efficiency for molecular motors and its applications. Europhys. Lett. 57:134–140. [Google Scholar]
- 21.Sekimoto, K. 1998. Langevin equation and thermodynamics. Prog. Theor. Phys. Suppl. 130:17–27. [Google Scholar]