Abstract
Fibrinogen is a blood plasma protein that, after activation by thrombin, assembles into fibrin fibers that form the elastic network of blood clots. We used atomic force microscopy to study the forced unfolding of engineered linear oligomers of fibrinogen, and we show that forced extension of the oligomers produces sawtooth patterns with a peak-to-peak length consistent with the independent unfolding of the coiled-coils in a cooperative two-state manner. In contrast with force plateaus seen for myosin coiled-coils that suggested rapid refolding of myosin, Monte Carlo simulations of fibrinogen unfolding confirm that fibrinogen refolding is negligible on experimental timescales. The distinct behavior of fibrinogen seems to be due to its topologically complex coiled-coils and an interaction between fibrinogen's αC-domains and its central region.
A blood clot needs to have the right degree of stiffness and plasticity for hemostasis and yet very stiff clots are not easily lysed and are associated with thrombosis and thromboembolism, but the origin of these mechanical properties is unknown (1). The elasticity of self-assembled networks of fibrin—the principal component of clots—also proves highly nonlinear (1) and is of likely importance to cell responses in remodeling such gels (2). Recent experiments have pushed mechanical measurements to the single fiber level (3) and a theory incorporating an enthalpic fiber stretch and entropic elasticity provides a better fit to macroscopic rheological data than one involving entropic elasticity alone (4). Despite these advances in understanding larger scales, the micromechanics of fibrinogen, the precursor of fibrin, remains unexplored.
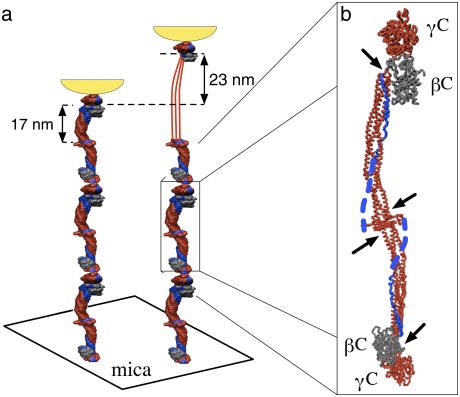
In this letter, we describe single-molecule atomic force microscopy (AFM) experiments on the extensibility of fibrinogen oligomers. As with previous single-molecule unfolding experiments, oligomers were required to generate reproducible, interpretable data (see for example, (5)). The fibrinogen oligomers used in this study were covalently cross-linked via the γC-modules located at the distal ends of adjacent fibrinogen molecules. Accordingly, when a fibrinogen oligomer is extended from the sample surface, the force is propagated only through the coiled-coils and the C-terminal portions of the γ-chains (Fig. 1 a), thus reducing the variety of potentially unfolded structures and possible force-extension curves.
FIGURE 1.
(a) Schematic of the pulling geometry. Fibrinogen oligomers adsorbed on mica were extended by the AFM tip shown in yellow. If a single coiled-coil is unfolded, the oligomer contour length increases by 23 nm. (b) Crystal structure of an individual fibrinogen molecule (6) with two globular modules, γC and βC, at each end separated by two triple-helical coiled-coils confined between clusters of disulfide bonds (indicated by arrows). The force-propagating coiled-coils, γC-modules, and the central region are shown in red. The C-terminal portions of the Aα-chains are shown in blue with the αC-domains, not visualized in the crystal structure, represented as dotted blue lines.
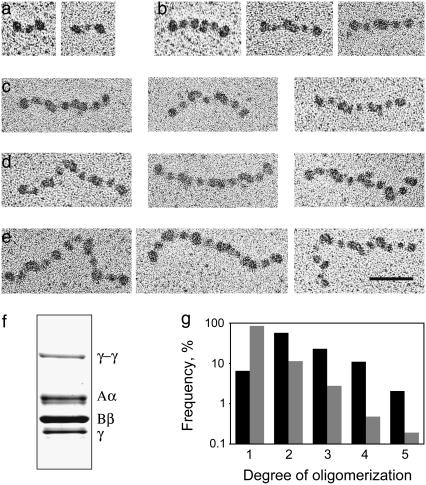
To induce end-to-end oligomerization of fibrinogen molecules, 10 mg/ml human fibrinogen (plasminogen-free, Hyphen BioMed, Andrésy, France) in 20 mM HEPES buffer (pH 7.4) containing 100 mM NaCl, 30 mM CaCl2, and 5 ATU/ml hirudin was mixed with human factor XIIIa (50 μg/ml final concentration) and incubated at room temperature until the beginning of gelation (∼30 min). At that point, the cross-linking reaction was stopped with 1 mM iodoacetimide and the “clot” was removed. For activation, a 0.8 mg/ml factor XIII solution (46 U/mg, glycerol/water, 0.5 mM EDTA and 2 mM CaCl2) was treated with 2 U/ml human thrombin (American Diagnostica, Greenwich, CT) for 1 h at room temperature, and the reaction was stopped by addition of hirudin (10 ATU/ml final concentration). Formation of single-stranded fibrinogen oligomers via crosslinking between γGln398 and γLys406 of the γC-modules was corroborated by transmission electron microscopy (TEM) (Fig. 2, a–e) and the presence of the γ-γ-chain band in SDS-PAGE of reduced samples of the fibrinogen preparation (Fig. 2 f). To separate nonligated monomers from oligomers, 0.5 ml of the cross-linked fibrinogen preparation was applied to a 1.5 × 15 cm Sepharose CL 6B column equilibrated with 20 mM HEPES buffer (pH 7.4) containing 100 mM NaCl and 3 mM CaCl2. As judged from TEM, the fraction collected in the void volume contained only ∼6% fibrinogen monomers and 94% di-, tri-, tetra-, and pentamers (Fig. 2 g).
FIGURE 2.
Transmission electron micrographs of fibrinogen monomers (a); di- (b); tri- (c); tetra- (d); and pentamers (e). The scale bar represents 50 nm. (f) The presence of the γ-γ band in SDS-PAGE confirms that the oligomers observed by TEM were covalently cross-linked. (g) Histograms of degree of oligomerization are plotted as the relative frequency of different species on a log scale. The fractionated sample contains only 6% monomer (solid bars, n = 291) compared to the unfractionated sample that contains 85% monomer (shaded bars, n = 1046).
For TEM, preparations of cross-linked fibrinogen were diluted with a volatile buffer (50 mM ammonium formate, pH 7.4, 25% glycerol) to a concentration of 20–40 μg/ml, immediately sprayed onto freshly cleaved mica, and rotary-shadowed with tungsten in a vacuum evaporator as previously described (7). Prepared specimens were observed in an FEI 400 electron microscope at 80 kV (FEI, Hillsboro, OR) and 60,000× magnification in many different areas of the preparations to obtain a random sample.
For the AFM experiments, 50 μl of a 50 μg/ml solution of the oligomerized fibrinogen were pipetted onto freshly cleaved mica and allowed to adsorb for 10 min before being rinsed gently with buffer. Force-extension curves were collected using a Digital Instruments Multimode AFM (Digital Instruments, Santa Barbara, CA) and Veeco silicon nitride cantilevers (Veeco, Woodbury, NY).
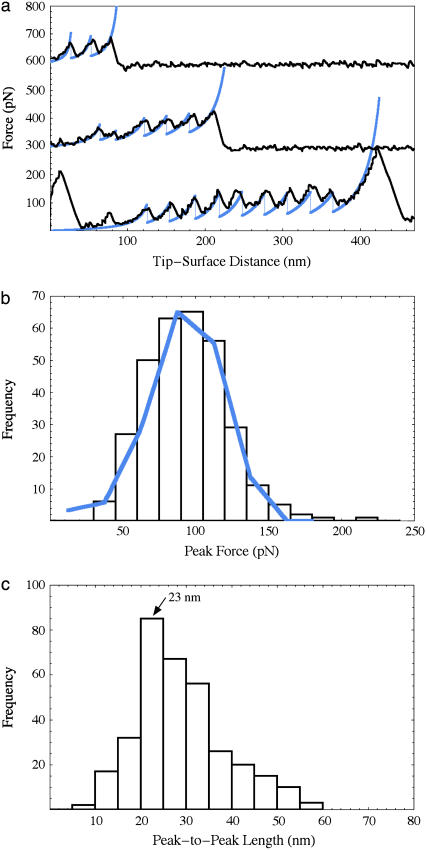
When unfolded under force, fibrinogen oligomers gave rise to a periodic sawtooth pattern (Fig. 3 a) with a length and regularity that was not observed in control experiments on monomers. Since the unfolding geometry is specified by γ-γ-crosslinking, the observed sawtooth patterns are most likely due to unfolding of either the coiled-coils or the globular C-terminal portions of the γ-chains. Each coiled-coil consists of 111 or 112 amino-acid residues of the Aα-, Bβ-, and γ-chains, which, when fully unfolded, form a thread 40-nm long (assuming a contour length per residue of 0.36 nm) (Fig. 1 a) corresponding to an expected peak-to-peak length of 23 nm (unfolded minus 17-nm folded length), in good agreement with the experimental data (Fig. 3 c). In contrast, the C-terminal γ-chains each consist of 215 amino-acid residues (not including the disulfide loops or the chain beyond the first crosslinking site) with an expected peak-to-peak length of 77 nm, significantly bigger than the average unfolding length observed in our experiments. The central region could also unfold but it is highly constrained by disulfide bonds and does not seem to contribute.
FIGURE 3.
(a) Force-extension curves (offset for clarity) observed for fibrinogen oligomers (black) with simulated overlays (blue). The traces have two, five, and ten coiled-coil unfolding peaks from oligomers with at least one, three, and five fibrinogen molecules, respectively (each molecule has two coiled-coils). The last peaks are due to protein desorption as is the first large peak in the lowest trace. (b) Histogram of peak forces (bars) is well approximated by the simulated data (blue line). (c) Histogram of peak-to-peak distances with a major peak at 23 nm.
Finally, we performed a Monte Carlo simulation that reproduces both the observed force-extension curves and the peak-force histogram. Reasonable agreement was obtained assuming negligible refolding and using a persistence length of 0.8 nm, an unfolding rate at zero force of 0.03 s−1, and a transition state distance of 0.31 nm. These parameters are in the same range as those observed previously for unfolding ubiquitin, a globular protein (8). It is interesting to note the difference between the unfolding of the triple-helical coiled-coils of fibrinogen and the double helical coiled-coil of myosin II (9). For myosin II, a force plateau is observed at 20 pN as opposed to the sawtooth behavior observed here for fibrinogen with an average peak force of 94 pN. The same two-state unfolding model has been shown to account for both force plateaus and sawtooth patterns in force-extension curves by changing two parameters: the length of the unit that unfolds in a two-state manner and that unit's refolding rate (10). There are several reasons to expect that these parameters are different for myosin and fibrinogen. The coiled-coils in a fibrinogen oligomer are divided into short segments 17 nm in length, whereas in myosin the coiled-coil is unbroken for 150 nm. Furthermore, the coiled-coils of fibrinogen are 1), interrupted by “stutters”; 2), contain a kink in the middle; and 3), are, in fact, partly quadruple-helical (6). This structure is significantly more complex than the coiled-coil of myosin II and it is possible that when it partly unfolds, the remainder is sufficiently destabilized to appear to unfold cooperatively on experimental timescales. Such an observation is not unprecedented: a helical linker has already been observed to propagate cooperative unfolding between adjacent globular domains in spectrin (11). Given these structural differences and its more complex topology, fibrinogen's refolding rate indeed seems likely to be considerably slower than that of myosin II. The C-terminal part of fibrinogen's Aα-chain that forms the fourth strand of the quadruple-helical portion of the coiled-coil (shown in blue, Fig. 1 b) is known to interact via the αC-domain with the central region (7). This interaction could contribute to the mechanical stability of fibrinogen and may also reduce the refolding rate. This possible new role for the αC-domains could be explored further using recombinant fibrinogens without these domains or by proteolytically cleaving them from oligomers prepared from wild-type fibrinogen.
This study identifies a new functional property of fibrinogen and suggests that the coiled-coil is more than a passive structural element of this molecule. Coiled-coil unfolding could account for up to a twofold strain in the recently observed large extensibility of fibrin fibers (12) but its role in the macroscopic properties of fibrin gels (1) remains to be determined. The constraints provided by our results will likely serve as a useful input for multiscale modeling efforts that will ultimately be required to fully understand blood clot mechanics.
Acknowledgments
We thank Chandrasekaran Nagaswami for electron microscopy and Yelena Baras for help with the early stages of data analysis.
This work was partially supported by National Institutes of Health grants to J.W.W. (grant No. HL30954) and D.E.D. (grant No. HL62352) and the Nano/Bio Interface Center through the National Science Foundation NSEC DMR-0425780. A.E.X.B. is supported by a scholarship from the Natural Sciences and Engineering Research Council of Canada.
References
- 1.Weisel, J. W. 2004. The mechanical properties of fibrin for basic scientists and clinicians. Biophys. Chem. 112:267–276. [DOI] [PubMed] [Google Scholar]
- 2.Discher, D. E., P. Janmey, and Y. Wang. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science. 310:1139–1143. [DOI] [PubMed] [Google Scholar]
- 3.Collet, J., H. Shuman, R. E. Ledger, S. Lee, and J. W. Weisel. 2005. The elasticity of a single fibrin fiber in a clot. Proc. Natl. Acad. Sci. USA. 102:9133–9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storm, C., J. J. Pastore, F. C. MacKintosh, T. C. Lubensky, and P. A. Janmey. 2005. Nonlinear elasticity in biological gels. Nature. 435:191–194. [DOI] [PubMed] [Google Scholar]
- 5.Dietz, H., and M. Rief. 2004. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 101:16192–16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, J. H., N. Volkmann, G. Jun, A. H. Henschen-Edman, and C. Cohen. 2000. The crystal structure of modified bovine fibrinogen. Proc. Natl. Acad. Sci. USA. 97:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veklich, Y. I., O. V. Gorkun, L. V. Medved, W. Nieuwenhuizen, and J. W. Weisel. 1993. Carboxyl-terminal portions of the α-chains of fibrinogen and fibrin. Localization by electron microscopy and the effects of isolated α-C fragments on polymerization. J. Biol. Chem. 268:13577–13585. [PubMed] [Google Scholar]
- 8.Schlierf, M., H. Li, and J. M. Fernandez. 2004. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA. 101:7299–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwaiger, I., C. Sattler, D. R. Hostetter, and M. Rief. 2002. The myosin coiled-coil is a truly elastic protein structure. Nature Mat. 1:232–235. [DOI] [PubMed] [Google Scholar]
- 10.Rief, M., J. M. Fernandez, and H. E. Gaub. 1998. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 81:4764–4767. [Google Scholar]
- 11.Law, R., P. Carl, S. Harper, P. Dalhaimer, D. W. Speicher, and D. E. Discher. 2003. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys. J. 84:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, W., L. M. Jawerth, E. A. Sparks, M. R. Falvo, R. R. Hantgan, R. Superfine, S. T. Lord, and M. Guthold. 2006. Fibrin fibers have extraordinary extensibility and elasticity. Science. 313:634. [DOI] [PMC free article] [PubMed] [Google Scholar]